ABSTRACT
The domestication of the dog from its wolf ancestors is perhaps the most complex genetic experiment in history, and certainly the most extensive. Beginning with the wolf, man has created dog breeds that are hunters or herders, big or small, lean or squat, and independent or loyal. Most breeds were established in the 1800s by dog fanciers, using a small number of founders that featured traits of particular interest. Popular sire effects, population bottlenecks, and strict breeding programs designed to expand populations with desirable traits led to the development of what are now closed breeding populations, with limited phenotypic and genetic heterogeneity, but which are ideal for genetic dissection of complex traits. In this review, we first discuss the advances in mapping and sequencing that accelerated the field in recent years. We then highlight findings of interest related to disease gene mapping and population structure. Finally, we summarize novel results on the genetics of morphologic variation.
Introduction
In July of 2004, the first high-quality draft (7.5×) sequence of the dog genome was made publicly available [1], providing an invaluable resource for navigating the genomes of more than 400 recognized dog breeds worldwide, as well as a number of closely related wild canids. This work represented the crowning achievement of the dog genomics community that has for years worked hard to demonstrate the utility of a mammalian genetic system outside of the commonly accepted rodent models [2–4]. In this review, we first briefly highlight some recent advances in our picture of the canine genome. With this background, we then focus on issues related to the identification of genes associated with complex traits, the dog as a model for advancement of human medical genetics, and the development of mapping tools that take advantage of features of population structure.
Canine Genome Maps and Sequence
The high-quality draft sequence of the dog (http://www.genome.ucsc.edu; http://www.ncbi.nlm.nih.gov; http://www.ensembl.org) has illuminated many qualities of the canine genome only hinted at from previous work [1]. The DAPI-banded karyotype of the dog's 40 chromosomes, combined with reciprocal chromosome paint studies from two independent groups, suggested that the canine genome was highly homologous to the human genome and comprised a limited number of conserved segments, intimating a low level of rearrangement between the two [5–7]. The size of the canine genome was initially estimated from maximum likelihood predictions to be 27 morgans in genetic distance [8]. Estimates based on flow sorting of chromosomes suggest a physical size of 2.8 gigabases [9,10]. Predictions based on sequence analysis of euchromatic sequence suggest a size of 2.3–2.4 gigabases [1,11]. Integrated linkage, radiation hybrid (RH), and cytogenetic maps of the dog genome confirmed these general conclusions [12–15].
Most recently a comparative RH map of the dog genome containing 10,000 canine gene sequences has proven invaluable for identifying small rearrangements within the canine genome (http://www-recomgen.univ-rennes1.fr/doggy.html) [16], as well as assisting in the ordering of contigs for the draft assembly [1]. The gene sequences used in construction of the 10,000-gene RH map were derived from a 1.5× sequence of the standard poodle. This initial sequence of the dog genome contained at least partial orthologs for 75% (18,473) of annotated human genes [11]. Portions of 9,850 individual genes or about half of all dog genes were localized on an RH panel with 200 kilobase resolution [16]. A total of 264 conserved segments less than 500 kilobases in size were identified from a comparison of the dog map and human genome sequence (National Center for Biotechnology Information Build 34), matching well with predictions from the draft assembly [1]. This provided a clear snapshot of the order of canine genes relative to the human genome, and precise information about breakpoint position. Subsequent studies on breakpoint reuse across mammalian systems, utilizing RH maps from a variety of mammalian species, verify these data [17].
The 7.5× draft assembly of the canine genome was derived from a female boxer, selected because of her apparent lack of heterozygosity (H.G. Parker, unpublished data). The assembled sequence spans most of the dog's 2.4 gigabases and is derived from 31.5 million sequence reads (http://www.genome.ucsc.edu) [1]. The quality of the assembly is extremely high compared to initial assemblies of other mammalian genomes [18–21]. Half of assembled bases (N50 contig size) are in contigs of 180 kilobases, and the N50 supercontig size is 45.0 megabases, which is considerably longer then the mouse genome at a similar point in its assembly. The reasons are 2-fold. First, technical advances in sequencing result in longer and higher-quality reads. Second, advances in bioinformatics have improved the accuracy with which genomes can be assembled. From a practical standpoint, this means that the majority of canine genes contain no sequence gaps and most canine autosomes are comprised of one to three supercontigs. The current gene count is listed as approximately 19,000, with about 75% representing 1:1:1 orthologs among dog, human, and mouse.
With the availability of the canine genome sequence, the research community is now ready to tackle goals stated nearly a decade ago, when the first arguments were put forth as to why the dog system offered unique advantages for mapping complex traits [22–24]. In general, researchers have focused on the dog as a system for advancing general medical knowledge. In addition, a small but growing number of groups have used the dog for tackling the genetics of morphology and behavior.
