Abstract
Repeats (27-nt) in intron 4 have been shown to play a cis-acting role in endothelial nitric oxide synthase (eNOS) promoter activity. We hypothesize that the 27-nt repeats could be the source of small nuclear RNA specifically regulating eNOS expression. In this study, we used synthesized 27-nt RNA duplex and found that the eNOS gene transcriptional efficiency was reduced 63% (0.047 ± 0.009 vs. 0.126 ± 0.015, P < 0.01) by nuclear run-on assay. In endothelial cells transfected with the 27-nt small RNA duplex, we found that the eNOS mRNA and protein levels were decreased by >64% (P < 0.01). Conversely, a randomly selected 27-nt from luciferase gene had no effect on the eNOS expression. Furthermore, this eNOS silencing effect appeared to be reversible under the stimulation of vascular endothelial growth factor (10 ng/ml), which is known to up-regulate eNOS expression. Using in situ hybridization and Northern blotting, we observed the presence of endogenous eNOS intron 4-derived 27-nt small RNA, which was confined to the nucleus. In summary, we demonstrated that intron-based microRNAs in eNOS can induce significant gene specific transcriptional suppression, which could be an effective negative feedback regulator for gene expression.
Keywords: intron, microRNA, negative feedback regulation, repeat polymorphism
Most eukaryotic genes are interrupted by one or more introns, which must be removed in the process of pre-mRNA splicing within the nucleus. The mature mRNA with intact ORF is then exported to the cytoplasm for translation. Since the discovery of introns as the noncoding sequences (1), they have mostly been regarded as redundant and useless genomic sequences. However, with the advancement in molecular technology, these intervening sequences have recently been recognized to play a significant role in gene evolution and expression (2-4). Introns can function as enhancer/repressor to regulate cell specific gene transcription; they can modulate the spliceosome conformation and affect the efficiency of pre-mRNA splicing (5, 6). A recent study has shown that the expression of human β-globin gene is highly intron-dependent, which regulates both pre-mRNA splicing and translational efficiency (7). Moreover, the intronic binding site for nuclear proteins mediates the p53-dependent transcriptional activation (8); intronic regulatory elements confer to control developmental and cell-specific gene expression (9). Thus, it is clear that introns are not only essential for genomic integrity, but are also important for controlling cell-specific gene expression.
Of particular interest, Hui et al. (10) have recently shown that the variable-length CA repeats in the endothelial nitric oxide synthase (eNOS) intron 13 can bind with hnRNP and affect eNOS splicing. Studies of ours and others have further suggested that the 27-nt repeats in the eNOS intron 4 could be potentially functional in regulating eNOS expression (11, 12). We have shown that the 27-nt repeats in intron 4 could act as a possible enhancer/repressor for the eNOS transcription (13). Theoretically, however, the 27-nt repeats spliced during eNOS pre-mRNA processing could be a source for small or microRNA that regulates eNOS expression.
Small RNAs, including small interfering RNAs (siRNAs), are commonly used for gene silencing. MicroRNAs, which are produced endogenously in eukaryotic or plant cells, can cause gene-specific silencing through either enhanced mRNA degradation or translational inhibition (14-17). MicroRNAs can be derived from either specific RNA gene transcription or from introns during pre-mRNA splicing (18-21). In addition to the characteristically described effects on mRNA stability and translation efficiency, nuclear small RNAs can affect chromatin remodeling, DNA methylation, and pre-mRNA splicing within the nucleus (22, 23). It is tempting to speculate that the small RNA-mediated gene regulation could be responsible for the self-regulatory mechanism, such as negative feedback regulations.
The human eNOS gene (7q35-36) contains 26 exons and 25 introns (24) and is expressed mainly in endothelial cells. In the form of homodimers, eNOS catalyzes l-arginine oxidation to generate l-citrulline and nitric oxide (NO) in the presence of several cofactors (25). Although enormous data have been accumulated with regard to the regulation of eNOS enzyme activity, relatively little information is available for molecular mechanisms regulating eNOS expression. In the current study, we have examined a hypothesis that the 27-nt repeat derived microRNA could regulate eNOS expression.
Materials and Methods
Supporting Information. For further details, see Supporting Text, which is published as supporting information on the PNAS web site.
siRNA and MicroRNA Preparation. We designed target-specific 19-nt (referred as siRNA) or 27-nt (referred as microRNA) RNA duplexes according to sequences of the type AA(N19)UU or AA(N27)UU (N, any nucleotide) from the eNOS genomic DNA. The sequence details of the small RNA duplexes are provided in Table 1. All siRNAs and microRNAs were chemically synthesized. To examine the subcellular location after transfection, the small RNAs were labeled with fluorescence (6-FAM). Before transfection with liposome, formation of siRNA or microRNA duplex (annealing) was performed according to the method described in ref. 26.
