Abstract
Endoplasmic reticulum aminopeptidase 1 (ERAP1) is an IFN-γ-induced aminopeptidase in the endoplasmic reticulum that trims longer precursors to the antigenic peptides presented on MHC class I molecules. We recently reported that purified ERAP1 trimmed N-extended precursors but spared peptides of 8-9 residues, the length required for binding to MHC class I molecules. Here, we show another remarkable property of ERAP1: that it strongly prefers substrates 9-16 residues long, the lengths of peptides transported efficiently into the ER by the transporter associated with antigen processing (TAP) transporter. This aminopeptidase rapidly degraded a model 13-mer to a 9-mer and then stopped, even though the substrate and the product had identical N- and C-terminal sequences. No other aminopeptidase, including the closely related ER-aminopeptidase ERAP2, showed a similar length preference. Unlike other aminopeptidases, the activity of ERAP1 depended on the C-terminal residue of the substrate. ERAP1, like most MHC class I molecules, prefers peptides with hydrophobic C termini and shows low affinity for peptides with charged C termini. Thus, ERAP1 is specialized to process precursors transported by TAP to peptides that can serve as MHC class I epitopes. Its “molecular ruler” mechanism involves binding the hydrophobic C terminus of the substrate 9-16 residues away from the active site.
Keywords: antigen presentation, antigen processing, proteases
MHC class I molecules bind tightly and display on the cell surface antigenic peptides that are derived from peptides generated during the degradation of intracellular proteins. If non-native peptides (e.g., from viral proteins) are presented, they are recognized by circulating cytotoxic T lymphocytes (1-4). To fit in the groove in most MHC class I molecules, these antigenic peptides must have a length of 8-10 residues (5, 6), although certain class I molecules can admit peptides up to 11 residues (7). It is now firmly established that the proteasome pathway is responsible for the generation of the great majority of antigenic peptides (8-10). Proteasomes generally degrade proteins to peptide fragments ranging from 2-25 residues long (11), but most are too short (<8 residues) for antigen presentation. Several studies using proteasome inhibitors have further shown that cleavages within proteasomes define the C-terminal residues of MHC class I-presented peptides (12). However, their N termini often are generated by aminopeptidases, which trim longer N-extended proteasome products to the mature epitopes (13). In fact, proteasomes, and especially immunoproteasomes (1), the forms found in immune tissues and induced by IFN-γ elsewhere, seem to preferentially generate such longer precursors (14), whose presentation requires N-terminal processing. Several cytosolic peptidases, including tripeptidyl peptidase II (TPPII) (15), bleomycin hydrolase and puromycin-sensitive aminopeptidase (16), and the IFN-γ-inducible enzyme leucine aminopeptidase (17), may play a role in trimming some precursors to antigenic peptides. However, trimming of many N-extended precursors occurs in the endoplasmic reticulum (ER).
After their generation in the cytosol, the N-extended precursors and antigenic peptides are translocated into the ER by the ATP-dependent TAP complex (transporter associated with antigen processing) (18-20). TAP preferentially transports peptides 8-16 residues long (21) and seems more efficient in transporting N-extended precursors than the mature epitopes. A variety of studies have demonstrated that these precursors are trimmed by aminopeptidases in the ER lumen to the correct lengths for presentation on MHC class I molecules (22-24). Recently, an aminopeptidase, ERAP1 (ER aminopeptidase 1; ERAAP1), has been isolated by our lab (25) and Serwold et al. (26) from the ER and shown to be responsible for trimming these N-extended precursors. Overexpressing this enzyme enhances, whereas decreasing its expression with small interfering RNAs (siRNAs) reduces, the presentation of many epitopes. Thus, processing by ERAP1 is essential or rate-limiting for the presentation of many antigenic precursors, especially in cells treated with IFN-γ (27). This potent stimulator of antigen presentation induces ERAP1 (25), together with many other key components of this process, including MHC class I molecules, the catalytic β-subunits found specifically in immunoproteasomes (10), which seem to generate N-extended antigenic precursors preferentially (14), and the TAP transporter (28), which transports such N-extended peptides into the ER.
It had been originally proposed that peptide precursors in the ER bind to class I molecules weakly and that the loose ends are then trimmed by exopeptidases until the tight-binding eight- or nine-residue epitopes are generated (29). However, a remarkable feature of ERAP1 is that, in the absence of any MHC class I molecules to serve as a template, it trims antigenic precursors but seems to lose activity when eight- or nine-residue epitopes are generated (27). These findings suggest that ERAP1 has unique catalytic properties that are particularly adapted to function in generating MHC class I epitopes. The present studies were undertaken to understand the basis of this intriguing ability to trim precursors to the appropriate size for MHC class I binding. Using synthetic peptides and natural antigenic precursors, we demonstrate that this enzyme also prefers substrates of the same lengths as are transported by TAP. We show that the preference of ERAP1 for substrates of specific lengths (i.e., 9-16 residues) is based upon its ability to monitor the nature of the C-terminal amino acid many residues away from the N-terminal cleavage site. These properties, especially its “molecular ruler” mechanism, distinguish ERAP1 from other aminopeptidases and must have evolved to facilitate antigen presentation.
Materials and Methods
Peptide Synthesis and Protein Purification. Peptides were synthesized by the University of Massachusetts Medical School Core Facility or Tufts University Core Facility and were purified by HPLC. The purity of peptides was >95% by MS analysis. Porcine kidney leucine aminopeptidase (Sigma) was further purified to single band on SDS/PAGE by UNO Q-1 column (Bio-Rad) and HiTrap Butyl Sepharose FF (Amersham Pharmacia Biosciences). Recombinant human puromycin-sensitive aminopeptidase was kindly provided by L. B. Hersh.