The Canine Genome and Molecular Mechanisms of Disease
Among the most well-studied elements of the canine genome sequence are the short interspersed nuclear elements (SINEs) [25–27]. These retrotransposons are implicated in genome evolution and include several families of well-recognized repeats, such as the Alu sequences in humans [18,19,28]. In dogs, the major family of SINEs is derived from a tRNA-Lys, and is distributed throughout the genome at about 126 kilobase spacing [26,29,30]. The frequency of bimorphic SINE elements is 10- to 100-fold higher than what is observed in humans, largely because of the expansion of a single subfamily, termed SINEC-Cf in the canine lineage [11].
As with human Alu repeats, a surprising number of SINEs seem to be located in positions that affect gene expression. A perfect example is the often cited SINEC-Cf element inserted into intron 3 of the gene encoding the hypocretin receptor, resulting in narcolepsy in the Doberman pinscher [31]. These data were the first to link the hypocretin gene family to sleep disorders, and a large body of work on molecular biology of sleep has evolved from these initial studies. Likewise, insertion of a SINE into the canine PTPLA gene leads to multiple splicing defects, causing an autosomal recessive centronuclear myopathy in the Labrador retriever [32].
By studying SINEs in the dog, genome researchers have learned about important disease mechanisms that have not been appreciated from the study of human families. Analysis of other canine diseases demonstrates this as well. For example, analysis of miniature wire-haired dachshunds in the United Kingdom revealed that recessive progressive myeloclonic epilepsy is due to expansion of a dodecamer repeat in the Epm2b gene [33]. Normal dogs carry two sequential copies of the repeat and a third slightly variant copy, while affected individuals carry up to 26 repeats, resulting in dramatic reduction of mRNA by about 900-fold. While simple mutations in the same gene cause Lafora disease in humans, this is the only report of a dodecamer repeat expansion associated with any mammalian disease.
Over 360 genetic disorders found in humans have been described in the dog [3,34], with 46% occurring largely in either one or a few breeds (http://www.vet.cam.ac.uk/idid). It has been said that the “low hanging fruit” of canine genetics is rapidly being plucked. That is, the genes that can be mapped using easily obtainable pedigrees or those caused by highly penetrant alleles are rapidly being identified. To some degree this is true. Loci have been mapped, and in some cases mutations found, for a multitude of common canine diseases (reviewed in [3,22,35,36]). In some cases, the biology of the underlying mutations has been helpful in understanding a comparable human disorder. In other cases, such as the identification of the CNGB3 gene for cone degeneration [37] or the folliculin gene for renal cystadenocarcinoma and nodular dermatofibrosis [38], the work has served primarily to highlight the power of canine genetics for dissecting genetic diseases common to humans and dogs.
Particularly challenging will be the identification of genes associated with complex diseases such as hip dysplasia, a common disease in dogs, affecting up to 50% of the large breeds. The disease is recognized radiographically as subluxation of the femoral head from the acetabulum of the hip joint [39,40], and is likely caused by a mixture of genetic [41–45] and environmental factors [44–48]. Two approaches have been used to try to identify causative genes.
Investigators at the University of Utah have looked for a genetic association in a population of well-characterized and densely genotyped Portuguese water dogs (PWD) using the Norberg angle, a highly heritable and quantitative radiographic measure of joint laxity. They report the presence of two unlinked quantitative trait loci (QTLs) on CFA1, located more than100 megabases apart, which demonstrated statistically significant associations [49]. A third locus on a different chromosome was found to be associated with osteoarthritis [50].
By comparison, Todhunter and colleagues have developed a large outcrossed pedigree of affected Labrador retrievers crossed with unaffected greyhounds [45,51,52]. A variety of measures, including age at detection of femoral capital epiphyseal ossification, distraction index, hip joint dorsolateral subluxation score, and hip joint osteoarthritis, are being used in a genome-wide scan for classical linkage [51]. While no gene has yet been found, pedigree analysis suggests that loci controlling these traits act additively, and that the distraction index may be controlled by a single major locus [45,53].
These studies represent two distinct methods for approaching a complex problem. Both highlight different advantages of using the canine system for genetic analysis. The first makes use of the availability of large controlled populations with limited genetic diversity. The second demonstrates the ability to cross populations showing extremes of phenotype in order to map genes. Each has the potential for success, and comparison of the two methods will improve the design of future studies.
The Canine System and Genetics of Complex Traits
The study of morphology in the PWD represents one of the most interesting stories from the canine genetics community to date. PWD of today are descended from a small number of dogs primarily from two kennels [54,55]. Six ancestors account for about 80% of the current gene pool of 10,000 dogs, and the breed is characterized, in part, by extensive consanguinity, with a range of 0–0.6 [56].
