Abstract
Lymphedema-distichiasis (LD) is an autosomal dominant disorder that classically presents as lymphedema of the limbs, with variable age at onset, and double rows of eyelashes (distichiasis). Other complications may include cardiac defects, cleft palate, extradural cysts, and photophobia, suggesting a defect in a gene with pleiotrophic effects acting during development. We previously reported neonatal lymphedema, similar to that in Turner syndrome, associated with a t(Y;16)(q12;q24.3) translocation. A candidate gene was not found on the Y chromosome, and we directed our efforts toward the chromosome 16 breakpoint. Subsequently, a gene for LD was mapped, by linkage studies, to a 16-cM region at 16q24.3. By FISH, we determined that the translocation breakpoint was within this critical region and further narrowed the breakpoint to a 20-kb interval. Because the translocation did not appear to interrupt a gene, we considered candidate genes in the immediate region that might be inactivated by position effect. In two additional unrelated families with LD, we identified inactivating mutations—a nonsense mutation and a frameshift mutation—in the FOXC2 (MFH-1) gene. FOXC2 is a member of the forkhead/winged-helix family of transcription factors, whose members are involved in diverse developmental pathways. FOXC2 knockout mice display cardiovascular, craniofacial, and vertebral abnormalities similar to those seen in LD syndrome. Our findings show that FOXC2 haploinsufficiency results in LD. FOXC2 represents the second known gene to result in hereditary lymphedema, and LD is only the second hereditary disorder known to be caused by a mutation in a forkhead-family gene.
Introduction
Lymphedema is a common chronic, debilitating condition that affects millions of people worldwide. It is usually secondary to filariasis, surgery, trauma, or infection. Genetic causes of lymphedema have been of much interest, partly due to the possibility of identification of molecular insights into diagnosis, prevention, or treatment. Congenital lymphedema can be found associated with Noonan and Turner syndromes. Hereditary lymphedema is heterogeneous and usually occurs as an autosomal dominant trait. The most common form of congenital, primary hereditary lymphedema—Milroy disease (MIM 153100)—is sometimes caused by mutations in the vascular endothelial growth–factor receptor-3 gene (VEGFR-3, or FLT4) (Karkkainen et al. 2000). Other forms of hereditary lymphedema include lymphedema-distichiasis syndrome (LD [MIM 153400]) in which lymphedema, primarily of the limbs, with variable age at onset, is seen together with distichiasis, or double rows of eyelashes. The extra eyelashes grow from the Meibomian glands and may protrude into the cornea, producing severe corneal abrasions. This combination was first described by Neel and Schull (1954, pp. 50–51) and by Falls and Kertesz (1964). A number of subsequent case reports have shown additional complications in some families, including extradural cysts, cardiac defects, and cleft palate (Falls and Kertesz 1964; Robinow et al. 1970; Schwartz et al. 1980; Corbett et al. 1982; Goldstein et al. 1985). This constellation of defects suggests that the gene involved would likely have pleiotrophic effects during development.
We previously reported neonatal lymphedema similar to that seen in Turner syndrome associated with a Y;16 translocation (Erickson et al. 1995). We searched for an “anti-Turner” gene, which might be involved in lymphedema on the Y-chromosome side of the translocation (Schmidt Drury et al. 1998). However, we were not able to find a candidate gene on this portion of the Y chromosome and turned our attention to the der(16) breakpoint region at 16q24.3. Shortly thereafter, LD was mapped to a 16-cM region on distal chromosome 16 (Mangion et al. 1999). We determined that the breakpoint was within this region and narrowed the breakpoint to a small, 20-kb segment. By analysis of neighboring genes, we identified inactivating mutations in the FOXC2 forkhead/winged-helix transcription factor in two families with autosomal dominant LD. We speculate that the lymphedema phenotype associated with the translocation breakpoint is due to position effect. These results show that mutations in the FOXC2 gene are responsible for LD.
