Abstract
Patients with cholestasis-lymphedema syndrome (CLS) suffer severe neonatal cholestasis that usually lessens during early childhood and becomes episodic; they also develop chronic severe lymphedema. The genetic cause of CLS is unknown. We performed a genome screen, using DNA from eight Norwegian patients with CLS and from seven unaffected relatives, all from an extended pedigree. Regions potentially shared identical by descent in patients were further characterized in a larger set of Norwegian patients. The patients manifest extensive allele and haplotype sharing over the 6.6-cM D15S979–D15S652 region: 30 (83.3%) of 36 chromosomes of affected individuals carry a six-marker haplotype not found on any of the 32 nontransmitted parental chromosomes. All Norwegian patients with CLS are likely homozygous for the same disease mutation, inherited from a shared ancestor.
Four proteins disrupted in forms of hereditary cholestasis have recently been identified. Lesions in JAG1, a ligand for the NOTCH1 receptor, underlie Alagille syndrome (MIM 118450) (Li et al. 1997; Oda et al. 1997). FIC1, a widely expressed P-type ATPase of unknown function, is altered in progressive familial intrahepatic cholestasis type 1 (PFIC1 [MIM 211600]) and in many cases of benign recurrent intrahepatic cholestasis, an episodic disorder (BRIC [MIM 243300]) (Bull et al. 1998). BSEP, a bile-acid transporter, is defective in progressive familial intrahepatic cholestasis type 2 (PFIC2 [MIM 601847]) (Gerloff et al. 1998; Strautnieks et al. 1998; Green et al. 2000). Defects in MDR3, a phospholipid flippase, are responsible for progressive familial intrahepatic cholestasis type 3 (PFIC3 [MIM 602347]) (de Vree et al. 1998; Jacquemin et al. 1999). The genetic basis of other hereditary cholestatic disorders remains to be elucidated (Knisely 2000). Among these is cholestasis-lymphedema syndrome (CLS).
Patients with CLS, or Aagenaes syndrome (MIM 214900), suffer severe neonatal cholestasis with hyperbilirubinemia, increased serum concentrations of bile acids, and giant-cell transformation of hepatocytes (Aagenaes et al. 1968, 1970; Aagenaes 1998). In most patients, manifestations of liver disease lessen during early childhood and become episodic, as in BRIC. In a few patients, however, the course of cholestasis resembles that in PFIC, with cirrhosis and death in childhood; other patients develop cirrhosis as adults. CLS is distinct from other forms of cholestatic liver disease in that patients manifest lymphedema, either at birth or during childhood; lymphedema becomes chronic and severe, mainly affecting the lower extremities but also the hands, scrotum, and periorbital soft tissues. Later in life, lymphedema can affect the small intestine and the thoracic soft tissue, with chylothorax (Ø. Aagenaes, unpublished data). Lymph vessels in patients with CLS are hypoplastic (Aagenaes et al. 1970).
Other forms of hereditary lymphedema, with or without other associated disease, exist; progress has recently been made in molecular genetic characterization of two of them. Some patients with autosomal dominant primary lymphedema (MIM 153100) carry alterations in FLT4 (vascular endothelial growth factor C receptor-3); evidence also supports locus heterogeneity for this disorder (Ferrell et al. 1998; Witte et al. 1998; Evans et al. 1999; Karkkainen et al. 2000). A locus for lymphedema-distichiasis syndrome (MIM 153200) has been mapped to chromosome 16q24 (Mangion et al. 1999). Hennekam syndrome (MIM 235510) demonstrates similarities to CLS, including autosomal recessive inheritance, lower-extremity lymphedema, and intestinal lymphangiectasia (Hennekam et al. 1989). Neither the Hennekam syndrome locus nor loci altered in other hereditary lymphedemas have been mapped. CLS is the only hereditary lymphedema associated with cholestasis.