Through a program termed the Georgie Project (http://www.georgieproject.com), investigators at the University of Utah, Gordon Lark, Kevin Chase, and collaborators, have collected extensive phenotypic data (five sets of X rays on over 500) and genotypic data (DNA samples of over 900) on registered PWDs. Ninety-one metrics describing aspects of canine morphology have been extracted from the X rays, and DNA samples have been genotyped with nearly 500 microsatellite markers that span the genome at about 5centimorgan density.
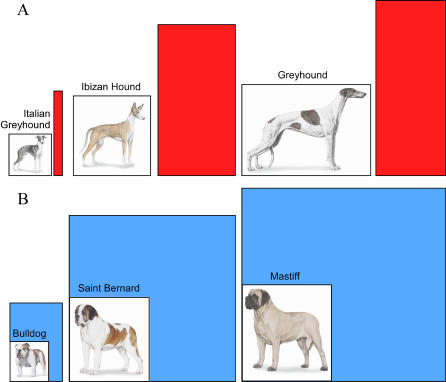
In 2002, a subset of the data was subjected to principal component analysis, which classifies variation of correlated traits into independent linear combinations. Principal component 4, for instance, demonstrates how skull and limb lengths are inversely correlated with the strength of the limb and axial skeletons. Bulldogs have proportionately shorter, wider bones designed to accommodate large body mass, while the greyhound's long, thin limbs are adapted for speed (see Figure 1). Analysis of these data has highlighted 44 putative QTLs on 22 chromosomes that are important for heritable skeletal phenotypes in the PWD [57]. Ongoing collaborations between our own laboratory and Gordon Lark, Kevin Chase, and collaborators are aimed at finding the specific variants responsible for each principal component.
Figure 1. Examples of Breed Morphology Representing a Trade-Off between Speed and Strength.
Dogs shown in (A)—Italian greyhound, Ibizan hound, and greyhound—have proportionately long, thin legs compared to those in (B)—bulldog, Saint Bernard, and mastiff—which have shorter, thicker bones. The colored box indicates the ratio of height at the shoulder in centimeters to weight of males in kilograms.
Among the most satisfying aspects of this work has been the ability to demonstrate how canine genetics allows us to unravel complex, but nonadditive, interactions between genetic loci, a problem which has proven difficult to approach using classical genetic methods. For example, 21% of the observed variation in skeletal size among PWDs results from differences between females and males. Analysis of the above dataset suggests that more than half of this sexual dimorphism results from an interaction between a QTL linked to canine Chromosome 15 and a locus adjacent to the CHM gene on the X chromosome [58]. In females, the haplotype associated with small size on canine Chromosome 15 is dominant, while in males it is the reverse—the haplotype associated with larger size is dominant. The introduction of the X chromosome locus complicates the story, but explains some curious observations as well. For instance, in any population of PWDs, there are always a small number of females who are as large as the largest males. Analysis of the X chromosome locus shows that females who are homozygous for both the CHM linked marker as well as the haplotype associated with large size on canine Chromosome 15 will be, on average, as large as the largest males [58]. Modifier genes such as the one on the X chromosome, which by themselves do not have a detectable effect, would not be identified using traditional QTL methods, highlighting again the value of this approach.
Phenotypic Variation in the Dog
Very recently, efforts have been made to understand the apparent plasticity of the canine genome [59]. Fondon and Garner propose that expansions and contractions of tandem repeats within coding sequences are a major source of phenotypic variation in dogs. As such, this mechanism serves to generate dogs with novel morphologies faster than would be otherwise predicted. To test their hypothesis, they sequenced 37 repeat-containing regions from 17 genes that were known or predicted to have a role in craniofacial development in 92 breeds of dogs. They found that the repeats in the dog were changing faster in terms of length than comparable repeats in humans. They also analyzed three-dimensional models of dog skulls from 20 breeds and some mixed-breed dogs, and found that variation in the number of repeats in the coding regions of the ALX4 and RUNX2 genes were quantitatively associated with significant differences in limb and skull morphology. The authors argue that the incremental effects of repeat length mutations would be an efficient way to generate the rapid yet morphologically conservative changes that distinguish various breeds of dog. If correct, this hypothesis would be in striking contrast to a commonly held view that variation arises largely from modification of gene regulatory sequences, such as transcriptional control elements. Several avenues of experimentation are suggested by this work, including studies of additional genes and more phenotypic measures. However, the initial data provide a starting point for relating novel features of the canine genome to repeated observations regarding rapid creation of morphologically distinct breeds.
Population Structure and Linkage Disequilibrium in the Domestic Dog
Investigation into the genetic relationships between dog breeds is an area of explosive recent growth that holds great promise. Initial studies of breed relationships were highly focused. In a study of five Finnish breeds, Koskinen et al. reported that the phylogenetic distances between breeds were greater than those typically seen between human populations [60]. In addition, individual dogs from these five breeds could be correctly assigned to their breed of origin by analyzing allele patterns associated with small numbers of microsatellites [61]. Irion et al. went a step further using 100 microsatellites and 28 breeds [62]. Their analysis methods, based largely on neighbor-joining trees, revealed the important fact that little higher order structure could be found to describe the relationships between most breeds.
