Abstract
To clarify the relationship between variation in mtDNA and the development of cardiomyopathy (CM), the complete sequences of mtDNAs of two brothers with dilated CM were compared with those of 181 patients who had CM and with those of 168 control subjects. Five patients with CM shared a novel homoplasmic point mutation (G12192A tRNAHis), and all of them demonstrated the evolutionarily related D-loop sequence. The results suggest that this novel mutation originated from the same ancestor and that its presence strongly predisposes carriers to CM.
DNA-based gene analysis has shown that more than half of hypertrophic cardiomyopathy (HCM [MIM 192600]) cases are the result of sarcomeric disease (Geisterfer-Lowrance et al. 1990; Spirito et al. 1997). However, in the remaining cases of HCM—and in cases of dilated cardiomyopathy (DCM) and of variable phenotype expression among members of the same family with a particular gene mutation (Graham and Owens 1999)—the pathogenesis remains unknown. Meanwhile, mtDNA mutation has emerged as a cause of hereditary cardiomyopathy (CM) (Wallace 1992, 1999; Ozawa 1997). Among several point mutations known to cause mitochondrial encephalomyopathy, frequent cardiac involvement was reported in patients with encephalopathy who have the A3243G transition mutation in mtDNA, which causes MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes) (Goto et al. 1990). A small but significant proportion (⩽2.5%) of patients with CM were found to have the A3243G transition mutation without encephalomyopathy (authors' unpublished data). To identify unknown mtDNA mutations causing CM, two brothers with maternally inherited CM were assessed.
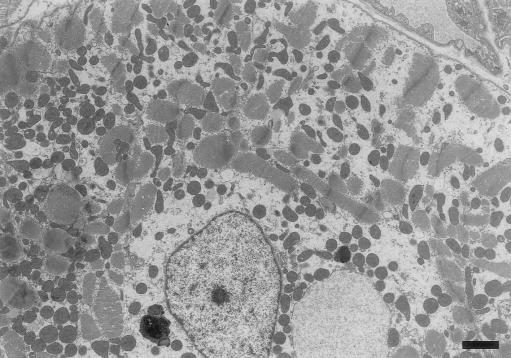
A 60-year-old Japanese man who had reduced contraction of the left ventricle (LV) (percent fractional shortening 20%–30%, by echocardiography) for 10 years was admitted for evaluation of his cardiac function. (His mother had died of congestive heart failure, and his younger brother [age 56 years] had been treated for acute exacerbation of DCM.) The echocardiography and electrocardiogram (EKG)-gated cardiac magnetic resonance imaging revealed a deformed LV cavity with decreased contraction (Suzuki et al. 1999). Results of the chest X-ray, EKG, cardiac catheterization, and coronary angiography were normal, except for the presence of diffuse hypokinesis of the LV wall (ejection fraction 51%). The endomyocardial biopsy specimen demonstrated marked loss or disruption of sarcomeres and massive accumulation of mitochondria of various sizes (fig. 1). Direct sequencing of the entire mtDNAs extracted from the endomyocardial biopsy sample of the elder brother and from white blood cells of both men revealed that the brothers share a G→A substitution at position 12192 in tRNAHis (MIM 590040). This mutation is located 2 bp from the 3′ end of the TΨC loop of the tRNA. Because the mutation adds an A:T base pair and shortens the loop itself, it may affect mitochondrial function.
Figure 1.
Electron-microscopic features of cardiomyocytes of the patient with the G12192A tRNAHis homoplasmic mutation. Mitochondria of various sizes heterogeneously accumulated within a single cell, and the sarcomere structure was disrupted or totally lost. Lysosomes were prominently accumulated at both sides of the nucleus. The round structure at the right of the nucleus has a double lumen and appears to be a greatly enlarged mitochondrion. (Scale bar = 2 μm.)
Searching for other patients with the same mutation, we screened 126 patients with HCM, 55 patients with DCM, and 168 control subjects without cardiac disease. After obtaining informed consent, we tested for single-strand conformation polymorphism. We detected four additional patients with the mutation. The first was a patient with HCM who had been resuscitated after cardiac arrest and whose mother and grandmother had died suddenly of DCM. The second patient had asymptomatic HCM; the third had DCM with frequent premature ventricular contractions; and the fourth had DCM with heart failure. No control subject had the mutation. To determine the risk of development of CM in persons carrying this mutation, an odds ratio was calculated from the G12192A frequency in patients with CM and in controls. The modified formula with small numbers of Woolf’s method (Schlesselman 1982) gave an odds ratio of 10.5 (95% confidence interval 1.0–110.0), indicating that persons with the G12192A mutation have a significantly higher mean risk of developing CM than do persons without the G12192A mutation.
All five subjects with the G12192A mutation showed homoplasmy. Because homoplasmic mutation can be a risk factor for slowly progressive disease (Wallace 2000), we performed phylogenetic analysis of mtDNAs by the neighbor-joining method (Saitou and Nei 1987) in the PHYLIP (phylogeny inference package), version 3.5c (Felsenstein 1993), using the 562-bp D-loop sequence (Shoffner et al. 1993; Hutchin and Cortopassi 1995) in the 5 patients with CM who had the G12192A mutation, in 16 of the patients with CM who did not have the mutation, and in 20 of the normal Asian individuals. To our surprise, analysis of the noncoding D-loops revealed that all patients with the G12192A mutation were related to one another and formed a single cluster in the phylogenetic tree (fig. 2). Furthermore, these patients with the G12192A mutation shared an A→T substitution at position 16318 in the D-loop, which was not detected in other patients with CM or in subjects without CM. These findings strongly support the close evolutionary relationship among the mtDNAs of the patients, as well as a single origin of the G12192A mutation.