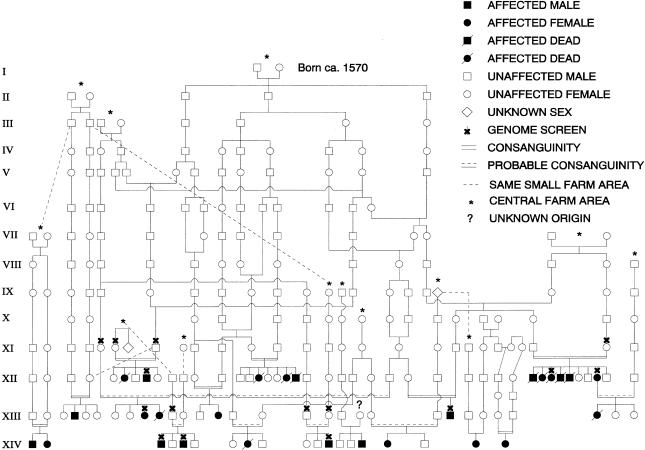
CLS was first described in a Norwegian pedigree (fig. 1), in which the disease demonstrates autosomal recessive inheritance (Aagenaes et al. 1968; Aagenaes 1998). Most reported cases have occurred in individuals of Norwegian descent, although CLS occurs as well in people of other ancestries (Sharp and Krivit 1971; Aagenaes 1974; Vajro et al. 1984; Pawlowska et al. 1994, 1995; Morris et al. 1997; Aagenaes 1998). Most Norwegian patients are from a single region in southwestern Norway and are known or likely descendants of a founding couple born circa 1570 (fig. 1).
Figure 1 .
Pedigree 1. Known and probable ancestral relationships between members are shown. Individuals whose DNA was included in the genome screen are indicated by an “X.” The affected sib pair in the lower-left corner was identified subsequent to performance of the genome screen.
Plans for this study were initiated by a group of patients with CLS and their relatives. Informed consent of participants was obtained using established protocols at the University of Oslo. This study was also approved by the Committee on Human Research at the University of California San Francisco. Blood was collected from Norwegian patients who met diagnostic criteria for CLS (Aagenaes 1998) and from their available unaffected parents.
To localize the CLS gene, we performed a whole-genome screen using DNA from members of pedigree 1 (fig. 1). Standard linkage analysis was not feasible because of the complex pedigree structure and lack of samples from many members. We therefore screened for genomic regions potentially shared identical by descent (IBD) by several patients. We genotyped eight affected members of the pedigree—including two sib pairs and one first-cousin pair—and seven unaffected relatives (fig. 1), with 385 autosomal microsatellite markers, most of which were from the PE Biosystems Linkage Mapping Set, version 2. We typed two additional markers in the FIC1 region. DNA was amplified in a GeneAmp 9700 apparatus (PE Biosystems), and reaction products were electrophoresed on a 377 or 3700 apparatus (PE Biosystems). Some follow-up genotyping in candidate regions was performed using radioactively labeled primers, as described elsewhere (Bull et al. 1999). All genotypes were determined independently by two observers. When possible, allele sizes were assigned relative to those reported or apparent in control sample 1347-2 (Centre d’Étude du Polymorphisme Humaine).
Under the assumptions of an autosomal recessive mode of inheritance for CLS and a single ancestral mutation in this pedigree, siblings should be IBD for both copies of the chromosomal region containing the disease gene, and cousins should be IBD for the copy of the region inherited from their shared grandparents. Therefore, regions consistent with potential IBD inheritance in all three of the two affected sib pairs and the affected cousin pair were considered to be promising CLS candidate regions. Fifty-seven genomic intervals, totaling 25% of the genome, were potentially shared IBD in each of the three pairs—that is, there were no apparent recombinations that would exclude such sharing.
To prioritize these 57 intervals for further analysis, we examined data from all eight patients, to identify potential sharing between the more distantly related chromosomes. Intervals containing a marker for which a single allele occurred with high frequency on chromosomes in affected individuals, as well as those in which a two-marker haplotype was shared by a number of chromosomes present in affected individuals, were considered to be particularly promising. Intervals of comparatively long genetic length and/or containing one or more markers were weighted as being more promising than those spanning very short genetic intervals and/or containing no typed markers. We judged 19 of the 57 potential candidate regions to be particularly promising.
We also evaluated regions containing genes mutated in forms of hereditary cholestasis or lymphedema. Data for the intervals containing BSEP, MDR3, FIC1, JAG1, and the lymphedema-distichiasis syndrome locus were inconsistent with potential IBD sharing in the three close-relative pairs; these intervals were not studied further. The region containing FLT4 was consistent with IBD sharing in the three close-relative pairs, but there was no evidence of allele or haplotype sharing of the region among chromosomes in affected individuals, in either initial or follow-up studies.
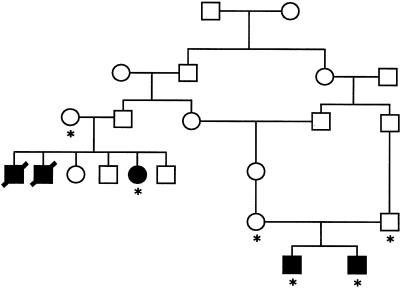
We further characterized the most promising regions, to distinguish the CLS region from the other, “false positive” regions, through genotyping of additional samples, including a second, recently identified Norwegian family containing an affected sib pair (pedigree 2; fig. 2); where necessary, we also typed these samples, as well as those included in the genome screen, for additional markers within the intervals.