Trimming Assays by RP-HPLC. For ERAP1, peptides (150 μM) were incubated with purified recombinant human ERAP1 (3.5 μg/ml) at 37°C in 50 mM Tris·HCl (pH 7.8) and 0.25 μg/ml protease-free BSA (Sigma). For other aminopeptidases, peptides (100 μM) were incubated with leucine aminopeptidase or puromycin-sensitive aminopeptidase (7 μg/ml, respectively) in 50 mM Tris·HCl (pH 7.5) and 0.25 μg/ml protease-free BSA at 37°C for 1 h. Reactions were terminated by adding 0.6% trifluoroacetic acid (J. T. Baker). The peptide-containing supernatant was analyzed by RP-HPLC on TS-0546-C183 (50 × 4.6 mm) column (Higgins Analytical, Mountain View, CA) and eluted with a linear 7-35% acetonitrile gradient in 10 mM sodium phosphate buffer (pH 6.6) or a linear gradient of 0.05% trifluoroacetic acid (TFA) to 50% acetonitrile in 0.03% TFA. The percentage of the substrate trimmed was calculated by integration of the area under every single peptide peak.
Trimming Assay by Fluorescence Detection. Peptides were incubated with purified ERAP1, leucine aminopeptidase or puromycin-sensitive aminopeptidase in 50 mM Hepes-KOH (pH 7.5) at 37°C. N termini of peptides and newly generated free amino acids by ERAP1 would interact with fluorescamine (30). After the incubation, 50 μl of each sample was mixed and rotated with 25 μl of fluorescamine solution in acetone (0.3 mg/ml) for 1 min and then diluted with 150 μl of 0.2 M sodium borate (pH 9.0). Two hundred microliters of each sample was transferred to the 96-well plate and measured with the FLUOstar Galaxy plate reader (BMG LABTECH, Durham, NC) at the excitation of 380 nm and the emission of 480 nm. The fluorescence unit was estimated by using an equimolar mixture of peptides of known concentration (EAA-NH2, AEAA-NH2, AAEAAG-NH2, AAVVAAG, and TTQRTRALV) as the standard. The degradation of peptide substrates was calculated by subtracting the initial fluorescence from the total fluorescence after incubation with the aminopeptidases.
Trypsin Digestion. QLESIINFEKL (200 μM) was incubated with TPCK (l-1-tosylamido-2-phenylethyl chloromethyl ketone)-treated trypsin at 37°C in 50 mM Tris·HCl buffer containing 1 mM CaCl2 (pH 7.8) for 12 h. Reactions were terminated by adding trypsin inhibitor (0.22 μg/μl) (ICN) and protease-free BSA (0.25 μg/μl). Purified ERAP1 (3.5 μg/ml) was then incubated with the untreated peptide or trypsin-treated peptide, and the supernatants were analyzed by RP-HPLC. Control experiments indicated that the presence of trypsin and the trypsin inhibitor did not affect trimming of other peptides by ERAP1 (not shown).
Results
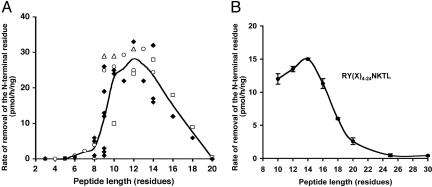
Like TAP, ERAP1 Prefers Substrates 9-16 Residues Long. TAP translocates preferentially peptides 8-16 aa long (21). To determine whether purified ERAP1 can process peptides of this length, we compared the rates of degradation of peptides up to 20 residues long. Recombinant human ERAP1, which was prepared as described (31), was found to degrade peptides of 9-16 residues (n = 29) much more rapidly than shorter (P < 0.0001, n = 9) or longer (P < 0.001, n = 4) ones (Fig. 1A). ERAP1 showed very little or no activity against 8- to 9-residue mature epitopes (n = 11) (in accord with our prior findings) and also no activity against shorter fragments (n = 7) (Fig. 6, which is published as supporting information on the PNAS web site). The ability of ERAP1 to trim substrates decreased sharply above 14 residues (Fig. 1), and little or no activity was seen against ones longer than 18 residues. These trends were clearly evident when the rates of hydrolysis of all available peptides were compared (Fig. 6). To further define the maximum length of substrates and to confirm that ERAP1 monitors length and not some feature of sequence, we compared the rates of trimming of peptide libraries ranging from 10 to 30 residues but having identical N- and C-terminal sequences. The general sequence was RY(X-n)NKTL (32), where X is an equimolar mixture of A, D, E, F, G, H, I, K, L, M, P, Q, R, S, T or V, and n ranges from 4 to 24 residues. As suggested above (Fig. 1A), ERAP1 strongly preferred substrates 9-16 residues long. It showed less activity against 18-mers and very little activity against peptides 20-30 residues long (Fig. 1B).
Fig. 1.
ERAP1 prefers substrates 9-16 residues long. (A) A variety of peptides were tested, including mature MHC class I-presented peptides (8-9 residues), 3- to 8-residue fragments of mature epitopes, and 9- to 20-residue N-extended precursors of MHC-binding peptides with the same epitope on their C termini and different N-terminal extensions (see Fig. 6 for identification of each peptide). Peptides (150 μM) were incubated with purified ERAP1 (3.5 μg/ml) at 37°C for 1 h. Each spot reflects the trimming of an individual peptide substrate. The line shows the mean of the degradation rates for different peptide substrates of the same length. The removal of their N-terminal residues was measured by RP-HPLC (see Materials and Methods). The trimming of the 14-mer, GLEQLESIINFEKL, the trimer, EKL, and all intervening peptides from 4-13 residues long, including SIINFEKL, the H2-Kb-binding antigenic peptide, are shown as open circle symbols. The degradation of N-extended FAPGNYPAL series is shown as open triangles, N-extended NANPDCKTIL series are shown as open squares, and other peptides are indicated as solid diamonds. (B) The maximum length of substrates was analyzed with a library of peptides of different lengths (10-30 residues) but identical N- and C-terminal sequences. The collection of peptides, RY(Xn)NKTL (100 μM), where X represents equimolar mixtures of A, D, E, F, G, H, I, K, L, M, P, Q, R, S, T or V, and n represents 4, 6, 8, 10, 12, 14, 19 or 24, was incubated individually with purified ERAP1 (3.5 μg/ml) at 37°C for 1 h and analyzed by the method of fluorescence detection (see Materials and Methods for details).