Subjects and Methods
Subjects
The proband with the t(Y;16) translocation was ascertained at the Department of Pediatrics at the University of Arizona and has been described elsewhere (Erickson et al. 1995). Family 1 (fig. 3) was ascertained at the Greenwood Genetic Center, in Greenwood, SC. Family 2 (fig. 3) was originally seen at the Pediatric Genetics Clinic at the University of Michigan. Informed consent for both participation and collection of tissue samples was obtained from all family members, in accordance with the procedures of the institutional review boards of either the University of Michigan Medical School or the University of Arizona.
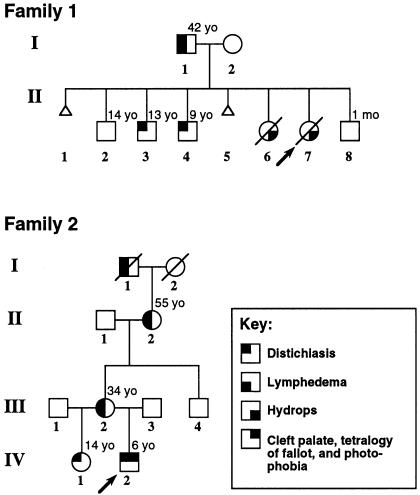
Figure 3.
Pedigrees of families with LD. All living members of both families were examined clinically, as were the two fetuses in family 1. The current ages of all relevant family members are indicated (yo = years old). Family 1 was ascertained via fetus II-7, which was electively aborted at 17 wk gestation, because of hydrops fetalis. Pathological examination showed severe nonimmune hydrops. No heart defects were noted. Additional family members presented with either lymphedema-distichiasis or distichiasis alone in younger individuals. Onset of lymphedema in I-1 was at 12 years of age. Fetus II-6 was electively aborted because of cystic hygroma and hydrops. The proband in family 2 was ascertained at age 4 mo, when he presented with cleft palate, cystic hygroma, tetralogy of Fallot, and distichiasis. Additional family members showed either lymphedema-distichiasis or distichiasis alone. No other family members had clinical signs of congenital heart disease, although echocardiography was not performed. Age at onset of lymphedema in individual II-2 was the early 30s, whereas that in individual III-2 was 17 years. Both pedigrees are consistent with an autosomal dominant mode of inheritance.
FISH
FISH was performed on metaphase spreads from an Epstein-Barr virus–transformed lymphoblastoid cell line that was established from the patient with the t(Y;16)(q11.2;q24;3) translocation, according to standard procedures. As described elsewhere (Dagenais et al. 1999), metaphase-chromosome slides were denatured and then hybridized overnight with biotin (biotin-14-dATP)-labeled probes. A BioNick Translation Kit (Gibco BRL) was used to label probes from YAC and bacterial artificial chromosome (BAC) clones and subclones of PCR products from BAC clone 58A18. Prior to hybridization, probes were denatured for 8 min at 70°C and were preannealed for 30–60 min at 37°C with human COT-1 DNA (Gibco BRL). FISH signals were visualized by incubation with two layers of FITC-conjugated avidin-DCS (Vector Laboratories) and fluorescin-conjugated anti-avidin IgG, whereas chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (Vector Laboratories). FISH results were analyzed with a Zeiss Axioscop epifluorescence microscope and were documented with a Photometrics SenSys camera and Vysis Quips imaging software (Applied Imaging). The PCR primers used for generating the FISH probes were as follows: PCR 58-1a, 5′-CATTCATTAGTCCATCCATCCGTC-3′; PCR 58-1b, 5′-AGAACTATG TTAGCCAGGGTA-3′; PCR 58-2a, 5′-GCAGGAAGAAGAAGCACAGTCC-3′; PCR 58-2b, 5′-TGTATTTTTTAGCGGAGACGGG-3′; PCR 58-3a, 5′-TAGAGGAGATGGGGAAGGGTAGC-3′; PCR 58-3b, 5′-TGATAGGCAGGTAAGCAGAGTTCG-3′; PCR 58-4a, 5′-CCCTAAGTTCCAGTGCAAGTATCC-3′; PCR 58-4b, 5′-ATGTGACCTGGAGCAAGTGACTAAC-3′; PCR 58-5a, 5′-GGGATAGCTAAGAAGAAAGCATGT-3′; PCR 58-5b, 5′-CAGATAGTTTCTGAAACCTCCGTT-3′; PCR 58-6a, 5′-GACAGGCTCCTCCACACAGAG-3′; PCR 58-6b, 5′-GCTGTCTTGAGAGGTTGAGATGC-3′; PCR 58-7a, 5′-AATACCTGTTTCTCAAGGACCCG-3′; PCR 58-7b, 5′-TGTGTGTGTGTGTGTGTGTGTTCG-3′; PCR 58-8a, 5′-CATGAGTAGTCTTGCATCTCTCTC-3′; PCR 58-8b, 5′-GACCTAACAGTAGTCAGGAACGCAC-3′; PCR 58-9a, 5′-AGAGATGGAGTTTCACCGTATTGG-3′; PCR 58-9b, 5′-CCGCACCCACTCGCCCCACAGAGCAC-3′; PCR 58-10a, 5′-GGTGCTCTGTGGGGCGAGTGGGTGCG-3′; PCR 58-10b, 5′-ATCCCAACAGAAGACCTTTCCGC-3′.