A subsequent and larger study from our own group that entailed genotyping five unrelated dogs from each of the 85 breeds with 96 microsatellite markers was undertaken in 2004 [63]. Assignment tests using the computer program Doh demonstrated that dogs could be correctly assigned to their breed of origin 99% of the time [63]. The majority of variation observed in the dog rests in the differences that separate breeds. In fact, 27% of genetic variation that exists in the dog is found when comparing breeds, whereas 5%–10% of all human variation is found between populations and races [63]. To determine how to best harness the power of canine population structure for mapping, studies of linkage disequilibrium (LD) in the dog have been undertaken [1,64–66]. Hyun et al. found LD extended for 33 centimorgans around the copper toxicosis disease locus in Australian Bedlington terriers. The authors suggest, however, that because the study focused on a single disease locus, already identified by linkage studies, the conclusions were probably not readily generalized to the rest of the genome. The study of Lou et al. is based on analysis of a 10 centimorgan microsatellite scan in a single crossbred pedigree, and identified LD spanning five to ten centimorgans. A particular strength of the paper is its clear description of the nuances of analyzing LD using multiallelic markers. The authors, however, suggest that since only a single pedigree was analyzed, much larger studies need to be undertaken involving larger numbers of both dogs and breeds to develop a clear picture of LD in the dog.
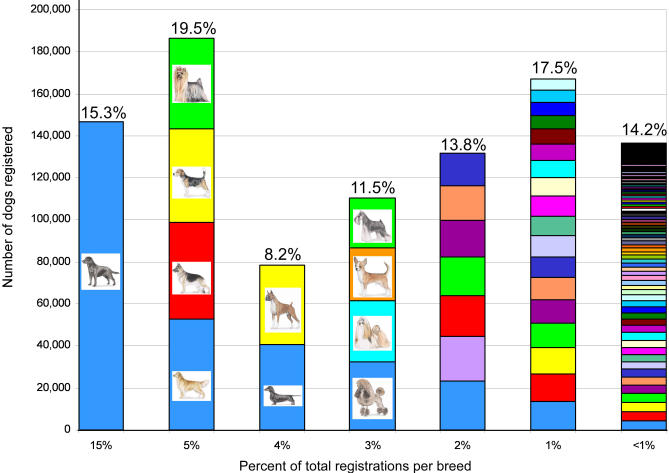
There are over 150 breeds recognized in the United States by the American Kennel Club. The top ten most popular breeds account for more than half of all registrations, while more than 100 of the more uncommon breeds account for less than 15% of the total (Figure 2). This range in population sizes is representative of a variety of breed histories. The LD studies of Sutter et al. and Lindblad-Toh, Wade, and collaborators were designed to be more widely applicable to the general population of purebred dogs [1,66]. Sutter and colleagues used 189 single nucleotide polymorphisms (SNPs) to examine 20 unrelated dogs from each of five breeds at five loci. They found a 10-fold difference in extent of LD in breeds that range from rare to popular, and whose population histories feature a range of popular sire and bottleneck effects [66]. These results were corroborated and extended in a much larger study by Lindblad-Toh et al. Using ten breeds and nearly 1,300 SNPs, the investigators were able to dissect the underlying haplotype structure of the dog genome in addition to measuring the extent of LD [1]. Both studies conclude that breed choice will have a profound effect on the number of markers required to complete whole genome association studies, and care should be taken when selecting breeds for the initial mapping stage. In addition, because of breed architecture, considerably fewer SNPs will be needed for mapping traits in dogs than in humans [1,66].
Figure 2. Distribution of American Kennel Club Registrations by Breed in 2004.
Nearly a million dogs are registered with the American Kennel Club each year. Though the total includes dogs from 154 breeds, most registrations represent a limited number of very popular breeds. The most popular breed, Labrador retriever, accounts for 15.3% of yearly registrations. This is greater than the 118 least popular breeds combined. Each breed on the chart above is represented by a colored block. The height on the y-axis indicates the number of dogs registered in 2004. The blocks are divided into six stacks indicating the percent of overall registrations acquired by that breed, as listed on the x-axis. Above each column is the percent of total registrations for all breeds in that category. Registration statistics can be found at http://www.akc.org/reg/dogreg_stats.cfm.