Table 1. Sequences for small RNA duplex used in the study.
| Names | Location | Sequences |
|---|---|---|
| 19 nt | 21268–21286 | 5′–AAGCGACGAGGUGCAGAACGCUU–3′ (sense) |
| eNOS exon 24 | 5′–AAGCGUUCUGCACCUCGUCGCUU–3′ (antisense) | |
| 19 nt | 5178–52196 | 5′–AAGUCUAGACCUGCUGCAGGGUU–3′ (sense) |
| eNOS intron 4 | 5′–AACCCUGCAGCAGGUCUAGACUU–3′ (antisense) | |
| 27 nt | 6370–6396 | 5′–AAAAAUGUUCACCUACAUCUGCAACCACAUU–3′ (sense) |
| eNOS exon 5 | 5′–AAUGUGGUUGCAGAUGUAGGUGAACAUUUUU–3′ (antisense) | |
| 27 nt | 1941–1967 | 5′–AAUAACAUGGGCAACUUGAAGAGCGUGGCUU–3′ (sense) |
| eNOS exon 1 | 5′–AAGCCACGCUCUUCAAGUUGCCCAUGUUUU–3′ (antisense) | |
| 27 nt | 5175–5201 | 5′–AAGAAGUCUAGACCUGCUGCAGGGGUGAGUU–3′ (sense) |
| eNOS intron 4 | 5′–AACUCACCCCUGCAGCAGGUCUAGACUUCUU–3′ (antisense) | |
| 27-nt one-base mutant | 5175–5201 | 5′–AAGAAGUCUUGACCUGCUGCAGGGGUGAGUU–3′ (sense) |
| eNOS intron 4 | 5′–AACUCACCCCUGCAGCAGGAUAAGACUUCUU–3′ (antisense) | |
| 27-nt three-base mutant | 5175–5201 | 5′–AAGAAGUCUUAUCCUGCUGCAGGGGUGAGUU–3′ (sense) |
| eNOS intron 4 | 5′–AACUCACCCCUGCAGCAGGAUAAGACUUCUU–3′ (antisense) | |
| 19 nt | 153–171 | 5′–AAGUCUGACAGUUACCAAUGCUU–3′ (sense) |
| Luciferase gene | 5′–AAGCAUUGGUAACUGUCAGACUU–3′ (antisense) | |
| 27 nt | 153–179 | 5′–AAGUCUGACAGUUACCAAUGCUUAAUCAGUU–3′ (sense) |
| Luciferase gene | 5′–AACUGAUUAAGCAUUGGUAACUGUCAGAC–3′ (antisense) |
Cell Culture, Treatment, and Transfection. Human aortic endothelial cells (HAECs) were cultured in EBM-2 Bulletkit containing 3% FBS. The siRNA or microRNA duplex was transfected with Lipofectamine 2000.
Isolation of Nuclei and Nuclear Run-On Assay. Nuclei were isolated from the cultured HAECs (Supporting Text) and immediately stored at -80°C until nuclear run-on transcription assay. Nuclear run-on assays were performed according to the method described by Greenberg (27) with minor modifications. Briefly, identical number of HAEC nuclei was used for preparation of nascent transcripts. To perform the nuclear run-on transcription, 5 × 106 nuclei were incubated in a reaction buffer (5 mmol/liter Tris·HCl, pH 8.0/2.5 mmol/liter MgCl2/150 mmol/liter KCl/2.5 mmol/liter each of ATP, GTP, CTP) and biotin-16-UTP at 30°C for 45 min in a final volume of 60 μl. The reaction was stopped by the addition of 1,000 units of RNase-free DNase and incubated for 10 min at 37°C. The nuclei were then lysed by the addition of lysis buffer containing 10 mmol/liter Tris·HCl, 1% SDS, and 5 mmol/liter EDTA. The reaction mixtures were treated with 20 μl of proteinase K (10 mg/ml). RNA was extracted with TRIzol reagent, ethanol-precipitated, and resuspended in 50 μl of RNase-free H2O. Biotinylated RNA was purified and subjected to quantitative real-time RT-PCR.