ERAP1 Monitors Minimum Substrate Length. Our prior data had suggested that ERAP1 has little activity against peptides of eight to nine residues. This apparent influence of length was clearly evident when ERAP1 was incubated with sequences of seven different lengths from ovalbumin that contain the immunodominant epitope SIINFEKL at their C terminus (open circle symbols in Fig. 1A). This final epitope and smaller fragments, like other eight-residue epitopes tested (Fig. 1A), were very poor substrates. The very low rate of hydrolysis of SIINFEKL occurs because it has a much lower affinity for the enzyme than the precursor, QLESIINFEKL. The Km for this 11-mer was 124 μM whereas that for SIINFEKL was at least 900 μM, but it could be higher because a clear saturation was not demonstrable and at high concentrations SIINFEKL has limited solubility (Fig. 7, which is published as supporting information on the PNAS web site). With two other such series of N-extended epitopes (FAPGNYPAL and NANPDCKTIL) and individual peptides tested, ERAP1 showed maximum activity toward 10- to 14-residue precursors and fell dramatically with 9-residue peptides or smaller (Fig. 1A). Thus, ERAP1 shows apparent length dependence because its activity for peptides decreased sharply when the distance between the C terminus and N terminus is <8 or 9 residues.
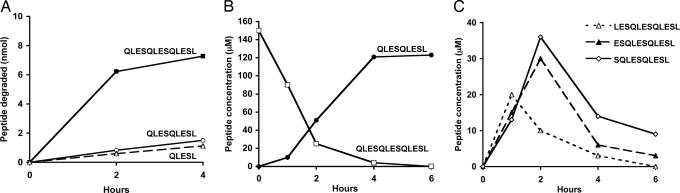
To test rigorously whether the activity of ERAP1 actually depends upon peptide length and not on some feature of the sequence of the substrate, we synthesized three peptides that contain the repeated element, QLES, 1-3 times and the same C-terminal residue (Leu), QLESL (QL5), QLESQLESL (QL9), and QLESQLESQLESL (QL13). This 13-residue peptide was by far the best substrate. In 2 h, ERAP1 cleaved the N-terminal Q from >80% of QL13, but from <10% of QL9 or of QL5 (Fig. 2A). Accordingly, when ERAP1 was incubated with QL13 for 4 h, this peptide was converted almost completely to QL9, after which the 9-residue product was very stable (Fig. 2B). The generation of this stable 9-mer occurred through sequential production of the 12-, 11-, and 10-residue intermediates. The levels of 12-, 11-, and 10-mer first increased and then decreased (Fig. 2C), which is consistent with their being metabolic intermediates and a nonprocessive mechanism. Also, because the amounts of these intermediates greatly exceeded enzyme levels, they must be released into the medium after each cleavage.
Fig. 2.
ERAP1 rapidly cleaves a 13-mer, but not a 9-mer or a 5-mer with identical N and C termini. (A) QLESQLESQLESL, QLESQLESL, and QLESL (150 μM) were incubated with purified ERAP1 (3.5 μg/ml). The peptide-containing supernatant was analyzed by RP-HPLC. (B) Quantitative conversion of the 13-mer to the 9-mer by ERAP1. QLESQLESQLESL was incubated with purified ERAP1 as described above, and an aliquot of the reaction was analyzed by HPLC at each time. (C) The 12-, 11- and 10-residue intermediates were generated from the 13-mer QLESQLESQLESL in a nonprocessive manner. LESQLESQLESL (12-mer), ESQLESQLESL (11-mer), and SQLESQLESL (10-mer) were clearly demonstrable before the product 9-mer was generated.
ERAP1 Slowly Trims Peptides with a Charged C-Terminal Residue. This remarkable length dependence strongly suggests that ERAP1 recognizes both the N and C termini of antigenic precursors. To test this possibility, we examined whether the nature of the C-terminal amino acid influences the activity of ERAP1. To change markedly the C-terminal residue while maintaining the N-terminal sequence, we initially used trypsin to remove the C-terminal Leu from the 11-residue precursor QLESIINFEKL. Although the 10-mer QLESIINFEK has a suitable length for rapid processing and identical N-terminal residues as QLESIINFEKL, it was trimmed much more slowly (Table 1). Thus, the presence of a C-terminal lysine markedly reduced trimming by ERAP1.
Table 1. ERAP1 poorly trims peptides with a charged C-terminal residue, which have high Kms.
| Peptide | Rate of removal of the N-terminal residue, pmol/h/ng | Km, μM | Vmax, pmol/min |
|---|---|---|---|
| QLESIINFEK | 3 | ND | ND |
| QLESIINFEKL | 30 | 124 | 110 |
| QLESIINFEKL-amide | 27 | 138 | 117 |
| QLESIINFEKA | 29 | 124 | 130 |
| QLESIINFEKY | 28 | 111 | 126 |
| QLESIINFELK | 13 | ND | ND |
| QLESIINFEKK | 6 | 800 | 120 |
| QLESIINFEKR | 8 | 910 | 106 |
| QLESIINFEKD | 5 | 700 | 160 |
| N-acetyl-QLESIINFEKL | 0 | ND | ND |
| EFAPGNYPAL | 27 | 148 | 89 |
| EFAPGNYPAK | 2 | 1,500 | 107 |
| EFAPGNYPAD | 3 | 1,325 | 85 |
| AYWANATRSGA | 27 | 90 | 95 |
| AYWANATRSGAD-form | 12 | 253 | 96 |
To measure the peptide hydrolysis, substrates (150 μM) were incubated with purified recombinant human ERAP1 (3.5 μg/ml) at 37°C for 1 h. The peptide-containing supernatant was further analyzed by RP-HPLC. To determine Km and Vmax, purified recombinant human ERAP1 (2 μg/ml) was incubated with varying amounts of the peptide substrate at 37°C for 30 min. AD-form, D-form alanine. ND, not determined.