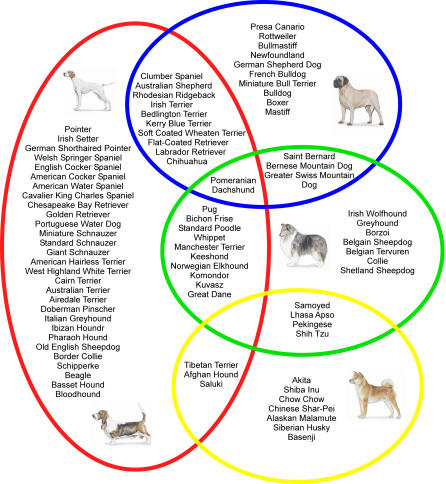
One additional way to improve power for fine mapping is to combine data across breeds. To determine the ancestral relationship between breeds, Parker et al. used the same dataset as described previously to perform an unsupervised clustering analysis with the computer program Structure [63]. The 85 breeds were ordered into four clusters, generating a new canine classification system for dog breeds based on similar patterns of alleles, presumably from a shared ancestral pool (Figure 3) [4]. Cluster one comprised dogs of Asian and African origin, as well as gray wolves. Cluster two was made up of mastiff-type dogs, largely sharing a common theme of big, boxy heads and strong, sturdy bodies. The third and fourth clusters split a group of herding dogs and sight hounds away from the general population of modern hunting dogs including terriers, hounds, and gun dogs.
Figure 3. The Population Structure of 85 Dog Breeds.
The dataset includes five unrelated dogs from each of the 85 breeds that have been genotyped using 96 (CA)n repeat-based microsatellites that spanned the dog genome at an average density of 30 megabases. Clusters were obtained using the computer program Structure [69], which implements a Bayesian model–based clustering algorithm that attempts to identify genetically distinct subpopulations based on patterns of allele frequencies. The work is described in detail in [63]. Four distinct clusters described by Parker et al. are depicted as colored circles: cluster one is yellow, cluster two is blue, cluster three is green, and cluster four is red. Breeds associated with each cluster are listed within the appropriate circle, and examples of breeds are shown in the pictures. Some breeds show patterning similar to more than one cluster, and are listed in the overlapping space. Analysis is ongoing to expand the number of breeds in the dataset and to refine the clusters.
The Parker clusters offered the first look at relationships between breeds, and in doing so, suggest study designs for trait mapping. For example, Modiano and colleagues have sought to determine the origin of B and T cell lymphomas in dogs [67]. They found that while B cell lymphomas are most common overall, rates of T cell lymphoma are significantly higher in breeds from the Parker cluster one, the Asian cluster, than any other group. This suggests an ancestral cause of T cell lymphoma in Asian dogs, while arguing against a single ancestor for B cell lymphoma in any other group. The optimal mapping study for T cell lymphomas would, therefore, focus on dogs from the Asian group. Also of interest is the work of Neff and colleagues who describe a single haplotype surrounding the multidrug-resistant gene MDR1 in nine breeds [68]. The nine breeds represented a range of herding dogs and sight hounds that presumably shared a single common ancestor, and again suggests a strategy for mapping studies involving this set of breeds.
While understanding the relationships between breeds will assist in minimizing the task of mapping multigenic diseases, moving from locus to gene remains a daunting task [66]. Both Sutter et al. and Lindblad-Toh, Wade, and collaborators have undertaken studies to determine how haplotype analysis can facilitate such efforts [1,66]. Using their respective datasets, both studies demonstrate high haplotype sharing between breeds and low haplotype diversity within breeds. Thus, disease alleles will be most easily identified by comparison of haplotypes that are identical by descent in affected dogs from two or more breeds. Data from additional breeds can then be used for fine resolution mapping. The recent availability of 2.1 million SNPs (http://www.broad.mit.edu/mammals/dog/snp/) from the canine genome sequencing project will greatly enhance such studies [1].
Conclusion
When describing a dog, Mark Twain once wrote, “The dog is a gentleman; I hope to go to his heaven, not man's.” Twain's simple comment reflects both our admiration for the loyalty, integrity, and devotion we have come to expect from our closest companions, and our desire to keep them ever at our side. In the last few years, as summarized here, the canine genome project has worked tirelessly to develop resources and paradigms that will lead to both the improvement of animal and human health and an understanding of the genetics that regulates variation between breeds. A great deal remains to be learned. We still don't know why Great Danes are big and Pekingese are small, or why herding dogs herd and pointing dogs point. But in another sense, we have succeeded beyond our wildest dreams, as the dog is now a viable system in which to tackle problems relating to the genetics of complex traits.
Acknowledgments
We thank Liz McNeil, Ed Giniger, Cheryl Maslen, K. Gordon Lark, and Kerstin Lindblad-Toh for their careful reading of the manuscript and useful comments. This work was supported in part by funds from the National Human Genome Research Institute Intramural Program. Finally, we gratefully acknowledge the many dog owners and breeders who continue to support our work by providing samples, pedigrees, and clinical information on their pets.
