Abstract
Primary microcephaly is a genetic disorder in which an affected individual has a head circumference >3 SDs below the age- and sex-related mean. A small but apparently normally formed brain is the reason for the reduced head circumference, and, probably because of this, all affected individuals are mentally retarded. The condition is genetically heterogeneous, and four loci have already been identified. We now report a fifth locus, MCPH5, which is an 8-cM region mapping to chromosome 1q31, defined by the markers GATA135F02 and D1S1678.
Primary microcephaly (MIM 251200) is defined as an autosomal recessive condition in which an individual’s head circumference is >3 SDs below the age- and sex-related mean at birth and thereafter. Affected individuals are usually moderately mentally retarded but have no other neurological, syndromic, or significantly dysmorphic features. Other causes of microcephaly, such as chromosomal abnormalities or environmental effects, must be rigorously eliminated, since primary microcephaly is, at present, a clinical “diagnosis of exclusion” (Ross and Frias 1977).
It has recently been shown that primary microcephaly is genetically heterogeneous, with four loci having been identified: MCPH1, at 8p (Jackson et al. 1998); MCPH2, at 19q (Roberts et al. 1999); MCPH3, at 9q (Moynihan et al. 2000); and MCPH4, at 15q (Jamieson et al. 1999). We now report a fifth locus, MCPH5, mapping to an 8-cM region, on chromosome 1q31, that is defined by the flanking markers GATA135F02 and D1S1678.
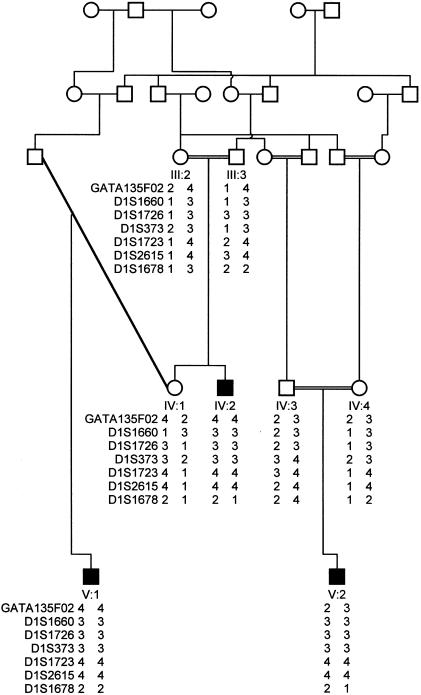
The MCPH5 locus was identified in a consanguineous family from Multan in Pakistan (fig. 1). In this family, there are three living affected individuals from whom DNA was available, as well as an additional child who, on the basis of medical history, had been identified as affected and who had died. All three affected children were microcephalic at birth, with head circumferences between −5 and −7 SD from the norm when they were examined at ages 4, 7, and 28 years (fig. 2). All had moderate mental retardation, as judged by a local pediatrician, the family, and ourselves, with no apparent diminution of abilities with age. They were all in good health and not dysmorphic, had no past medical history worthy of note, and, with the exception of minor language delay, had normal developmental milestones. All were affable, followed instructions well, and hence had learned good self-help skills. None had undergone neuroimaging studies. The parents had normal intelligence and head-circumference measurements. Although it has been stated that carriers of primary microcephaly have diminished intelligence (Qazi and Reed 1975), this has not been seen in any of the families in which linkage to MCPH loci has been reported.
Figure 1.
Pedigree structure of family studied, and genotypes for the five markers on 1q31 that encompass the MCPH5 locus; for clarity, unaffected siblings have been omitted. Marker order is that of the map published by the Centre for Medical Genetics, Marshfield Medical Research Foundation.
Figure 2.
Photograph of individual IV-2 at age 28 years, illustrating a small head size but otherwise normal appearance.
DNA was extracted from peripheral blood lymphocytes by a standard nonorganic method. A high-resolution genome search was performed on seven family members, by use of the CHLC/Weber Human Screening set, version 8 (Research Genetics). This screening set contains 386 fluorescently labeled microsatellite markers, 365 of which cover the 22 autosomal chromosomes, with an average distance of 10 cM between markers.
PCR reactions were performed according to the manufacturer’s instructions. The PCR products were separated by gel electrophoresis on an ABI Prism 377 sequencer and were analyzed by the ABI GENESCAN and GENOTYPER 1.1.1 analysis packages. A region on chromosome 1 was examined further, by use of additional markers—GATA135F02, D1S1726, D1S373, D1S1723, and D1S2615—that were by use of The Genome Database and the Center for Medical Genetics, Marshfield Medical Research Foundation. Allele frequencies for the markers were determined on the basis of data from 30 unrelated nonmicrocephalic individuals from northern Pakistan.