To examine further the influence of the C-terminal amino acid, we replaced the C-terminal Leu of QLESIINFEKL with a variety of residues. ERAP1 trimmed peptides with C-terminal Lys, Arg, or Asp much more slowly than ones with Ala, Tyr, or Leu in that position (Table 1). Also, when the last two C-terminal residues of QLESIINFEKL were reversed (i.e., from KL to LK), the degradation of QLESIINFELK was also much slower than that of QLESIINFEKL but faster than QLESIINFEKK. A similar analysis of the effects of the C terminus was carried out by using EFAPGNYPAL, the precursor of the dominant epitope FAPGNYPAL from Sendai virus nucleoprotein. Under conditions where >60% of the EFAPGNYPAL was processed (Table 1), EFAPGNYPAK and EFAPGNYPAD were hardly altered. Furthermore, measurement of the Km and Vmax for these variants of QLESIINFEKL and EFAPGNYPAL with purified ERAP1 showed that the peptides with a charged C-terminal amino acid have 8-10 times higher Kms than peptides with a hydrophobic C-terminal residue, but all of them have very similar Vmax (Table 1), which is presumably determined by the nature of the N-terminal cleavage site. Thus, ERAP1 binds peptides with a charged C-terminal residue with quite low affinity.
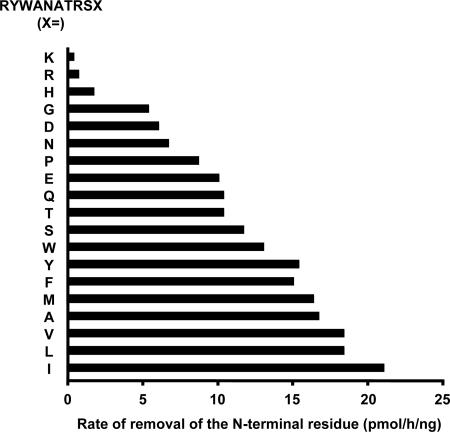
ERAP1 Prefers Peptides with a Large Hydrophobic C-Terminal Residue. To characterize the C-terminal preference of ERAP1 more systematically, we compared the rates of removal of the N-terminal residue from 19 variants of RYWANATRSX (33), where X represents each of the naturally occurring 20 aa (except Cys) (Fig. 3). The rates of cleavage were much faster for peptides with hydrophobic C-terminal residues. ERAP1 trimmed this sequence very poorly when it had a basic C-terminal residue K, R, or H. Hydrophilic neutral amino acids (G, N, P, Q, T, S) in this position allowed some activity, as did the acidic residues (D and E), but ERAP1 strongly prefers peptides with hydrophobic or aromatic C-terminal residues (W, Y, F, M, A, V, L, or I). This preference for hydrophobic C-terminal amino acids resembles the peptides bound selectively by all MHC class I molecules in rodents and most in humans (7).
Fig. 3.
ERAP1 prefers peptides with a hydrophobic C terminus. The library of 19 peptides of RYWANATRSX (150 μM respectively), where X represents each of the naturally occurring 20 aa (except Cys), was incubated with purified ERAP1 (3.5 μg/ml) at 37°C for 1 h. Samples were analyzed by RP-HPLC with a linear gradient of 0.05% trifluoroacetic acid (TFA) to 50% acetonitrile in 0.03% TFA.
Although the nature of the C-terminal side chain is important, the presence of a free carboxyl group on its C terminus is not critical, because replacing the Leu on QLESIINFEKL by Leuamide did not reduce its processing (Table 1). However, like typical aminopeptidases, ERAP1 requires a free amino group on the N terminus of its substrate. The acetylation of N terminus of QLESIINFEKL blocks its processing by ERAP1 (Table 1). We also investigated whether an d-amino acid in this position would affect trimming by ERAP1. Although the antigenic precursor derived from influenza A virus NP, AYWANATRSGA, is a good substrate for ERAP1, when its C terminus was replaced by d-Ala, its processing was reduced by 55% (Table 1). Therefore, the C-terminal preference of ERAP1 is a unique property, which clearly differentiates it from other aminopeptidases, whose activity is solely determined by the N-terminal sequence.