DNA Sequencing
To amplify the single coding exon of FOXC2, genomic DNA isolated from peripheral blood or cultured fibroblasts of probands and family members was used as template in PCR reactions. We used the GC Rich System (Roche) and primers 5′-TCTCTCGCGCTCTCTCGCTC-3 and 5′-CTTTTTTGCGTCTCTGCAGCCC-3′ to generate the PCR products. Before sequencing, PCR products were purified by treatment with Exonuclease I (Amersham Pharmacia Biotech and shrimp alkaline phosphatase (Amersham Pharmacia Biotech) for 15 min at 37°C. Enzymes were heat inactivated by incubating the reaction at 80°C for 15 min. Sequencing was performed by the Sequencing Core Laboratory at the University of Michigan, by the dideoxy termination method and an ABI 377 automated sequencer. Results were verified by sequencing both the sense and the antisense strands.
Restriction-Enzyme Analysis
Sequence analysis of the proband in family 1 revealed the TAC→TAG mutation creating a novel BfaI restriction site. This site was used to confirm the presence of the mutation in all additional affected family members. A 439-bp region of the FOXC2 gene was PCR amplified from genomic DNA, by means of the GC-rich protocol described by Baskaran et al. (1996), with primers 5′-TCTCTCGCGCTCTCTCGCTC-3′ and 5′-TGCCAGCCCTGCTTGTTCTCC-3′. PCR products were column purified (Qiagen) and were digested with BfaI (New England Biolabs) at 37°C for 1 h prior to gel electrophoresis. In family 2, the 4-nucleotide insertion creates a novel NaeI site. A region surrounding the insertion was PCR amplified using primers 5′-CGAGCGATGAGCCTGTACACC-3′ and 5′-GCGAGGTTGAGAGCGCTCAGG-3′. PCR products were purified and digested with NaeI at 37°C for 2 h, and fragments were resolved by gel electrophoresis on a 4% agarose gel.
Results
The region of the Y-chromosome translocation breakpoint in our patient with congenital lymphedema and a t(Y;16)(q11.2;q24.3) translocation contained low-level chromosome Y–specific repeats, making its exact localization by FISH—and, hence, isolation of sequences for “hopping” to the chromosome 16 side of the breakpoint—problematic. We thus obtained a number of YAC clones from the 16q24.3 region, to identify this breakpoint. By FISH, YAC 733F10 was found to cross the translocation breakpoint. This YAC maps within the 16-cM interval reported, by Mangion et al. (1999), to contain a gene for hereditary LD. YAC 733F10 contains marker WI-15115, which was used to identify BAC 463O9, which, in turn, was used to identify BAC 58A18 from the GenBank databases (GenBank Overview). BAC clone 58A18 crossed the translocation breakpoint by FISH (fig. 1). By assembling the previously unassembled sequence of BAC 58A18, we developed PCR-generated probes for FISH and narrowed the breakpoint to a 20-kb interval. The closest candidates were expressed sequence tag (EST) AW138879, located 42 kb distally, and EST AW182066, located 40 kb proximally, neither of which contained open reading frames. We did not detect expression of these ESTs by RT-PCR in multiple tissues, including fibroblasts, lymphoblastoid cells, and a number of cancer cell lines. Expression was also not detected in endothelial, lymph node (Clontech), or heart (Clontech) cDNA libraries or by hybridization with EST-specific probes to the Clontech multiple-tissue array, which contains RNA from 76 different tissues.