In Vitro Transcription Assay. HelaScribe Nuclear Extract in vitro Transcription System and the endothelial nuclei were applied in these reactions. A standard transcription reaction was carried out in a volume of 25 μl containing 3.4 μl of the endothelial nuclear extract (8.0 units per reaction). The reaction mixture also contained 7.6 μl of reaction buffer, 3.0 mmol/liter MgCl2, 10 μmol/liter of each ATP, CTP, UTG, and GTP. The RNA duplexes were added to the reaction as indicated in Fig. 2 and Table 1. Typical reaction was conducted at 30°C for 45 min and stopped by the addition of 100 units of RNase-free DNase I with an incubation of 10 min at 37°C. This was followed with the addition of a 175-μl stop solution provided by the kit.
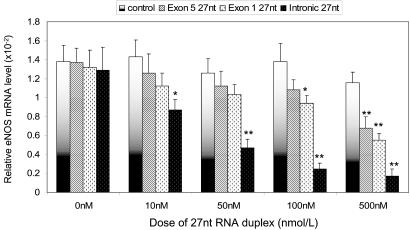
Fig. 2.
Effects of small RNAs eNOS gene transcription. Nuclei of HAECs were isolated and used in the nuclear run-on transcription reaction containing 2.5 mmol/liter of ATP, CTP, GTP, biotin-16-UPT, and various small RNAs as indicated in the legend. After the reaction at 30°C for 45 min, the biotinylated RNA was separated from total RNA, and used for cDNA synthesis and real-time PCR. The transcription efficiency was presented by the relative mRNA levels of the eNOS and β-actin. Data presented were obtained from three independent experiments; *, P < 0.05; **, P < 0.01 by independent Student's t test when compared to the control.
Northern Blotting for Small RNAs. Because the conventional method for RNA extraction normally discarded RNAs of small sizes, we used a mirVana miRNA Isolation kit (Ambion, catalog no. 1560), which is specifically designed for microRNA extraction. In addition to the RNAs from the total cell extracts, we also prepared the RNAs from nuclear and cytosolic fractions to explore the subcellular location of the 27-nt microRNA. Small RNAs from the HAECs (10 μg) were loaded on a 12.5% denaturing polyacrylamide gel, and electrophoresized with 130 V for 2 h at room temperature. The resolved RNAs were transferred to the positively charged Nylon membrane (Ambion, catalog no. 10102). The synthesized antisense 27-nt oligonucleotide probe according to the 27-nt repeat sequence in the eNOS was 5′ end-labeled with [γ-32P]ATP by the T4 kinase. This was then followed with the standard Northern blotting procedures.
In Situ Hybridization. In situ hybridization was performed by using the complementary 27-nt single-strand DNA as the probe (5′-CTCACCCCTGCAGCAGGTCTAGACTTC-3′ complementary to the sense strand), which was labeled with digoxigenin by using the DIG Oligonucleotide 3-End Labeling kit (Roche Diagnostics catalog no. 03353575910). In the control probe, we used a same sized DIG-labeled 27-nt single-strand DNA (5′-CTGATTAAGCATTGGTAACTGTCAGAC-3′), which is complementary to the sense sequence of luciferase gene.
cDNA Array for mRNA Expression in Endothelial Cells. To explore whether the 27-nt microRNA also affected the expression of genes other than eNOS in endothelia cells, we used GEArray Q Series Human Endothelial Cell Biology Gene Array (SuperArray Bioscience, catalog no. HS-036) to document mRNA changes in endothelial cells. The biotin-labeled cDNA probes were generated from the RNAs extracted from endothelial cells treated with or without the 27-nt microRNAs. The hybridization, visualization, and data interpretation were carried out according to the manufacturer's instruction.
Statistical Analyses. Data are expressed as mean ± SEM of the three separate experiments. Independent Student's t test was used for the comparisons of between group differences; and the one-way ANOVA model was applied for multiple group comparisons with post hoc Bonferroni correction for multiple comparisons using spss 12.0 for Windows (SPSS, Chicago). Outcomes were considered statistically significant with two-tailed P < 0.05.