Figure 2 .
Pedigree 2. Asterisks (*) indicate individuals from whom we obtained DNA.
The 21.7-cM D15S205–D15S130 interval was among the most interesting regions, on the basis of results from the genome screen; however, it was not unequivocally identifiable as the CLS region until the follow-up analysis was performed. In the initial screen, 15 of the 16 copies of chromosome 15 present in affected individuals carried an identical allele of marker D15S127, which lies between D15S205 and D15S130. For only one other marker in the genome screen, D3S1565, did this many disease chromosomes share a single allele. Suggestive evidence for some haplotype sharing over the D15S205–D15S130 interval was also obtained from the genome screen.
Eleven microsatellite markers within the D15S205–D15S130 region were typed in a total of 23 Norwegian patients with CLS and in their unaffected relatives. These 23 patients include 17 affected members of pedigree 1, including a recently identified additional affected sib pair (fig. 1), as well as 3 affected members from pedigree 2 (fig. 2) and 3 Norwegian patients with CLS who are the only known affected individuals in their families. We focused particularly on typing of markers near D15S127. Table 1 summarizes the results of this work; for each of the 11 markers, the allele most enriched on chromosomes of patients, compared with nontransmitted parental chromosomes, is shown, as are the percentages of affected and nontransmitted chromosomes that carry this allele. (For this analysis, data from one member of each affected sib pair are excluded; when two sibs differ, because of recombination, in the allele for a given marker that is inherited from a parent, each allele is counted as 0.5 copies.) To a striking degree, affected chromosomes share alleles in this region, particularly at the markers from D15S996 to D15S963; for five of these six markers, >90% of the affected chromosomes carry the same allele. A disease-associated haplotype can be defined for this six-marker interval. These results indicate that the CLS gene lies within the 6.6-cM D15S979–D15S652 interval (genetic distances are from the Center for Medical Genetics, Marshfield Medical Research Foundation map.)
Table 1.
Allele Sharing in the D15S205–D15S130 Region[Note]
|
Proprotion of Chromosomes(%) |
|||
| Markera | Alleleb | CLSc | NTd |
| D15S205 | 158 | 15.8 | 0 |
| D15S655 | 234 | 34.2 | 21.9 |
| D15S979 | 157 | 15.8 | 0 |
| D15S996 | 196 | 92.1 | 44.4 |
| D15S127 | 119 | 86.8 | 22.6 |
| FESe | 16 | 94.7 | 50.0 |
| IP15M9 | 69 | 100 | 43.3 |
| D15S158 | 95 | 94.7 | 7.1 |
| D15S963 | 271 | 100 | 56.3 |
| D15S652 | 305 | 32.9 | 18.3 |
| D15S130 | 288 | 34.2 | 27.6 |
Note.— Data from 36–38 of the 38 affected chromosomes included were available for each marker, as were data from 27–32 of the 32 nontransmitted chromosomes.
Listed in order from centromere to telomere.
Allele most enriched on copies of the region present in affected individuals, relative to the nontransmitted chromosomes of patients’ parents (alleles rarely present on disease chromosomes are not included).
Percentage of chromosomes present in affected individuals who carry this enriched allele.
Percentage of nontransmitted chromosomes carrying this allele.
Alleles present in 1347-2 were arbitrarily designated “24-20.”
Some alleles present on the shared disease haplotype are also relatively common on the nontransmitted chromosomes; many of the individuals studied are known to be at least distantly related; and no genetic distance between D15S996 and D15S963 is currently detectable. Therefore, to exclude the possibility that background, rather than disease-associated, linkage disequilibrium (LD) makes a major contribution to sharing of this haplotype, we determined the frequency of the six-marker haplotype on affected and nontransmitted chromosomes. The full haplotype is present on 30 (83.3%) of 36 affected chromosomes for which complete, phase-known data are available (again, one member of each sib pair was excluded from this analysis). In contrast, none of the 32 nontransmitted chromosomes carried the full haplotype. This finding confirms that the haplotype sharing in this region is due to the presence of a disease mutation rather than to non–disease-associated LD. Data from each of the four affected sib pairs are consistent with linkage to this region, under an autosomal recessive model; recombination events in two of these four families place the CLS gene within the D15S979–D15S652 interval, the same interval in which haplotype analysis places the gene.