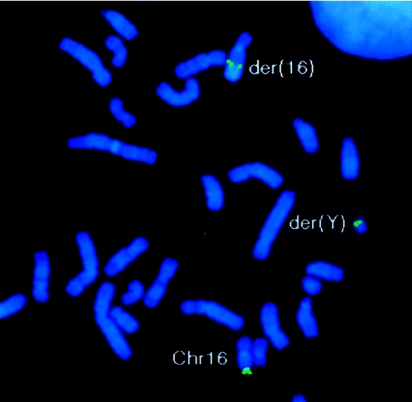
Figure 1.
Results of FISH. BAC clone 58A18 (green), which maps to 16q24.3, crosses the t(Y;16)(q11.2;q24.3) translocation breakpoint and hybridizes to the der(Y), der(16), and normal chromosome 16. Metaphase spreads of lymphoblastoid cells from the patient with the t(Y;16) translocation were hybridized with a biotin-labeled BAC 58A18 probe, as described in the Subjects and Methods section.
Further database searches of available sequences from BAC 58A18 and overlapping proximal BAC 463O9 revealed the presence of the FOXF1 (FREAC-1, or FKHL-5) forkhead-family-member gene, ∼200 kb proximal to the translocation breakpoint. Although this gene seemed to be a good candidate for LD, the distance from the breakpoint was quite large. However, further analysis of sequences from BAC 463O9 showed the presence of two additional forkheadlike-family-member genes—FOXC2 and the human orthologue of mouse FKH-6 (FREAC-7). FOXC2 and FREAC-7 (now termed “FOXL1” [Kaestner et al. 2000]) had previously been shown to map to 16q24 in humans and to be tightly linked in both mouse and humans (Kaestner et al. 1996). We determined that the two genes map ∼60 kb distal to FOXF1 and ∼120 kb proximal to the translocation breakpoint that we studied (fig. 2). Thus, three forkhead-family homologues cluster within a 65-kb interval at 16q24.3. The complete coding sequence (GenBank accession number AF315075 [GenBank Overview]) of FOXL1 was determined on the basis of the genomic sequence. We first investigated FOXC2 for involvement in LD, because of both its role in heart, craniofacial, and vertebral development in the mouse and the fact that it is strongly expressed in developing mouse blood vessels (Kaestner et al. 1996; Iida et al. 1997).
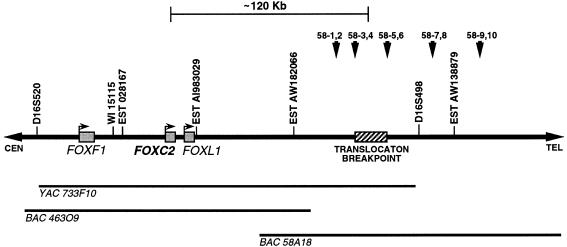
Figure 2.
Map of the 16q24.3 region containing the FOXC2 gene. The positions of several sequence-tagged sites and ESTs are indicated. The shaded boxes represent three forkhead genes—FOXF1, FOXC2, and FOXL1. The hatched box indicates the position of the translocation breakpoint identified in the patients with the t(Y;16) translocation and lymphedema. The translocation breakpoint is ∼120 kb distal to the FOXC2 gene. The positions of YAC 733F10, BAC 463O9, and BAC 58A18 are indicated below the map. The arrowheads indicate the positions of five PCR-based FISH probes used to fine-map the translocation breakpoint
The FOXC2 gene produces a 2.2-kb transcript with a 1.5-kb single exon coding region that is highly (∼70%) GC rich (Miura et al. 1997). Primers were developed for sequencing the gene from PCR-amplified genomic DNA. We first sequenced DNA from a fibroblast line established from a fetus (II-7 in family 1; see fig. 3) that, because of hydrops fetalis, was electively aborted at 17 wk gestation. The fetal karyotype was 46,XX. The father was diagnosed with hereditary LD, and two sons have distichiasis. An earlier pregnancy (II-6 in family 1) was electively aborted because of the presence of hydrops and presumed Turner syndrome, although subsequent pathological examination did not show internal abnormalities compatible with Turner syndrome. A karyotype was not performed. The family history suggested to us that the hydrops fetalis seen in the two fetuses was a result of the lymphedema gene mutation in this family.