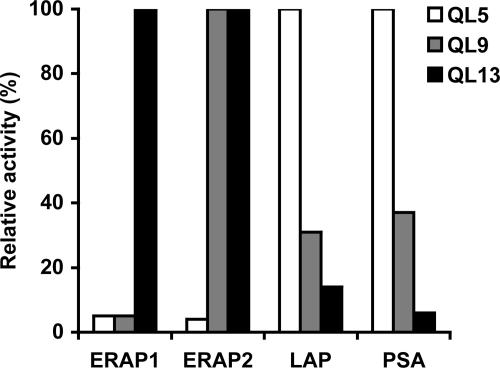
Other Aminopeptidases, Including ERAP2, Do Not Show a Similar Length Preference. These results strongly suggest that length determination by ERAP1 is through its ability to interact with both the N- and C-terminal residues of the longer precursors. Specifically, the hydrophobicity of the side chain of the C-terminal residue is an important factor determining the ability of ERAP1 to remove the N-terminal residue, even though it may be 9-16 residues away. Even though no such properties have been reported, we examined whether the major cytosolic aminopeptidases might perhaps show a similar length preference. We therefore incubated QLESIINFEKL and analogs with different C-terminal residues with leucine aminopeptidase and puromycin-sensitive aminopeptidase, both of which also have been implicated in trimming some antigenic precursors in the cytosol (16, 17) but are probably primarily involved in degrading short peptides to amino acids released by proteasomes (34). Unlike ERAP1, these aminopeptidases degraded the 10-residue peptides at identical slow rates that were independent of their C termini (Table 2, which is published as supporting information on the PNAS web site). Moreover, these aminopeptidases show a very different length preference from ERAP1 (Fig. 4). Although ERAP1 digested the 13-residue QL13 many times faster than QL9 and QL5 (Fig. 2A), these cytosolic aminopeptidases digested the 5-residue QL5 most rapidly and the 9- and 13-mer much more slowly. The finding that leucine aminopeptidase and puromycin-sensitive aminopeptidase degrade shorter peptides preferentially is consistent with the prior findings (35) that cytosolic aminopeptidases, by destroying very short peptides (2-5 residues), catalyze the final steps in the ubiquitin-proteasome pathway (34).
Fig. 4.
ERAP1 shows an opposite length preference from ERAP2 and cytosolic aminopeptidases, leucine aminopeptidase (LAP), and puromycin-sensitive aminopeptidase (PSA). QLESQLESQLESL (QL13), QLESQLESL (QL9), and QLESL (Q5) (100 μM, respectively) were incubated with purified ERAP1, ERAP2, LAP, or PSA (3.5 μg/ml, respectively) at 37°C for 1 h. The aminopeptidase activity against different peptide substrates was measured by the method of fluorescence detection.
Recently, Tanioka et al. (36) have cloned from human leukocytes an aminopeptidase that they termed l-RAP. This zinc-metallopeptidase displays 49% amino acid identity to ERAP1 and is also localized in the ER of many cells and is induced by IFN-γ. Consequently, we believe it should be designated “ERAP2,” a name also proposed by Saveanu et al. (37). These extensive similarities suggest that ERAP2 also plays a role in processing longer precursors to 8- to 9-residue antigenic peptides. We therefore tested whether ERAP2/L-RAP showed a similar preference for substrates of 9-16 residues and hydrophobic C termini. Surprisingly, ERAP2 digested the model 13-mer QLESQLESQLESL and the 9-mer QLESQLESL at similar rates and actually spared the 5-mer QLESL (Fig. 4). ERAP2, unlike ERAP1, also rapidly digested other 8- to 9-residue peptides, including Class I epitopes SIINFEKL and FAPGNYPAL (Table 3, which is published as supporting information on the PNAS web site). Also, in contrast to ERAP1 (Table 1), ERAP2 showed no preference for a hydrophobic C-terminal residue when the C-terminal leucine of the antigenic precursors QLESIINFEKL or EFAPGNYPAL was systemically altered, and it readily trimmed ones with basic C termini (Table 3). Thus, these two ER aminopeptidases, despite their many similarities, function in distinct manners, and the binding by ERAP1 of the C-terminal residue of the substrate seems to be a unique property, probably related to its other unique property, its preference for substrates of 9-16 residues.
Discussion
Unique Properties of ERAP1. The present studies have uncovered several properties of ERAP1 that distinguish it from other peptidases and that seem to have evolved to enhance the efficiency of antigen presentation. Its strong preference for substrates longer than eight residues clearly distinguishes ERAP1 from the closely homologous members of the M1 family of aminopeptidases [e.g., aminopeptidase B (38) and leukotriene A4 hydrolase (39)], which strongly prefer di- or tripeptides, and from the major cytosolic aminopeptidases (Fig. 4), leucine aminopeptidase and puromycin-sensitive aminopeptidase (also a member of the M1 family), and from the several aminopeptidase activities present in mammalian cell extracts (34). These intracellular enzymes strongly prefer peptides shorter than five residues, which is consistent with their carrying out the final steps in the complete degradation of proteins in the cytosol (35). Also, the other aminopeptidase in the ER, ERAP2/L-RAP (36, 37), despite its close homology to ERAP1, does not show a similar length preference. Thus, this property of ERAP1 seems to be truly exceptional.
Like TAP, which efficiently transports peptides 8-16 aa long (21), ERAP1 shows a strong preference for peptides of 9-16 residues (Fig. 1) and has no activity against peptides 20 residues or longer. One other metallopeptidase is known that also prefers substrates 8-16 residues long, thimet oligopeptidase. However, it is a cytosolic endopeptidase that cleaves most proteasomal products of this length (34). By destroying cytosolic precursors, this activity limits the supply of antigenic peptides to the ER (40). Although this enzyme and ERAP1 bear no structural homologies and catalyze quite different reactions (N-terminal trimming vs. endoproteolytic cleavage), these enzymes and TAP seem to have evolved similar constraints on maximal substrate length.
Like other aminopeptidases, ERAP1 requires an unblocked amino group on the N terminus of its substrate (Table 1), as does TAP (33, 41). Thus, the substrate-binding groove on these coordinately regulated, sequentially acting proteins may have structural similarities, although no homologies in primary sequence have been found (unpublished observations). One clear difference, however, is that TAP strongly prefers substrates with a C-terminal carboxyl group (33, 41), whereas ERAP1 is equally active against peptides with C-terminal amides. Thus, ERAP1 does not bind the C-terminal residue by interacting with the carboxyl group but instead associates with its hydrophobic side chain (Table 1). This substrate-recognition mechanism contrasts with that of certain related M1 aminopeptidases (e.g., leukotriene A4 hydrolase) that cleave di- and tripeptides by binding their carboxyl group through conserved Arg and Lys residues (42). ERAP1, accordingly, lacks these conserved basic residues.