Abbreviations
- PWD
Portuguese water dog
- LD
linkage disequilibrium
- RH
radiation hybrid
- SINE
short interspersed nuclear element
- SNP
single nucleotide polymorphism
- QTL
quantitative trait locus
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Public Domain declaration which stipulates that, once placed in the public domain, this work may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose.
References
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005. In press. [DOI] [PubMed]
- Sutter NB, Ostrander EA. Dog star rising: The canine genetic system. Nat Rev Genet. 2004;5:900–910. doi: 10.1038/nrg1492. [DOI] [PubMed] [Google Scholar]
- Patterson D. Companion animal medicine in the age of medical genetics. J Vet Internal Med. 2000;14:1–9. [PubMed] [Google Scholar]
- Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005. In press. [DOI] [PubMed]
- Breen M, Bullerdiek J, Langford CF. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res. 1999;7:401–406. doi: 10.1023/a:1009224232134. [DOI] [PubMed] [Google Scholar]
- Yang F, O'Brien PC, Milne BS, Graphodatsky AS, Solanky N, et al. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- Sargan DR, Yang F, Squire M, Milne BS, O'Brien PC, et al. Use of flow-sorted canine chromosomes in the assignment of canine linkage, radiation hybrid, and syntenic groups to chromosomes: Refinement and verification of the comparative chromosome map for dog and human. Genomics. 2000;69:182–195. doi: 10.1006/geno.2000.6334. [DOI] [PubMed] [Google Scholar]
- Neff MW, Broman KW, Mellersh CS, Ray K, Acland GM, et al. A second-generation genetic linkage map of the domestic dog, Canis familiaris . Genetics. 1999;151:803–820. doi: 10.1093/genetics/151.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford CF, Fischer PE, Binns MM, Holmes NG, Carter NP. Chromosome-specific paints from a high-resolution flow karyotype of the dog. Chromosome Res. 1996;4:115–123. doi: 10.1007/BF02259704. [DOI] [PubMed] [Google Scholar]
- Breen M, Langford CF, Carter NP, Holmes NG, Dickens HF, et al. FISH mapping and identification of canine chromosomes. J Hered. 1999;90:27–30. doi: 10.1093/jhered/90.1.27. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, et al. The dog genome: Survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, et al. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:1–11. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon R, Kirkness EF, Lorentzen TD, Hitte C, Comstock KE, et al. Building comparative maps using 1.5x sequence coverage: Human chromosome 1p and the canine genome. Cold Spring Harb Symp Quant Biol. 2003;68:171–177. doi: 10.1101/sqb.2003.68.171. [DOI] [PubMed] [Google Scholar]
- Guyon R, Lorentzen TD, Hitte C, Kim L, Cadieu E, et al. A 1-Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci U S A. 2003;100:5296–5301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M, Jouquand S, Renier C, Mellersh CS, Hitte C, et al. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitte C, Madeoy J, Kirkness EF, Priat C, Lorentzen TD, et al. Survey sequencing combined with dense radiation hybrid gene mapping facilitates genome navigation. Nat Rev Genet. 2005. In press. [DOI] [PubMed]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Galibert F, Andre C, Cheron A, Chuat JC, Hitte C, et al. The importance of the canine model in medical genetics. Bull Acad Natl Med. 1998;182:811–821. [PubMed] [Google Scholar]
- Ostrander EA, Giniger E. Semper fidelis: What man's best friend can teach us about human biology and disease. Am J Hum Genet. 1997;61:475–480. doi: 10.1086/515522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DF, Haskins ME, Jezyk PF. Models of human genetic disease in domestic animals. Adv Hum Genet. 1982;12:263–339. doi: 10.1007/978-1-4615-8315-8_4. [DOI] [PubMed] [Google Scholar]
- Minnick MF, Stillwell LC, Heineman JM, Stiegler GL. A highly repetitive DNA sequence possibly unique to canids. Gene. 1992;110:235–238. doi: 10.1016/0378-1119(92)90654-8. [DOI] [PubMed] [Google Scholar]
- Bentolila S, Bach JM, Kessler JL, Bordelais I, Cruaud C, et al. Analysis of major repetitive DNA sequences in the dog (Canis familiaris) genome. Mamm Genome. 1999;10:699–705. doi: 10.1007/s003359901074. [DOI] [PubMed] [Google Scholar]
- Vassetzky NS, Kramerov DA. CAN—A pan-carnivore SINE family. Mamm Genome. 2002;13:50–57. doi: 10.1007/s00335-001-2111-1. [DOI] [PubMed] [Google Scholar]