A nonsense mutation was found at nucleotide 297 of the FOXC2 coding region in fibroblasts from fetus II-7 in family 1, by both forward and reverse sequencing. This mutation creates a novel BfaI restriction site and was verified in additional affected family members, by sequence analysis and restriction digestion of PCR products from genomic DNA (fig. 4a). The mutation was subsequently confirmed in archival tissue from fetus II-6, by restriction-digestion analysis of PCR products. The mutation creates a stop codon that would result in truncation of the predicted protein at amino acid 99 within the conserved forkhead domain. A second family diagnosed with LD, in which the additional features of cystic hygroma, arachnoid cysts, and cleft palate were seen in the proband, was subsequently analyzed (fig. 3). A 4-nucleotide (GGCC) duplication at position 1093 of the coding region was detected by direct sequencing (fig. 4b). The mutation, which would create 98 novel amino acids before truncating the protein, lies in the carboxy-terminal region after the forkhead domain. This mutation creates a novel Nae I restriction site, which allowed verification of the mutation in all family members (fig. 4b). Both mutations would result in haploinsufficiency of the FOXC2 protein and are consistent with a frequently seen mechanism for dominantly inherited disorders with variable phenotype.
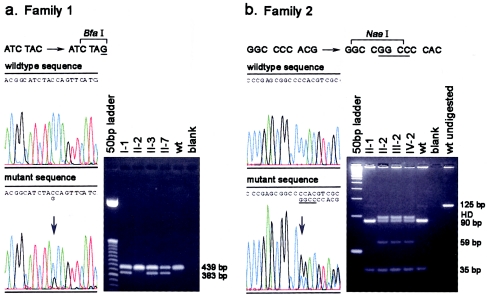
Figure 4.
Mutation analysis in two families with hereditary lymphedema. Mutations in the FOXC2 gene were detected by direct sequencing of the PCR-amplified coding region of the gene. Results were confirmed by sequencing the reverse strand and by restriction-enzyme analysis. a, Sequencing revealed a TAC→TAG nonsense mutation (C297G) (arrow, left panel) in the proband (II-7) in family 1, creating a novel BfaI restriction site. The right panel shows the cosegregation of the novel restriction site with the LD phenotype. Fetus II-6 was also shown to have the mutation, by restriction digest analysis (not shown). A 439-bp region of the FOXC2 gene was PCR amplified and digested with BfaI. Products from normal alleles are not cut and remain 439 bp, whereas the mutant allele is cut into 383- and 53-bp fragments (not retained on gel). b, Sequence analysis revealing a 4-bp insertion in the proband (IV-2) in family 2. The GGCC insertion creates a shifted sequence beginning at the arrow (left panel). A novel NaeI restriction site is created by the insertion. The right panel shows the cosegregation of the NaeI fragments with the LD phenotype in family 2. PCR was used to amplify a 125-bp region of the FOXC2 gene (129 bp for the mutant allele). NaeI digestion of the PCR products results in 35- and 90-bp fragments for normal alleles, because of the presence of a naturally occurring NaeI site in the product. In alleles with the insertion, the 129-bp product is cut twice, generating two 35-bp fragments and a 59-bp fragment. Bands labeled “HD” show uncut heteroduplexes.
