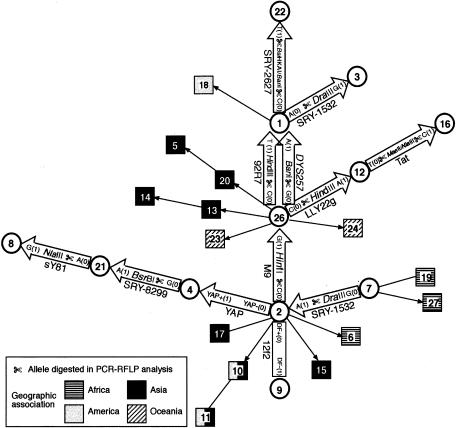
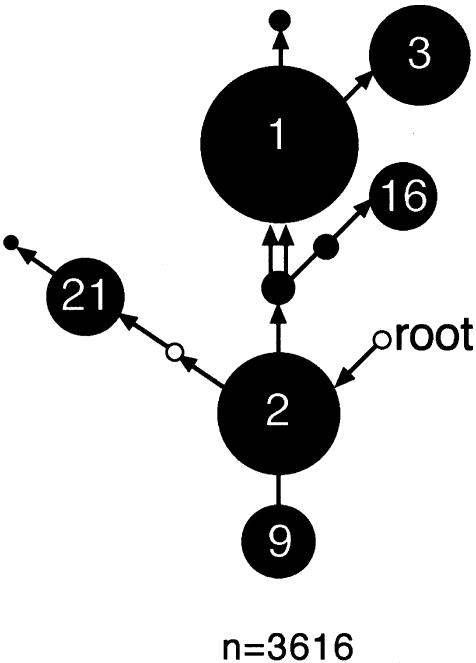
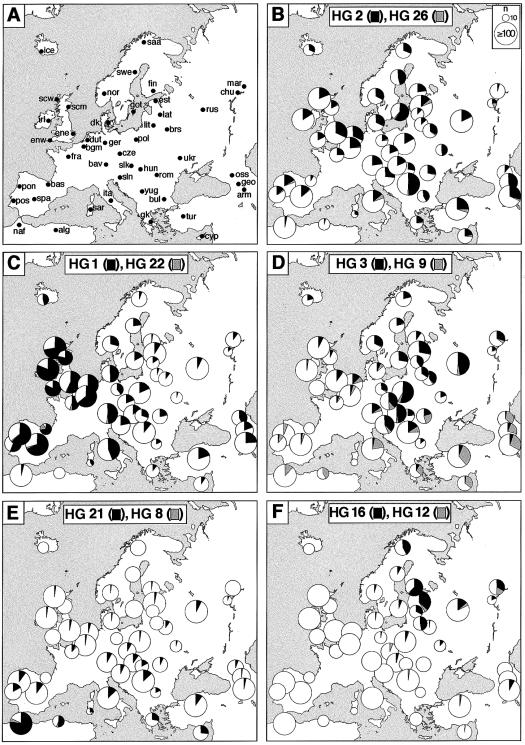
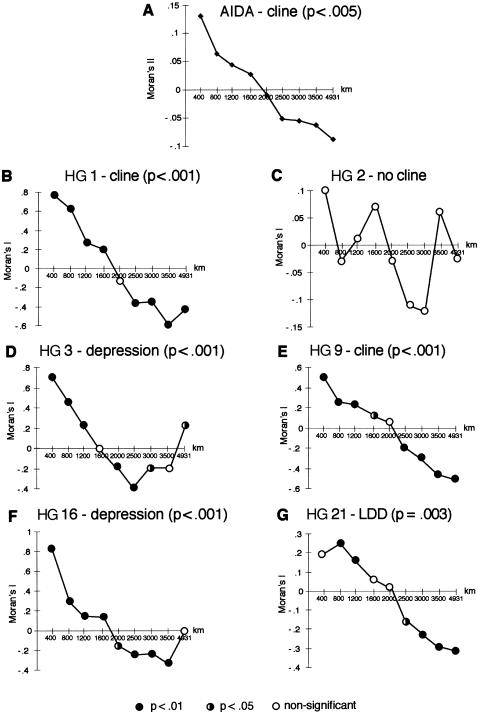
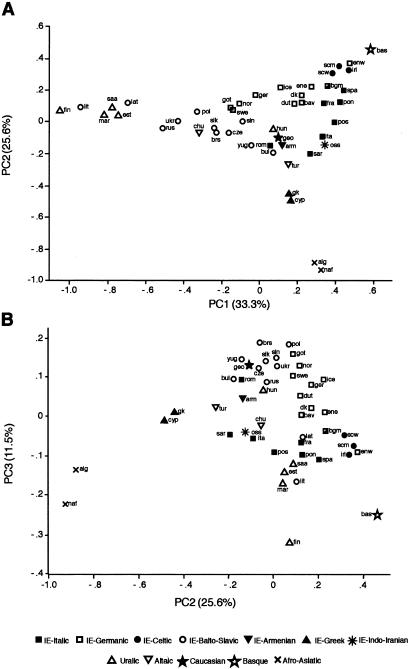
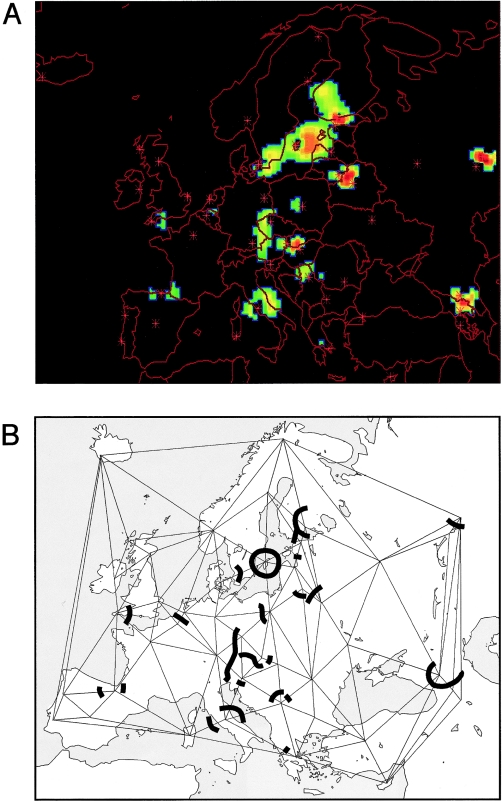
Zoë H Rosser
Zoë H Rosser
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
1,
Tatiana Zerjal
Tatiana Zerjal
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
2,
Matthew E Hurles
Matthew E Hurles
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
1,,*,
Maarja Adojaan
Maarja Adojaan
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
5,
Dragan Alavantic
Dragan Alavantic
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
6,
António Amorim
António Amorim
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
7,
William Amos
William Amos
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
8,
Manuel Armenteros
Manuel Armenteros
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
9,
Eduardo Arroyo
Eduardo Arroyo
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
10,
Guido Barbujani
Guido Barbujani
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
11,
Gunhild Beckman
Gunhild Beckman
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
12,
Lars Beckman
Lars Beckman
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
12,
Jaume Bertranpetit
Jaume Bertranpetit
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
13,
Elena Bosch
Elena Bosch
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
13,,†,
Daniel G Bradley
Daniel G Bradley
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
14,
Gaute Brede
Gaute Brede
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
15,
Gillian Cooper
Gillian Cooper
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
8,
Helena B S M Côrte-Real
Helena B S M Côrte-Real
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
16,
Peter de Knijff
Peter de Knijff
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
17,
Ronny Decorte
Ronny Decorte
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
18,
Yuri E Dubrova
Yuri E Dubrova
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
1,
Oleg Evgrafov
Oleg Evgrafov
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
19,
Anja Gilissen
Anja Gilissen
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
18,
Sanja Glisic
Sanja Glisic
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
6,
Mukaddes Gölge
Mukaddes Gölge
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
20,
Emmeline W Hill
Emmeline W Hill
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
14,
Anna Jeziorowska
Anna Jeziorowska
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
21,
Luba Kalaydjieva
Luba Kalaydjieva
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
22,
Manfred Kayser
Manfred Kayser
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
23,,‡,
Toomas Kivisild
Toomas Kivisild
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
3,
Sergey A Kravchenko
Sergey A Kravchenko
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