ERAP1's Molecular Ruler Mechanism. Of special interest is the monitoring by ERAP1 of substrate length and its failure to digest rapidly peptides of eight to 9 residues, the length necessary for tight binding to most MHC class I molecules. This property can account for its ability to process longer precursors to mature epitopes in the absence of an MHC class I molecule as a template. Thus, sequential removal of N-terminal residues from the 11-mer QLESIINFEKL [or the 13-mer QLESQLESQLESL (Fig. 2)] by ERAP1 ceased with the generation of the 8-mer SIINFEKL (or the 9-mer QLESQLESL), which was not further trimmed and therefore accumulated (25). These effects of substrate length and the C-terminal residue on susceptibility to ERAP1 are due to differences in substrate affinity for the enzyme. For example, the 8-residue epitope SIINFEKL has a much higher Km for ERAP1 than the 11-residue precursor QLESIINFEKL (Fig. 7). Because ERAP1 trims longer precursors in a nonprocessive manner [i.e., after each cleavage, it releases peptides of intermediate lengths, which are in turn trimmed further (25, 27) (Fig. 2C)], the final accumulation of 8- to 9-residue epitopes seems to be due to their inability to bind strongly to the enzyme (i.e., their high Kms) (see below).
The precise minimum length where the activity of ERAP1 decreases sharply varies between 8 and 9 residues, presumably depending on the sequence and its length. The length requirements for peptide binding to MHC class I molecules is also not strictly a function of the number of residues. Certain 10-residue peptides (e.g., ones containing internal prolines) can bind tightly to the groove in Class I molecules that primarily present 8- to 9-mers (7). One such 10-residue epitope (NANPDCKTIL) was studied here, and, interestingly, ERAP1 also digested it poorly, as if it were an 8- to 9-mer. Thus, when peptides associate with ERAP1 or Class I molecules, they probably assume similar conformations and are bound in extended configurations in a long pocket, with key anchoring residues at each terminus.
As the present studies demonstrate, the molecular ruler mechanism used in substrate selection by ERAP1 involves recognition of the N-terminal residue to be cleaved and the side chain on the C-terminal residue, 9-16 residues away. These findings indicate a very long substrate-binding groove in which these 2 terminal residues are of primary importance (Fig. 5), as is also true for peptide binding to TAP and MHC class I molecules. Like most MHC-bound peptides (7), ERAP1 shows a strong preference for peptides with C-terminal hydrophobic residues. A basic side chain in this position drastically decreases peptide processing by reducing Km, although acidic or hydrophilic side chains have clear but smaller inhibitory effects (depending on the peptide).
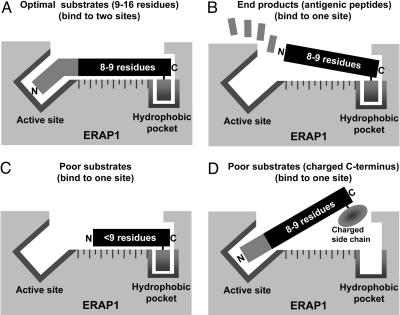
Fig. 5.
Proposed model based on present observations to explain the substrate preference of ERAP1 (molecular ruler). (A) High-affinity binding of optimal substrates through the C-terminal hydrophobic residue and active site. (B) Release of antigenic products (eight to nine residues) after removal of additional N-terminal residues. Affinity is low because it is unable to reach to the active site. (C) Poor substrates (fewer than nine residues) have low affinity because they can interact only with the active site, or if they bind to the hydrophobic pocket, their N-terminal residues cannot reach the active site. (D) A poor substrate has low affinity because its C-terminal residue has a charged side chain.
This requirement for a hydrophobic C-terminal residue for substrate binding (Fig. 5) would predict that short peptides (fewer than eight residues) with hydrophobic C termini should weakly inhibit the trimming of antigenic precursors. However, it has proven impossible to demonstrate such competitive inhibition because of another surprising feature of ERAP1 that we have uncovered: that short peptides, by binding to a regulatory site, distinct from the active site, allosterically activate its peptidase activity (S.-H.C. and A.L.G., unpublished results). The physiological significance of this intriguing property for antigen processing is unclear.
The present findings suggest a simple structural explanation for the length preference of ERAP1 as shown in Fig. 5. The high-affinity binding of substrates to ERAP1 probably involves a hydrophobic pocket that binds the hydrophobic side chain of the C-terminal residue, in addition to the active site of the enzyme (nine or more residues away), which binds the unblocked N-terminal residue that precedes the epitope. Because the affinity of substrates falls when these N-extended residues are removed, and the 8- to 9-mers are generated, it seems likely that peptides eight residues or shorter are not able to stretch from the C-terminal-binding pocket to the active site. Although additional residues may also contribute to substrate binding (as they do for MHC class I binding), this simple model (Fig. 5) can account for its unusual molecular ruler mechanism. Solution of the structure of ERAP1 should indicate whether this simple model is valid.
Significance of the Properties of ERAP1 in Antigen Presentation. This strong preference for peptides with large hydrophobic C termini is likely to have important immunological implications. Interestingly, murine TAP and rat TAP2u prefer a C-terminal hydrophobic residue (33, 41), and all peptides found associated with murine MHC class I molecules have a hydrophobic side chain on their C termini (7). However, rat TAP2a and human TAP have high affinities for basic as well as hydrophobic C termini (33, 41), and a number of human MHC class I molecules present peptides with C-terminal lysines or arginines, especially those presented by HLA-A3 (43), HLA-Aw68 (44), and HLA-B27 (45). Accordingly, the immunoproteasomes show a greater capacity to generate peptides with hydrophobic or basic residues than constitutive proteasomes (14). Thus, in mice, ERAP1 can process all TAP-transported precursors to epitopes that bind to all MHC molecules, but, in humans, ERAP1 is adapted to generate efficiently only MHC class I epitopes with hydrophobic C-terminal residues that constitute the great majority. Those human epitopes with basic C termini presumably therefore are generated independently of ERAP1, either directly by proteasomes, or are trimmed by aminopeptidase(s) in the cytosol (16, 17), or by another aminopeptidase in the ER. ERAP2 is an attractive candidate for such a role because it is also induced by IFN-γ (36) and because it does not show a requirement for a hydrophobic C terminus. Interestingly, mice, which transport into the ER and present antigenic peptides only with hydrophobic C termini, express ERAP1 but lack ERAP2 (unpublished observations). Although ERAP2 can trim antigenic precursors with basic C termini, it lacks the length specificity of ERAP1 and must process such epitopes by a distinct mechanism.