Discussion
The forkhead “winged-helix” family of transcription factors now includes >80 members in various species, with ⩾17 homologues currently known in humans. Forkhead proteins bind DNA via a highly conserved 100-amino-acid forkhead motif (Kaestner et al. 1993), a variant of the helix-turn-helix motif, and were first identified in the Drosophila gene forkhead (fkh) (Weigel et al. 1989). The forkhead DNA–binding domain is widely conserved among species, and its developmental role has been defined by mutations in Drosophila, mice, and other eukaryotes (Kaufmann and Knochel 1996). The human forkhead genes generally map throughout the genome, with the exception of 16q24.3, where three forkhead genes—FOXF1 (FKHL-5, or FREAC-1), FOXL1 (FREAC-7), and FOXC2 (MFH-1)—are located. This clustering was previously noted in both humans and mice, and we have found that these genes lie within a 65-kb region in humans. FOXC2 and FOXL1 are tightly linked, suggesting coordinate regulation (Kaestner et al. 1996).
No mutations in FOXC2 have been previously described in humans. Mutations in other forkhead-family genes have been found in association with chromosomal translocations in acute lymphocytic leukemia (Parry et al. 1994), secondary acute leukemia (Hillion et al. 1997), and alveolar rhabdomyosarcoma (Galili et al. 1993) but, to date, in only one other hereditary disorder. Mutations in the FOXC1 (FKHL-7) gene have been identified in a number of families with dominantly inherited glaucoma phenotypes, including Axenfeld anomaly, Rieger anomaly, iris hypoplasia, and Rieger syndrome (Nishimura et al. 1998; Lehmann et al. 2000; Mirzayans et al. 2000).
Mice with targeted disruptions of the FOXC2 gene have a variety of developmental abnormalities. The gene is expressed in the developing mesodermal mesenchyme of the head, kidney, and bones and may play a role in somite formation. It is also expressed in the developing heart, blood vessels, and limbs and is essential for normal development of the aortic arch and axial skeleton (Kaestner et al. 1996; Yang et al. 2000). Mice deficient in FOXC2 die either ∼12.5 d postconception or shortly after birth and show abnormalities of the heart, aorta, palate, and vertebrae, abnormalities that are sometimes seen in LD. The most commonly found abnormality was interruption of the aortic arch and ventricular-septal defect, similar to that seen in DiGeorge syndrome (Iida et al. 1997). No lymphedema has been reported in FOXC2-deficient mice, and heterozygotes have been noted either to be normal (Iida et al. 1997) or to exhibit ocular abnormalities, including abnormalities in ocular drainage structures, hypoplastic iris, and thin iris and pigment epithelium (Smith et al. 2000). These abnormalities are thought to arise from abnormal mesodermal development of the iris. The abnormalities in anterior-chamber drainage of aqueous humor are distantly related to lymphedema. Detailed examination for lymphedema and distichiasis in FOXC2-deficient mice has not been reported but is warranted on the basis of our findings, as is detailed examination of ocular structures in patients with LD.
Both mutations seen in the two families described in the present study would be predicted to truncate and inactivate one allele of FOXC2 in affected patients. No mutations were detected in the normal allele, by forward and reverse sequencing of the entire coding region. The results are consistent with an autosomal dominant mode of inheritance due to haploinsufficiency. This mechanism of mutation and mode of inheritance are known to result in variability in phenotype in a number of human genetic disorders, especially when found in a developmental gene with pleiotrophic effects. This is likely to account for the variable phenotype seen in LD, which can include cleft palate, extradural cysts, and heart defects. As we have found in family 1, fetal cystic hygroma and hydrops can also be associated with FOXC2 mutation and should be considered as a further phenotypic manifestation.
We hypothesize that the translocation in our index case inactivates the FOXC2 gene—and, perhaps, other genes in the region—by a position effect such as has been observed with PAX6 and SOX9 inactivation in humans (Kleinjan and van Heyningen 1998). SOX9 inactivation from breakpoints as far as 850 kb away from the gene has been observed. Interestingly, it has also been shown that primary congenital glaucoma in one of the patients described by Nishimura et al. (1998) was caused by inactivation of FOXC1, as a result of a translocation position effect. The breakpoint described in the present study is ∼120 kb 3′ of the FOXC2 gene. Since the FOXL1 gene maps between the two, it could also be inactivated and have phenotypic effects in this patient. Unfortunately, the patient was lost to long-term follow-up at infancy, and clinical outcome is unknown. We examined this hypothesis by measuring FOXC2 mRNA levels in lymphoblastoid cells from the translocation patient, compared with those in normal control cell lines. However, no expression of FOXC2 was found in these cells by either northern blot analysis or reverse-transcriptase PCR.