- Schmid CW. Alu: Structure, origin, evolution, significance and function of one-tenth of human DNA. Prog Nucleic Acid Res Mol Biol. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- Coltman DW, Wright JM. Can SINEs: A family of tRNA-derived retroposons specific to the superfamily Canoidea. Nucleic Acids Res. 1994;22:2726–2730. doi: 10.1093/nar/22.14.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF. SINEs of canine genomic diversity. In: Ostrander EA, Giger U, Lindblad-Toh K, editors. The dog and its genome. Cold Spring Harbor (New York): Cold Spring Harbor Press; 2006. In press. [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Pele M, Tiret L, Kessler JL, Blot S, Panthier JJ. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 2005;14:1417–1427. doi: 10.1093/hmg/ddi151. [DOI] [PubMed] [Google Scholar]
- Lohi H, Young EJ, Fitzmaurice SN, Rusbridge C, Chan EM, et al. Expanded repeat in canine epilepsy. Science. 2005;307:81. doi: 10.1126/science.1102832. [DOI] [PubMed] [Google Scholar]
- Sargan D. IDID: Inherited diseases in dogs: Web-based information for canine inherited disease genetics. Mamm Genome. 2004;15:503–506. doi: 10.1007/s00335-004-3047-z. [DOI] [PubMed] [Google Scholar]
- Switonski M, Szczerbal I, Nowacka J. The dog genome map and its use in mammalian comparative genomics. J Appl Genet. 2004;45:195–214. [PubMed] [Google Scholar]
- Ostrander EA, Galibert F, Patterson DF. Canine genetics comes of age. Trends Genet. 2000;16:117–124. doi: 10.1016/s0168-9525(99)01958-7. [DOI] [PubMed] [Google Scholar]
- Sidjanin DJ, Lowe JK, McElwee JL, Milne BS, Phippen TM, et al. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum Mol Genet. 2002;11:1823–1833. doi: 10.1093/hmg/11.16.1823. [DOI] [PubMed] [Google Scholar]
- Lingaas F, Comstock KE, Kirkness EF, Sorensen A, Aarskaug T, et al. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Hum Mol Genet. 2003;12:3043–3053. doi: 10.1093/hmg/ddg336. [DOI] [PubMed] [Google Scholar]
- Riser W. The dysplastic hip joint: It's radiographic and histologic development. J Am Vet Radiol Soc. 1973;14:35–50. [Google Scholar]
- Olsson SE, Marshall JL, Story E. Osteophytosis of the knee joint in the dog. A sign of instability. Acta Radiol Suppl. 1972;319:165–167. [PubMed] [Google Scholar]
- Maki K, Janss LL, Groen AF, Liinamo AE, Ojala M. An indication of major genes affecting hip and elbow dysplasia in four Finnish dog populations. Heredity. 2004;92:402–408. doi: 10.1038/sj.hdy.6800434. [DOI] [PubMed] [Google Scholar]
- Bliss S, Todhunter RJ, Quaas R, Casella G, Wu R, et al. Quantitative genetics of traits associated with hip dysplasia in a canine pedigree constructed by mating dysplastic Labrador Retrievers with unaffected Greyhounds. Am J Vet Res. 2002;63:1029–1035. doi: 10.2460/ajvr.2002.63.1029. [DOI] [PubMed] [Google Scholar]
- LaFond E, Breur GJ, Austin CC. Breed susceptibility for developmental orthopedic diseases in dogs. J Am Anim Hosp Assoc. 2002;38:467–477. doi: 10.5326/0380467. [DOI] [PubMed] [Google Scholar]
- Smith GK, Mayhew PD, Kapatkin AS, McKelvie PJ, Shofer FS, et al. Evaluation of risk factors for degenerative joint disease associated with hip dysplasia in German Shepherd Dogs, Golden Retrievers, Labrador Retrievers, and Rottweilers. J Am Vet Med Assoc. 2001;219:1719–1724. doi: 10.2460/javma.2001.219.1719. [DOI] [PubMed] [Google Scholar]
- Todhunter RJ, Bliss SP, Casella G, Wu R, Lust G, et al. Genetic structure of susceptibility traits for hip dysplasia and microsatellite informativeness of an outcrossed canine pedigree. J Hered. 2003;94:39–48. doi: 10.1093/jhered/esg006. [DOI] [PubMed] [Google Scholar]
- Kealy RD, Lawler DF, Ballam JM, Lust G, Biery DN, et al. Evaluation of the effect of limited food consumption on radiographic evidence of osteoarthritis in dogs. J Am Vet Med Assoc. 2000;217:1678–1680. doi: 10.2460/javma.2000.217.1678. [DOI] [PubMed] [Google Scholar]
- Kealy RD, Lawler DF, Ballam JM, Lust G, Smith GK, et al. Five-year longitudinal study on limited food consumption and development of osteoarthritis in coxofemoral joints of dogs. J Am Vet Med Assoc. 1997;210:222–225. [PubMed] [Google Scholar]
- Leighton EA, Linn JM, Willham RL, Castelberry MW. A genetic study of canine hip dysplasia. Am J Vet Res. 1977;38:241–244. [PubMed] [Google Scholar]