Results
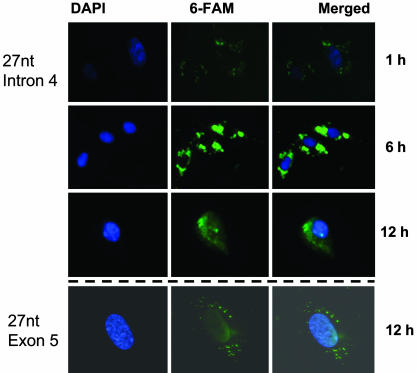
Distribution of the Exogenous 27-nt Small RNA within Endothelial Cells After Transfection. To observe the intracellular distribution of the small RNA after transfection, HAECs were incubated in the chamber slide and grown up to 50% confluence. Cells were then transfected with the 27-nt RNA duplex labeled with 6-FAM using Lipofectamine 2000. After the transfection for the indicated duration, cells were fixed and stained blue with DAPI (10 ng/ml) for nuclei. The transfected 27-nt RNA duplex was stained green by 6-FAM. At 1 h after the transfection, only a small amount of labeled 27-nt RNA was detectable in the cytoplasm of the HAECs (Fig. 1). Six hours after transfection, a large amount of 27-nt RNA accumulated in the cytoplasm, as shown by the bright green light. However, at 12 h after transfection, some of the 27-nt RNA started migrating to the nuclei (Fig. 1). To exclude the possible nonspecific fluorescence, the same procedure was performed by using unlabeled RNA duplex instead of 6-FAM-labeled RNA duplex. Only nuclei were stained with DAPI (blue), no green fluorescence was detectable in the control cells under the same conditions of visualization (data not shown). Furthermore, the exogenously transfected 27-nt RNA of the eNOS exon 5 showed a similar subcellular distribution as that of the 27-nt intron 4 RNA (Fig. 1). A similar pattern was also shown for the 27-nt RNA duplex of the luciferase gene (data not shown). These findings suggest that the nuclear import of the exogenously transfected small RNA is not sequence specific.
Fig. 1.
Intracellular distribution of the transfected exogenous 27-nt RNA duplex in cultured HAECs. HAECs were plated onto chamber sliders, transfected with the 6-FAM-labeled 27-nt RNA duplex complimentary to the sequence of the eNOS intron 4 (Top) or eNOS exon 5 (Bottom) with liposome. After the indicated duration of transfection, cells were fixed and stained with DAPI. The samples were mounted and detection performed under fluorescence microscopy.
27-nt MicroRNA Reduced the eNOS Transcription. We next investigated the effects of the 27-nt exogenous microRNA on eNOS transcription efficiency by the nuclear run-on assay. Nuclei were isolated from the HAECs, and used with the reaction containing 2.5 mmol/liter biotin-16-UTP. The 27-nt microRNA was added into the reactions. We used a 27-nt microRNA complementary to the coding sequence of luciferase gene as a negative control because it should not specifically affect any human gene. Meanwhile, the 27-nt microRNA homologous to a partial sequence of the exon 5 and a partial sequence of the exon 1 (exonic 27-nt microRNA) were used to compare the effects with the intron 4-based 27-nt microRNA (intronic 27-nt microRNA). After transcriptional reaction, the biotinylated RNA was isolated and quantitative real-time RT-PCR was conducted. The relative eNOS mRNA levels reflected the eNOS gene transcription efficiency. As shown in Fig. 2, intronic 27-nt microRNA significantly decreased the eNOS gene transcription efficiency in a dose-dependent manner. With the increased concentrations of intronic 27-nt microRNA at 10, 50, 100, and 500 nmol/liter, the eNOS mRNA levels were dose-dependently decreased by 40.0% (0.087 ± 0.011 vs. 0.143 ± 0.018, P < 0.05), 62.4% (0.047 ± 0.009 vs. 0.126 ± 0.015, P < 0.01), 81.8% (0.025 ± 0.006 vs. 0.138 ± 0.019, P < 0.01), and 85.3% (0.017 ± 0.008 vs. 0.116 ± 0.011, P < 0.01), respectively. Our data clearly showed that the intronic 27-nt microRNA can significantly depress eNOS transcription efficiency. On the other hand, the exonic 27-nt microRNA only showed a depressing effect at higher concentration of 500 nmol/liter for a 42% inhibition (0.068 ± 0.012 vs. 0.116 ± 0.011, P < 0.05). There was no effect at the lower doses (Fig. 2). In addition, we also tested the possible differences between the designs of the 19-nt siRNA and 27-nt microRNA based on the same sequence (Table 1). There was no obvious difference in eNOS transcription efficiency between the 19-nt and 27-nt RNA duplexes (data not shown). To examine the relative specificity of the 27-nt microRNA homologous to the 27-nt repeat in intron 4 of the eNOS gene, we further designed two mutants with either one or three bases altered in the intronic 27-nt microRNA (Table 1). Although the single base mutation at the position of 10th nucleotide did not appear to affect the eNOS depressing effect (74.5% reduction as compared to 81.8% reduction by the wild-type 27-nt microRNA at 100 nM), the three-base mutation (from 10th to 12th nucleotide) appeared to considerably attenuated the inhibiting effect (9.5% reduction vs. 81.8% reduction by the wild-type 27-nt microRNA).