The unique mechanism of ERAP1 (Fig. 5) and the resulting specificity for 9- to 16-residue substrates, like its subcellular localization and regulation by IFN-γ, must have evolved to facilitate immune surveillance. Much, however, remains to be learned about its precise role in processing different types of epitopes, the possible immunological significance of the human polymorphisms in ERAP1 (46), and the structural basis for its molecular ruler mechanism.
Supplementary Material
Acknowledgments
We thank K. L. Rock and I. A. York (University of Massachusetts Medical School, Worcester, MA) for several peptide substrates and many valuable discussions; Ellen Bishop and Mary Dethavong for their assistance in preparing this manuscript; A. Hattori and M. Tsujimoto (RIKEN, Saitama, Japan) for recombinant human ERAP1 and ERAP2; L. B. Hersh (University of Kentucky, Lexington, KY) for providing human puromycin-sensitive aminopeptidase; and T. Saric for helpful advice. This work was supported by National Institute of General Medical Sciences Grant 5 R01 GM46147-12 (to A.L.G.).
Author contributions: S.-C.C. and A.L.G. designed research; S.-C.C. and N.B. performed research; F.M. contributed new reagents/analytic tools; S.-C.C., N.B., and A.L.G. analyzed data; and S.-C.C. and A.L.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; ERAP1, ER aminopeptidase 1; TAP, transporter associated with antigen processing.
References
- 1.Rock, K. L. & Goldberg, A. L. (1999) Annu. Rev. Immunol. 17, 739-779. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell, J. W. & Bennink, J. R. (2001) Curr. Opin. Immunol. 13, 13-18. [DOI] [PubMed] [Google Scholar]
- 3.Shastri, N., Schwab, S. & Serwold, T. (2002) Annu. Rev. Immunol. 20, 463-493. [DOI] [PubMed] [Google Scholar]
- 4.Rock, K. L., York, I. A., Saric, T. & Goldberg, A. L. (2002) Adv. Immunol. 80, 1-70. [DOI] [PubMed] [Google Scholar]
- 5.Madden, D. R. (1995) Annu. Rev. Immunol. 13, 587-622. [DOI] [PubMed] [Google Scholar]
- 6.Jones, E. Y. (1997) Curr. Opin. Immunol. 9, 75-79. [DOI] [PubMed] [Google Scholar]
- 7.Rammensee, H. G., Friede, T. & Stevanoviic, S. (1995) Immunogenetics 41, 178-228. [DOI] [PubMed] [Google Scholar]
- 8.Rock, K. L., Gramm, C., Rothstein, L., Clark, K., Stein, R., Dick, L., Hwang, D. & Goldberg, A. L. (1994) Cell 78, 761-771. [DOI] [PubMed] [Google Scholar]
- 9.Lehner, P. J. & Cresswell, P. (1996) Curr. Opin. Immunol. 8, 59-67. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg, A. L., Cascio, P., Saric, T. & Rock, K. L. (2002) Mol. Immunol. 39, 147-164. [DOI] [PubMed] [Google Scholar]
- 11.Kisselev, A. F., Akopian, T. N., Woo, K. M. & Goldberg, A. L. (1999) J. Biol. Chem. 274, 3363-3371. [DOI] [PubMed] [Google Scholar]
- 12.Mo, X. Y., Cascio, P., Lemerise, K., Goldberg, A. L. & Rock, K. (1999) J. Immunol. 163, 5851-5859. [PubMed] [Google Scholar]
- 13.Saveanu, L., Fruci, D. & van Endert, P. (2002) Mol. Immunol. 39, 203-215. [DOI] [PubMed] [Google Scholar]
- 14.Cascio, P., Hilton, C., Kisselev, A. F., Rock, K. L. & Goldberg, A. L. (2001) EMBO J. 20, 2357-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reits, E., Neijssen, J., Herberts, C., Benckhuijsen, W., Janssen, L., Drijfhout, J. W. & Neefjes, J. (2004) Immunity 20, 495-506. [DOI] [PubMed] [Google Scholar]
- 16.Stoltze, L., Schirle, M., Schwarz, G., Schroter, C., Thompson, M. W., Hersh, L. B., Kalbacher, H., Stevanovic, S., Rammensee, H. G. & Schild, H. (2000) Nat. Immunol. 1, 413-418. [DOI] [PubMed] [Google Scholar]
- 17.Beninga, J., Rock, K. L. & Goldberg, A. L. (1998) J. Biol. Chem. 273, 18734-18742. [DOI] [PubMed] [Google Scholar]
- 18.Androlewicz, M. J., Anderson, K. S. & Cresswell, P. (1993) Proc. Natl. Acad. Sci. USA 90, 9130-9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neefjes, J. J., Momburg, F. & Hammerling, G. J. (1993) Science 261, 769-771. [DOI] [PubMed] [Google Scholar]
- 20.Gromme, M. & Neefjes, J. (2002) Mol. Immunol. 39, 181-202. [DOI] [PubMed] [Google Scholar]