The finding of inactivating mutations in one allele of FOXC2 in two unrelated families with LD, in conjunction with a likely position-effect inactivation in a third individual, provides strong evidence that these mutations are responsible for the disorder. Hereditary lymphedema is a heterogeneous disorder, and mutations in the FLT4 (VEGFR-3) gene at 5q35 have been shown to be one cause of hereditary lymphedema type I, or Milroy disease (Karkkainen et al. 2000). The phenotype in LD can include not only lymphedema and distichiasis but also abnormalities of cardiac and skeletal development and, perhaps, of other tissues. This is consistent with the predicted pleiotrophic effects of mutations in a forkhead gene such as FOXC2, which plays a key role in multiple developmental processes involving mesenchymal cell interactions. The identification of the gene responsible for LD will make molecular diagnoses possible and will allow a better definition of both this syndrome and its frequency. It also provides further insight into the mechanisms of primary lymphedema and the role of FOXC2 and forkhead genes, in general, in human development and genetic disease.
Acknowledgments
We are grateful to the family members who participated in this study. We thank Charles Jackson and Janet Ulm, for help in ascertaining and locating the families; Robert Lyons and the University of Michigan Sequencing Core, for assistance; N. Reede Cooly, Jr., for fetal pathology assistance with fetus II-7 in family 1; and Cynthia Gaffney, for manuscript preparation. This work was supported in part by National Institutes of Health grant CA43222 (to T.W.G.) and a Muscular Dystrophy Association grant (to R.P.E.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for FOXC2 [accession number NM_005251] and FOXL1 [accession number AF315075])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for Milroy disease [MIM 153100] and LD [MIM 153400])
References
- Baskaran N, Kandpal RP, Bhargava AK, Glynn MW, Bale A, Weissman SM (1996) Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res 6:633–638 [DOI] [PubMed] [Google Scholar]
- Corbett CRR, Dale RF, Coltart DJ, Kinmonth JB (1982) Congenital heart disease in patients with primary lymphoedemas. Lymphology 15:85–90 [PubMed] [Google Scholar]
- Dagenais SL, Guevara-Fujita M, Loechel R, Burgess AC, Miller DE, Yuzbasiyan-Gurkan V, Brewer GJ, Glover TW (1999) The canine copper toxicosis locus is not syntenic with ATP7B or ATX1 and maps to a region showing homology to human 2p21. Mamm Genome 10:753–756 [DOI] [PubMed] [Google Scholar]
- Erickson RP, Hudgins L, Stone JF, Schmidt S, Wilke C, Glover TW (1995) A “balanced” Y;16 translocation associated with Turner-like neonatal lymphedema suggests the location of a potential anti-Turner gene on the Y chromosome. Cytogenet Cell Genet 71:163–167 [DOI] [PubMed] [Google Scholar]
- Falls HF, Kertesz ED (1964) A new syndrome combining pterygium colli with developmental anomalies of the eyelids and lymphatics of the lower extremities. Trans Am Ophthalmol Soc 62:248–275 [PMC free article] [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ III, Emanuel BS, Rovera G, Barr FG (1993) Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 5:230–235 [DOI] [PubMed] [Google Scholar]
- Goldstein S, Qazi QH, Fitzgerald J, Goldstein J, Friedman AP, Sawyer P (1985) Distichiasis, congenital heart defects and mixed peripheral vascular anomalies. Am J Med Genet 20:283–294 [DOI] [PubMed] [Google Scholar]
- Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard OA (1997) AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood 90:3714–3719 [PubMed] [Google Scholar]