Figure 2.
Phylogenetic analysis of mitochondrial D-loop sequence in patients with CM who have the G12192A mutation, in other patients with CM, and in control subjects. The phylogenetic tree was constructed by the neighbor-joining method in the PHYLIP package (Felsenstein 1993). White circles = subjects without CM; green circles = patients with CM; and red circles = patients with both CM and the G12192A mutation.
All five patients with the G12192A mutation showed signs of cardiac abnormalities but no evidence of neurological abnormalities or of diabetes, which usually appear (singly or together) in patients with MELAS due to the heteroplasmic A3243G mutation (MIM 540000) and may also appear in a small percentage of patients with CM (Ozawa 1997; Wallace 1999). In contrast to homoplasmy of mtDNAs, heteroplasmic mutations in mitochondrial genes are evolutionarily unrelated and cause severe clinical symptoms in children and young adults (Wallace 2000). Taken together with this homoplasmic mutation and the tight clonal evolutionary relationship of the mtDNAs carrying the mutation, this particular transition mutation belongs to a class of mitochondrial disorders that are characterized by late onset and that therefore escape negative selection (e.g., A4336G [late-onset Alzheimer disease; Hutchin and Cortopassi 1995] or A1555G [maternally inherited susceptibility to antibiotic-induced deafness; Prezant et al. 1993]).
We have investigated in mtDNA a novel point mutation that is associated with cardiomyopathy. Therefore, this entity does not belong to any mutations known to be associated with CM. It is not connected with idiopathic mitochondrial DCM (MIM 510000), because that mutation is restricted to DCM, whereas the point mutation of the present study can result in either DCM and HCM.
Morphological alteration of cardiac mitochondria and structural change, of the mutant tRNAHis, combined with the unique D-loop sequence therefore suggest that patients with the G12192A mutation may constitute a group with genetically determined high risk for the gradual development of HCM or DCM. Screening of blood samples of patients with CM, for homoplasmic mutation in mtDNAs, promises detection of all patients with the mutation and therefore may be useful both for early diagnosis and for prevention of a potentially lethal disease.
Acknowledgments
We thank Drs. Katsushi Tokunaga and Satoru Fukuda (University of Tokyo) for helpful discussions and for the electron-microscopic examination, respectively. This study was supported by the Ministries of Education, Science and Culture and of Health and Welfare, Japan.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for idiopathic DCM, mitochondrial [MIM 510000], RNA, mitochondrial, histidine [MIM 590040], MELAS [MIM 540000], and familial HCM 1 [MIM 192600]).
References
- Felsenstein J (1993) PHYLIP (phylogeny inference package) version 3.5c. Department of Genetics, University of Washington, Seattle [Google Scholar]
- Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG (1990) A molecular basis for familiar hypertrophic cardiomyopathy: a β cardiac myosin heavy chain gene missense mutation. Cell 62:999–1006 [DOI] [PubMed] [Google Scholar]
- Goto Y, Nonaka I, Horai S (1990) A mutation in the tRNALEU(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348:651–653 [DOI] [PubMed] [Google Scholar]
- Graham RM, Owens WA (1999) Pathogenesis of inherited forms of dilated cardiomyopathy. N Engl J Med 341:1759–1762 [DOI] [PubMed] [Google Scholar]
- Hutchin T, Cortopassi G (1995) A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci USA 92:6892–6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T (1997) Genetic and functional changes in mitochondria associated with aging. Physiol Rev 77:425–464 [DOI] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, Shohat M, Fischel-Ghodsian N (1993) Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 4:289–294 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Schlesselman J (1982) Case control studies: design, conduct, analysis. Oxford University Press, Oxford [Google Scholar]
- Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MF, Mirra SS, Beal MF, Yang CC, Gearing M, Salvo R, Watts RL, Juncos JL, Hansen LA, Crain BJ, Fayad M, Reckord CL, Wallace DC (1993) Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics 17:171–184 [DOI] [PubMed] [Google Scholar]
- Spirito P, Seidman CE, McKenna WJ, Maron BJ (1997) The management of hypertrophic cardiomyopathy. N Engl J Med 336:775–785 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Shimamoto R, Nishikawa J, Yamazaki T, Tsuji T, Nakamura F, Shin WS, Nakajima T, Toyo-Oka T, Ohotomo K (1999) Morphological onset and early diagnosis in apical hypertrophic cardiomyopathy: a long-term analysis with nuclear magnetic resonance imaging. J Am Coll Cardiol 33:146–151 [DOI] [PubMed] [Google Scholar]
- Wallace DC (1992) Diseases of the mitochondrial DNA. Annu Rev Biochem 61:1175–1212 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Mitochondrial diseases in man and mouse. Science 283:1482–1488 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am Heart J Suppl 139:S70–S85 [DOI] [PubMed]