- Chase K, Lawler DF, Adler FR, Ostrander EA, Lark KG. Bilaterally asymmetric effects of quantitative trait loci (QTLs): QTLs that affect laxity in the right versus left coxofemoral (hip) joints of the dog (Canis familiaris) . Am J Med Genet A. 2004;124:239–247. doi: 10.1002/ajmg.a.20363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Lawler DF, Carrier DR, Lark KG. Genetic regulation of osteoarthritis: A QTL regulating cranial and caudal acetabular osteophyte formation in the hip joint of the dog (Canis familiaris) . Am J Hum Genet. 2005;135:334–335. doi: 10.1002/ajmg.a.30719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todhunter R, Acland G, Olivier M, Williams A, Vernier-Singer M, et al. An outcrossed canine pedigree for linkage analysis of hip dysplasia. J Hered. 1999. pp. 83–92. [DOI] [PubMed]
- Bliss S, Todhunter RJ, Quaas R, Casella G, Wu R, et al. Quantitative genetics of traits associated with hip dysplasia in a canine pedigree constructed by mating dysplastic Labrador Retrievers with unaffected Greyhounds. Am J Vet Res. 2002;63:1029–1035. doi: 10.2460/ajvr.2002.63.1029. [DOI] [PubMed] [Google Scholar]
- Todhunter RJ, Casella G, Bliss SP, Lust G, Williams AJ, et al. Power of a Labrador Retriever-Greyhound pedigree for linkage analysis of hip dysplasia and osteoarthritis. Am J Vet Res. 2003;64:418–424. doi: 10.2460/ajvr.2003.64.418. [DOI] [PubMed] [Google Scholar]
- Braund K, Miller DF. The complete Portuguese water dog. New York: Howell Book House; 1986. 304. p. [Google Scholar]
- Molinari C. The Portuguese water dog. Portugal: ELO-Publicidade; 1993. 156. p. [Google Scholar]
- Chase K, Adler FR, Miller-Stebbings K, Lark KG. Teaching a new dog old tricks: Identifying quantitative trait loci using lessons from plants. J Hered. 1999;90:43–51. doi: 10.1093/jhered/90.1.43. [DOI] [PubMed] [Google Scholar]
- Lark KG, Chase K, Carrier DR, Adler FR, editors. Genetic analysis of the canid skeleton: Morphological loci in the Portuguese water dog population. Cold Spring Harbor (New York): Cold Spring Harbor Press. 2006. In press.
- Chase K, Carrier DF, Adler FR, Ostrander EA, Lark KG. Size sexual dimorphism in Portugese water dogs: Interaction between an autosome and the X chromsome. Genome Res. 2005. In press. [DOI] [PMC free article] [PubMed]
- Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci U S A. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen MT, Bredbacka P. Assessment of the population structure of five Finnish dog breeds with microsatellites. Anim Genet. 2000;31:310–317. doi: 10.1046/j.1365-2052.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- Koskinen MT. Individual assignment using microsatellite DNA reveals unambiguous breed identification in the domestic dog. Anim Genet. 2003;34:297–301. doi: 10.1046/j.1365-2052.2003.01005.x. [DOI] [PubMed] [Google Scholar]
- Irion DN, Schaffer AL, Famula TR, Eggleston ML, Hughes SS, et al. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J Hered. 2003;94:81–87. doi: 10.1093/jhered/esg004. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Hyun C, Filippich LJ, Lea RA, Shepherd G, Hughes IP, et al. Prospects for whole genome linkage disequilibrium mapping in domestic dog breeds. Mamm Genome. 2003;14:640–649. doi: 10.1007/s00335-003-3006-0. [DOI] [PubMed] [Google Scholar]
- Lou XY, Todhunter RJ, Lin M, Lu Q, Liu T, et al. The extent and distribution of linkage disequilibrium in a multi-hierarchic outbred canine pedigree. Mamm Genome. 2003;14:555–564. doi: 10.1007/s00335-003-2272-1. [DOI] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, et al. Extensive and breed-specific linkage disequilibrium in Canis familiaris . Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modiano JF, Breen M, Burnett RC, Parker HG, Inusah S, et al. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65:5654–5661. doi: 10.1158/0008-5472.CAN-04-4613. [DOI] [PubMed] [Google Scholar]
- Neff MW, Robertson KR, Wong AK, Safra N, Broman KW, et al. Breed distribution and history of canine mdr1-1Delta, a pharmacogenetic mutation that marks the emergence of breeds from the collie lineage. Proc Natl Acad Sci U S A. 2004;101:11725–11730. doi: 10.1073/pnas.0402374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inferrence of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]