On all but two of the affected chromosomes, alleles of the six-marker shared D15S996–D15S963 haplotype occur at four or more contiguous markers; the two chromosomes on which alleles of the disease haplotype occur at fewer than four contiguous markers are in members of pedigree 1. These findings indicate that all of the Norwegian patients with CLS in our study carry the same founder mutation and that all CLS cases in Norway are likely due to this mutation.
We used several criteria to evaluate the data from our genome screen; in retrospect, the data on sharing of alleles at single markers, rather than on sharing of two-marker haplotypes, most clearly highlighted the candidate region in the results of the genome screen. Because dramatic haplotype sharing in the CLS region extends over <7 cM, either a higher-density marker set or a patient sample size large enough to make the application of statistical approaches to data analysis appropriate would have been needed for clear identification of the shared haplotype in the genome screen. However, sharing of a single allele at D15S127 on 15 of the 16 chromosomes present in affected individuals in the genome screen clearly marked this region as being very promising.
Because several disease chromosomes in our data set carry only a portion of the conserved haplotype, genotyping these patients with additional markers should further refine the localization of the CLS gene. Additional families, of Scandinavian and non-Scandinavian descent, with CLS are now under study, to determine whether the Norwegian CLS haplotype is shared by patients of other backgrounds and whether evidence for locus homogeneity or heterogeneity exists. Since significant migrations occurred from southwestern Norway to the Netherlands (during the 17th and 18th centuries) and to the United States (during the 19th and 20th centuries), the Norwegian CLS mutation may well be present in these populations (Sharp and Krivit 1971).
The cause of cholestasis in patients with CLS is unknown. It has been speculated both that an abnormal lymphatic system serves a normal liver, with cholestasis arising secondarily (Sigstad et al. 1970) and, alternatively, that both bile ducts and lymphatic vessels develop abnormally in CLS (Shneider 1999). Although lymphedema progresses with age in patients with CLS, cholestasis decreases (Aagenaes 1998); the liver in patients with CLS may become able to compensate for the CLS defect as the patient matures. (A study of liver biopsy specimens, obtained at different ages, from most of the Norwegian patients is under way and may provide insight.) Because it is difficult to predict what type of protein the gene altered in CLS will encode and numerous genes lie within the CLS interval, further refinement of the CLS gene location will greatly facilitate its identification.
Acknowledgments
We would like to thank Line Syversen, Jasmine Nguyen, and Terry Song for technical assistance; Sue Service for assistance with data analysis; Victoria Carlton for assistance with database design; Julie Vargas for assistance with data entry; and the patients and their families for their participation in this study. This work was supported by National Institutes of Health grant R01 DK50697 to N.B.F.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics.
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/ (for Alagille syndrome [MIM 118450], PFIC1 [MIM 211600], BRIC [MIM 243300], PFIC2 [MIM 601847], PFIC3 [MIM 602347], CLS [MIM 214900], primary lymphedema [MIM 153100], lymphedema-distichiasis syndrome [MIM 153200], and Hennekam syndrome [MIM 235510])
References
- Aagenaes O (1974) Hereditary recurrent cholestasis with lymphoedema—two new families. Acta Paediatr Scand 63:465–471 [DOI] [PubMed] [Google Scholar]
- ——— (1998) Hereditary cholestasis with lymphoedema (Aagenaes syndrome, cholestasis-lymphoedema syndrome): new cases and follow-up from infancy to adult age. Scand J Gastroenterol 33:335–345 [DOI] [PubMed] [Google Scholar]
- Aagenaes O, Sigstad H, Bjorn-Hansen R (1970) Lymphoedema in hereditary recurrent cholestasis from birth. Arch Dis Child 45:690–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagenaes O, van der Hagen CB, Refsum S (1968) Hereditary recurrent intrahepatic cholestasis from birth. Arch Dis Child 43:646–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull LN, Juijn JA, Liao M, van Eijk MJ, Sinke RJ, Stricker NL, DeYoung JA, et al (1999) Fine-resolution mapping by haplotype evaluation: the examples of PFIC1 and BRIC. Hum Genet 104:241–248 [DOI] [PubMed] [Google Scholar]