24,
Astrida Krumina
Astrida Krumina
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
25,
Vaidutis Kučinskas
Vaidutis Kučinskas
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
26,
João Lavinha
João Lavinha
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
16,
Ludmila A Livshits
Ludmila A Livshits
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
24,
Patrizia Malaspina
Patrizia Malaspina
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
27,
Syrrou Maria
Syrrou Maria
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
28,
Ken McElreavey
Ken McElreavey
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
29,
Thomas A Meitinger
Thomas A Meitinger
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
30,
Aavo-Valdur Mikelsaar
Aavo-Valdur Mikelsaar
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
4,
R John Mitchell
R John Mitchell
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
31,
Khedoudja Nafa
Khedoudja Nafa
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
32,
Jayne Nicholson
Jayne Nicholson
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
3,
Søren Nørby
Søren Nørby
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
33,
Arpita Pandya
Arpita Pandya
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
2,
Jüri Parik
Jüri Parik
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
5,
Philippos C Patsalis
Philippos C Patsalis
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
28,
Luísa Pereira
Luísa Pereira
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
7,
Borut Peterlin
Borut Peterlin
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
34,
Gerli Pielberg
Gerli Pielberg
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
5,
Maria João Prata
Maria João Prata
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
7,
Carlo Previderé
Carlo Previderé
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
35,
Lutz Roewer
Lutz Roewer
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
23,
Siiri Rootsi
Siiri Rootsi
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
5,
D C Rubinsztein
D C Rubinsztein
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
36,
Juliette Saillard
Juliette Saillard
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
33,
Fabrício R Santos
Fabrício R Santos
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
2,,§,
Gheorghe Stefanescu
Gheorghe Stefanescu
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
37,
Bryan C Sykes
Bryan C Sykes
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
32,
Aslihan Tolun
Aslihan Tolun
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
38,
Richard Villems
Richard Villems
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
5,
Chris Tyler-Smith
Chris Tyler-Smith
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
2,
Mark A Jobling
Mark A Jobling
1Department of
Genetics, University of Leicester, Leicester; 2CRC Chromosome
Molecular Biology Group, Department of Biochemistry,
and 3Institute of Molecular Medicine, University of