- 21.van Endert, P. M., Tampe, R., Meyer, T. H., Tisch, R., Bach, J. F. & McDevitt, H. O. (1994) Immunity 1, 491-500. [DOI] [PubMed] [Google Scholar]
- 22.Lobigs, M., Chelvanayagam, G. & Mullbacher, A. (2000) Eur. J. Immunol. 30, 1496-1506. [DOI] [PubMed] [Google Scholar]
- 23.Komlosh, A., Momburg, F., Weinschenk, T., Emmerich, N., Schild, H., Nadav, E., Shaked, I. & Reiss, Y. (2001) J. Biol. Chem. 276, 30050-30056. [DOI] [PubMed] [Google Scholar]
- 24.Fruci, D., Niedermann, G., Butler, R. H. & van Endert, P. M. (2001) Immunity 15, 467-476. [DOI] [PubMed] [Google Scholar]
- 25.Saric, T., Chang, S. C., Hattori, A., York, I. A., Markant, S., Rock, K. L., Tsujimoto, M. & Goldberg, A. L. (2002) Nat. Immunol. 3, 1169-1176. [DOI] [PubMed] [Google Scholar]
- 26.Serwold, T., Gonzalez, F., Kim, J., Jacob, R. & Shastri, N. (2002) Nature 419, 480-483. [DOI] [PubMed] [Google Scholar]
- 27.York, I. A., Chang, S. C., Saric, T., Keys, J. A., Favreau, J. M., Goldberg, A. L. & Rock, K. L. (2002) Nat. Immunol. 3, 1177-1184. [DOI] [PubMed] [Google Scholar]
- 28.Ma, W., Lehner, P. J., Cresswell, P., Pober, J. S. & Johnson, D. R. (1997) J. Biol. Chem. 272, 16585-16590. [DOI] [PubMed] [Google Scholar]
- 29.Serwold, T., Gaw, S. & Shastri, N. (2001) Nat. Immunol. 2, 644-651. [DOI] [PubMed] [Google Scholar]
- 30.Udenfriend, S., Stein, S., Bohlen, P., Dairman, W., Leimgruber, W. & Weigele, M. (1972) Science 178, 871-872. [DOI] [PubMed] [Google Scholar]
- 31.Hattori, A., Kitatani, K., Matsumoto, H., Miyazawa, S., Rogi, T., Tsuruoka, N., Mizutani, S., Natori, Y. & Tsujimoto, M. (2000) J. Biochem. (Tokyo) 128, 755-762. [DOI] [PubMed] [Google Scholar]
- 32.Koopmann, J. O., Post, M., Neefjes, J. J., Hammerling, G. J. & Momburg, F. (1996) Eur. J. Immunol. 26, 1720-1728. [DOI] [PubMed] [Google Scholar]
- 33.Momburg, F., Roelse, J., Howard, J. C., Butcher, G. W., Hammerling, G. J. & Neefjes, J. J. (1994) Nature 367, 648-651. [DOI] [PubMed] [Google Scholar]
- 34.Saric, T., Graef, C. I. & Goldberg, A. L. (2004) J. Biol. Chem. 279, 46723-46732. [DOI] [PubMed] [Google Scholar]
- 35.Botbol, V. & Scornik, O. A. (1989) J. Biol. Chem. 264, 13504-13509. [PubMed] [Google Scholar]
- 36.Tanioka, T., Hattori, A., Masuda, S., Nomura, Y., Nakayama, H., Mizutani, S. & Tsujimoto, M. (2003) J. Biol. Chem. 278, 32275-32283. [DOI] [PubMed] [Google Scholar]
- 37.Saveanu, L., Carroll, O., Lindo, V., Del Val, M., Lopez, D., Lepelletier, Y., Greer, F., Schomburg, L., Fruci, D., Niedermann, G. & van Endert, P. M. (2005) Nat. Immunol. 6, 689-697. [DOI] [PubMed] [Google Scholar]
- 38.Soderling, E. (1983) Arch. Biochem. Biophys. 220, 1-10. [DOI] [PubMed] [Google Scholar]
- 39.Orning, L., Gierse, J. K. & Fitzpatrick, F. A. (1994) J. Biol. Chem. 269, 11269-11273. [PubMed] [Google Scholar]
- 40.York, I. A., Mo, A. X., Lemerise, K., Zeng, W., Shen, Y., Abraham, C. R., Saric, T., Goldberg, A. L. & Rock, K. L. (2003) Immunity 18, 429-440. [DOI] [PubMed] [Google Scholar]
- 41.Schumacher, T. N., Kantesaria, D. V., Heemels, M. T., Ashton-Rickardt, P. G., Shepherd, J. C., Fruh, K., Yang, Y., Peterson, P. A., Tonegawa, S. & Ploegh, H. L. (1994) J. Exp. Med. 179, 533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudberg, P. C., Tholander, F., Andberg, M., Thunnissen, M. M. & Haeggstrom, J. Z. (2004) J. Biol. Chem. 279, 27376-27382. [DOI] [PubMed] [Google Scholar]
- 43.Jaramillo, A., Majumder, K., Manna, P. P., Fleming, T. P., Doherty, G., Dipersio, J. F. & Mohanakumar, T. (2002) Int. J. Cancer. 102, 499-506. [DOI] [PubMed] [Google Scholar]
- 44.Guo, H. C., Jardetzky, T. S., Garrett, T. P., Lane, W. S., Strominger, J. L. & Wiley, D. C. (1992) Nature 360, 364-366. [DOI] [PubMed] [Google Scholar]
- 45.Jardetzky, T. S., Lane, W. S., Robinson, R. A., Madden, D. R. & Wiley, D. C. (1991) Nature 353, 326-329. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto, N., Nakayama, J., Yamakawa-Kobayashi, K., Hamaguchi, H., Miyazaki, R. & Arinami, T. (2002) Hum. Mutat. 19, 251-257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.