- Bull LN, van Eijk MJ, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, Klomp LW, Lomri N, Berger R, Scharschmidt BF, Knisely AS, Houwen RH, Freimer NB (1998) A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet 18:219–224 [DOI] [PubMed] [Google Scholar]
- de Vree JM, Jacquemin E, Sturm E, Cresteil D, Bosma PJ, Aten J, Deleuze JF, Desrochers M, Burdelski M, Bernard O, Oude Elferink RP, Hadchouel M (1998) Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc Natl Acad Sci USA 95:282–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AL, Brice G, Sotirova V, Mortimer P, Beninson J, Burnand K, Rosbotham J, Child A, Sarfarazi M (1999) Mapping of primary congenital lymphedema to the 5q35.3 region. Am J Hum Genet 64:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell RE, Levinson KL, Esman JH, Kimak MA, Lawrence EC, Barmada MM, Finegold DN (1998) Hereditary lymphedema: evidence for linkage and genetic heterogeneity. Hum Mol Genet 7:2073–2078 [DOI] [PubMed] [Google Scholar]
- Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, Meier PJ (1998) The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem 273:10046–10050 [DOI] [PubMed] [Google Scholar]
- Green RM, Hoda F, Ward KL (2000) Molecular cloning and characterization of the murine bile salt export pump. Gene 241:117–123 [DOI] [PubMed] [Google Scholar]
- Hennekam RCM, Geerdink RA, Hamel BCJ, Hennekam FAM, Kraus P, Rammeloo JA, Tillemans AAW (1989) Autosomal recessive intestinal lymphangiectasia and lymphedema, with facial anomalies and mental retardation. Am J Med Genet 34:593–600 [DOI] [PubMed] [Google Scholar]
- Jacquemin E, Cresteil D, Manouvrier S, Boute O, Hadchouel M (1999) Heterozygous non-sense mutation of the MDR3 gene in familial intrahepatic cholestasis of pregnancy. Lancet 353:210–211 [DOI] [PubMed] [Google Scholar]
- Karkkainen MJ, Ferrell RE, Lawrence EC, Kimak MA, Levinson KL, McTigue MA, Alitalo K, Finegold DN (2000) Missense mutations interfere with VEGFR-3 signalling in primary lymphoedema. Nat Genet 25:153–159 [DOI] [PubMed] [Google Scholar]
- Knisely AS (2000) Progressive familial intrahepatic cholestasis: a personal perspective. Pediatr Dev Pathol 3:113–125 [DOI] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB (1997) Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet 16:243–251 [DOI] [PubMed] [Google Scholar]
- Mangion J, Rahman N, Mansour S, Brice G, Rosbotham J, Child AH, Murday VA, Mortimer PS, Barfoot R, Sigurdsson A (1999) A gene for lymphedema-distichiasis maps to 16q24.3. Am J Hum Genet 65:427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AAM, Sequeira JSS, Malone M, Slaney SF, Clayton PT (1997) Parent-child transmission of infantile cholestasis with lymphoedema (Aagenaes syndrome). J Med Genet 34:852–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC (1997) Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet 16:235–242 [DOI] [PubMed] [Google Scholar]
- Pawlowska J, Rujner J, Socha J, Majchrzyk-Ossowska T, Piontek E, Wróblewska M, Wozniewicz B (1994) Two cases of Aagenaes syndrome with different clinical courses (in Polish). Hepatol Polska 1:43–52 [Google Scholar]
- Pawlowska J, Socha J, Wróblewska M, Wozniewicz B (1995) Aagenaes syndrome mimicking extrahepatic biliary atresia. Surg Child Int 1:54–60 [Google Scholar]
- Sharp HL, Krivit W (1971) Hereditary lymphedema and obstructive jaundice. J Pediatr 78:491–496 [DOI] [PubMed] [Google Scholar]
- Shneider BL (1999) Genetic cholestasis syndromes. J Pediatr Gastroenterol Nutr 28:124–131 [DOI] [PubMed] [Google Scholar]
- Sigstad H, Aagenaes O, Bjorn-Hansen R, Rootwelt K (1970) Primary lymphedema combined with hereditary recurrent intrahepatic cholestasis. Acta Med Scand 188:213–219 [DOI] [PubMed] [Google Scholar]
- Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V, Tanner MS, Kagalwalla AF, Nâemeth A, Pawlowska J, Baker A, Mieli-Vergani G, Freimer NB, Gardiner RM, Thompson RJ (1998) A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet 20:233–238 [DOI] [PubMed] [Google Scholar]
- Vajro P, Romano A, Fontanello A, Oggero V, Vecchione R, Shmerling DH (1984) Aagenaes's syndrome in an Italian child. Acta Paediatr Scand 73:695–696 [DOI] [PubMed] [Google Scholar]
- Witte MH, Erickson R, Bernas M, Andrade M, Reiser F, Conlon W, Hoyme HE, Witte CL (1998) Phenotypic and genotypic heterogeneity in familial Milroy lymphedema. Lymphology 31:145–155 [PubMed] [Google Scholar]