Oxford, Oxford; 4Institute of General and Molecular
Pathology, University of Tartu and 5Estonian Biocentre, Tartu,
Estonia; 6Laboratory for Radiobiology and Molecular Genetics,
Institute of Nuclear Sciences “Vinca," Belgrade; 75
IPATIMUP and Faculdade de Ciências, Universidade do Porto, Porto,
Portugal; 8Department of Zoology, University of Cambridge,
Cambridge; 9Centro de Investigacion y Criminalistica,
Laboratorio de ADN, Policia Judicial, Guardia Civil, and
10Laboratorio de Biología Forense, Departamento de
Toxicología y Legislación Sanitaria, Universidad Complutense,
Madrid; 11Dipartimento di Biologia, Universita di Ferrara,
Ferrara, Italy; 12Umeå University, Department of
Medical Genetics, Umeå, Sweden; 13Unitat de Biologia
Evolutiva, Facultat de Ciecies de la Salut I de la Vida, Universitat Pompeu
Fabra, Barcelona; 14Department of Genetics, Trinity College,
Dublin; 15University of Oslo, Centre for Biotechnology,
Oslo; 16Instituto Nacional de Saúde Dr. Ricardo
Jorge, Lisbon; 17Forensic Laboratory for DNA
Research, MGC-Department of Human and Clinical Genetics, Leiden
University Medical Center, Leiden, The Netherlands;
18Laboratory for Forensic Genetics and Molecular Archaeology,
Center for Human Genetics, K.U. Leuven, Leuven, Belgium;
19Research Centre for Medical Genetics, Russian Academy of
Medical Sciences, Moscow; 20Department of Physiology,
University of Kiel, Kiel; 21Department of Medical Genetics,
Institute of Endocrinology, Medical University of Lódz, Lódz,
Poland; 22Department of Human Biology, Edith Cowan
University, Joondalup Campus, and Western Australian Institute for Medical
Research, Royal Perth Hospital, Perth; 23Genetic
Research Laboratory, Institute of Legal Medicine, Medical Faculty
(Charité), Humboldt-University Berlin, Berlin;
24Institute of Molecular Biology and Genetics, National
Academy of Science of Ukraine, Kiev; 25Department of Medical
Biology and Genetics, Medical Academy of Latvia, Riga;
26Center of Human Genetics, University of Vilnius, Vilnius,
Lithuania; 27Department of Biology, University of Rome
“Tor Vergata,” Rome; 28The Cyprus Institute of
Neurology and Genetics, Nicosia; 29Unité
d'Immunogénétique Humaine, Institut Pasteur, Paris;
30Institute of Human Genetics, GSF-National Research
Centre, Munich; 31La Trobe University, School of Genetics and
Human Variation, Bundoora, Australia 32Department of Human
Genetics, Memorial Sloan-Kettering Cancer Center, New York;
33Laboratory of Biological Anthropology, Institute of Forensic
Medicine, University of Copenhagen, Copenhagen; 34Division of
Medical Genetics, Department of Obstetrics and Gynaecology, Ljubljana,
Slovenia; 35Dipartimento di Medicina Legale e Sanita Pubblica,
Pavia, Italy; 36Department of Medical Genetics, Wellcome Trust
Centre for Molecular Mechanisms in Disease, Cambridge Institute for
Medical Research, Addenbrooke's Hospital, Cambridge;
37I.C. Biologice, Iasi, Romania; and 38Bogazici
University, Department of Molecular Biology and
Genetics, Istanbul
1