- Iida K, Koseki H, Kakinuma H, Kato N, Mizutani-Koseki Y, Ohucki H, Yoshioka H, Noji S, Kawamura K, Kataoka Y, Ueno F, Taniguchi M, Yoshida N, Sugiyama T, Miura N (1997) Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development 124:4627–4638 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Bleckmann SC, Monaghan AP, Schlöndorff J, Mincheva A, Lichter P, Schütz G (1996) Clustered arrangement of winged helix genes fkh-6 and MFH-1: possible implications for mesoderm development. Development 122:1751–1758 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knöchel W, Martínez DE (2000) Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 14:142–146 [PubMed] [Google Scholar]
- Kaestner KH, Lee K-H, Schlöndorff J, Hiemisch H, Monaghan AP, Schütz G (1993) Six members of the mouse forkhead gene family are developmentally regulated. Proc Natl Acad Sci USA 90:7628–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkkainen MJ, Ferrell RE, Lawrence EC, Kimak MA, Levinson KL, McTigue MA, Alitalo K, Finegold DN (2000) Missense mutations interfere with VEGFR-3 signalling in primary lymphoedema. Nat Genet 25:153–159 [DOI] [PubMed] [Google Scholar]
- Kaufmann E, Knochel W (1996) Five years on the wings of fork head. Mech Dev 57:3–20 [DOI] [PubMed] [Google Scholar]
- Kleinjan D-J, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 [DOI] [PubMed] [Google Scholar]
- Lehmann OJ, Ebenezer ND, Jordan T, Fox M, Ocaka L, Payne A, Leroy BP, Clark BJ, Hitchings RA, Povey S, Khaw PT, Bhattacharya SS (2000) Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am J Hum Genet 67 (electronically published; print version pending) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangion J, Rahman N, Mansour S, Brice G, Rosbotham J, Child AH, Murday VA, Mortimer PS, Barfoot R, Sigurdsson A, Edkins S, Sarfarazi M, Burnand K, Evans AL, Nunan TO, Stratton MR, Jeffery S (1999) A gene for lymphedema-distichiasis maps to 16q24.3. Am J Hum Genet 65:427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans F, Gould DB, Heon E, Billingsley GD, Cheung JC, Mears AJ, Walter MA (2000) Axenfeld-Rieger syndrome resulting from mutation of the FKHL-7 gene on chromosome 6p25. Eur J Hum Genet 8:71–74 [DOI] [PubMed] [Google Scholar]
- Miura N, Iida K, Kakinuma H, Yang XL, Sugiyama T (1997) Isolation of the mouse (MFH-1) and human (FKHL-14) mesenchyme fork head-1 genes reveals conservation of their gene and protein structures. Genomics 41:489–492 [DOI] [PubMed] [Google Scholar]
- Neel JV, Schull WJ (1954) The dominant gene in man. In: Human heredity. University of Chicago Press, Chicago, pp 41–56 [Google Scholar]
- Nishimura D, Swiderski R, Alward W, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC (1998) The forkhead transcription factor gene FKHL-7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19:140–147 [DOI] [PubMed] [Google Scholar]
- Parry P, Wei Y, Evans G (1994) Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer 11:79–84 [DOI] [PubMed] [Google Scholar]
- Robinow M, Johnson GF, Verhagen AD (1970) Distichiasis-lymphoedema. A hereditary syndrome of multiple congenital defects. Am J Dis Child 119:343–347 [PubMed] [Google Scholar]
- Schmidt Drury S, Erickson RP, Glover TW (1998) Y;16 translocation breakpoint associated with a partial Turner phenotype identifies a foamy virus insertion. Cytogenet Cell Genet 80:199–203 [DOI] [PubMed] [Google Scholar]
- Schwartz JF, O'Brien MS, Hoffman JS Jr (1980) Hereditary spinal arachnoid cysts, distichiasis, and lymphedema. Ann Neurol 7:340–343 [DOI] [PubMed] [Google Scholar]
- Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, Nishimura DY, Alward WLM, Hogan BLM, John SWM (2000) Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet 9:1021–1032 [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H (1989) The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57:645–658 [DOI] [PubMed] [Google Scholar]
- Yang X-L, Matsuura H, Fu Y, Sugiyama T, Miura N (2000) MFH-1 is required for bone morphogenetic protein-2-induced osteoblastic differentiation of C2C12 myoblasts. FEBS Lett 470:29–34 [DOI] [PubMed] [Google Scholar]