The LINKAGE analysis programs (Lathrop et al. 1984) were used for two-point analysis assuming an autosomal recessive mode of inheritance, full penetrance, a disease-gene frequency of .003, and marker-allele frequencies that we had derived in the manner discussed above. Multipoint analysis was performed by GENEHUNTER 2.0 beta (Kruglyak et al. 1996), with the same parameters as have been noted above. However, because of both the computational limitations of GENEHUNTER 2.0 beta and the size of the family, the pedigree was split into two parts, for multipoint-LOD-score analysis (the uncle and nephew [IV-2 and V-1] were analyzed together, and individual V-2 was analyzed separately), with the same coefficient of inbreeding being used for all three of these affected individuals. Marker distances were calculated by use of the Marshfield linkage map (Center for Medical Genetics, Marshfield Medical Research Foundation).
Analysis of the results of the genome search identified an area of interest on chromosome 1q31. A common homozygous region of 8 cM was found in the three affected individuals, defined by markers GATA135F02 and D1S1678 (fig 1). Two-point analysis gave a LOD score of 3.28 (recombination fraction [θ] 0) at D1S1723 (table 1). A maximum multipoint LOD score of 3.7 was calculated for marker D1S1726 on chromosome 1q31. The locus is registered at the HUGO Gene Nomenclature Committee Web site, as “MCPH5.”
Table 1.
Two-Point LOD Scores, at θ = 0, for Each Marker at 1q31, versus the Primary Microcephaly Phenotype in the Family Studied
| Marker | LOD Score at θ = 0 |
| GATA135F02 | −2.99 |
| D1S1660 | 2.07 |
| D1S1726 | 1.22 |
| D1S373 | 2.65 |
| D1S1723 | 3.28 |
| D1S2615 | 2.56 |
| D1S1678 | −∞ |
Haplotype analysis and a LOD score >3 both suggest that region 1q31 contains a gene causing primary microcephaly. The MCPH5 locus is <8 cM in size and contains no obvious candidate gene (GeneMap’99). It will be interesting to see how many autosomal recessive gene loci are found for primary microcephaly and by what mechanism those genes cause a reduction in cerebral cortex size. Deletions encompassing the chromosome 1q31 region have been described that invariably cause mental retardation and that usually are associated with congenital microcephaly (Schinzel 1994). Whether the microcephaly seen in such patients is caused by hemizygosity for the MCPH5 gene, resulting in either a gene-dosage effect or the exposure of the recessive-gene mutation by the deletion, is unclear (Perez-Castillo et al. 1984).
Acknowledgments
We would like to thank the family for their help and the Wellcome Trust, Action Research, and West Riding Fund for funding.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield, Medical Research Foundation, http://research.marshfieldclinic.org/genetics (for marker order and distances)
- GeneMap'99, http://ncbi.nlm.nih.gov/genemap99 (for radiation-hybrid database of mapped genes and expressed sequence tags)
- Genome Database, The, http://www.gdb.org (for primer sequences for fine-mapping markers)
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature (for MCPH5)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for autosomal recessive primary microcephaly [MIM 251200])
References
- Jackson AP, McHale DP, Campbell DA, Jaffri H, Rashid Y, Mannan J, Karbani G, Corry P, Levene MI, Mueller RF, Markham AF, Lench NJ, Woods CG (1998) Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet 63:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Govaerts C, Abramowicz MJ (1999) Primary autosomal recessive microcephaly: homozygosity mapping of MCPH4 to chromosome 15. Am J Hum Genet 65:1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalovel J-M, Julier C, Ott J (1984) Strategies for multilocus analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan L, Jackson AP, Roberts E, Karbani G, Lewis I, Corry P, Turner G, Mueller RF, Lench NJ, Woods CG (2000) A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am J Hum Genet 66:724–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castillo A, Martin-Lucas MA, Abrisqueta JA (1984) Is a gene for microcephaly located on chromosome 1? Hum Genet 67: 230–232 [DOI] [PubMed] [Google Scholar]
- Qazi QH, Reed TE (1975) A possible major contribution to mental retardation in the general population by the gene for microcephaly. Clin Genet 7: 85–90. [DOI] [PubMed] [Google Scholar]
- Roberts E, Jackson AP, Carradice AC, Deeble VJ, Mannan J, Rashid Y, McHale DP, Markham AF, Lench NJ, Woods CG (1999) The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1-13.2. Eur J Hum Genet 7:815–820 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Frias JL (1977) Microcephaly. In: Vinken PJ, Bruyn GW (eds) Congenital malformations of the brain and skull. Vol 30: Handbook of clinical neurology. Elsevier Holland Biomedical, Amsterdam, pp 507–524 [Google Scholar]
- Schinzel A (1994) Human cytogenetics database, version 1. Oxford University Press, Oxford [Google Scholar]