Using cDNA array, we also found that the effects of the intronic 27-nt microRNA appeared to be specific on the eNOS gene. We showed that the expression of eNOS was decreased by the intronic 27-nt RNA duplex treatment comparing to the control treatment (luciferase 27-nt RNA duplex) and no treatment (data not shown). However, the house-keeping genes, e.g., β-actin, showed little change. In addition, a few other genes also showed changes in expression. For example, the mRNA levels of Annexin-5 and IL1B were increased, but the levels of the Bax and CRADD (CASP2 and RIPK1 domain containing adaptor with death domain) were decreased by the intronic 27-nt RNA treatment. The alteration in the expression of the apoptosis related genes is more likely to be the result of reduced eNOS expression and NO production rather than the direct effect of the 27-nt RNA on the transcription of these genes. The experiments were repeated three times.
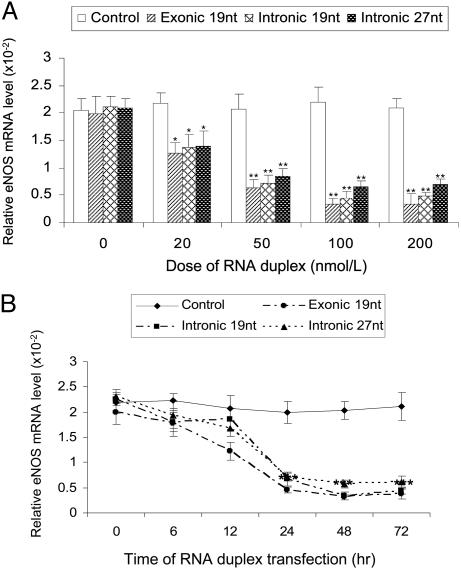
Effects of the 19-nt siRNA and the 27-nt microRNA on eNOS Expression. To examine the number of nucleotides of the small RNAs that are required for the effective eNOS suppression, we compared the effects of the 19- and 27-nt small RNA, in which the 19-nt had the same RNA sequence as the nucleotides 4-23 in the 27-nt microRNA (Table 1). Cells were cultured in six-well plates and grown up to 90% confluence before they were transfected with 19-nt exonic siRNA (exon 24), 19-nt intronic siRNA (19 nt of the 27-nt repeats sequence in intron 4), and the 27-nt microRNA (27-nt repeat sequence in intron 4) with concentration ranging from 0 to 200 nM. At the end of the 24-h posttransfection, total RNA was isolated from the cells for quantitative real-time RT-PCR. As shown in Fig. 3A, exonic 19-nt siRNA, intronic 19-nt siRNA and the 27-nt microRNA all induced a dose-dependent reduction in the eNOS mRNA levels. The efficiency of the inhibition on eNOS expression appeared to be the same among all three different types of the small RNAs. At the dose of 50 nmol/liter, we also tested the time-course of the inhibiting effects. As shown in Fig. 3B, the decrease started from the 6 h and continued to the lowest level at 48 h with significant reduction by 85%, 84%, and 72% with the treatments of exonic 19-nt siRNA, intronic 19-nt siRNA, and intronic 27-nt microRNA, respectively (Fig. 3B).
Fig. 3.
Effect of small RNAs on eNOS mRNA in HAECs. HAECs were plated onto six-well plates and grown up to 90% confluence, then transfected with indicated synthetic 27-nt RNA duplexes by liposome. After indicated duration of transfection, total RNA was isolated and used to cDNA synthesis, followed by real-time PCR. The eNOS mRNA level was presented by the relative ratio of eNOS copies to β-actin. (A) A dose-dependent effect. (B) Time course changes. ***, P < 0.001 by the Student t test comparing to the levels at 0 and 6 h.
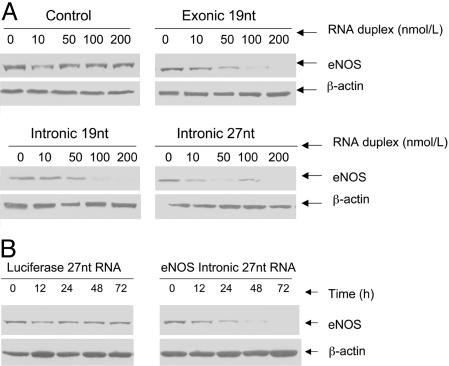
Repression of eNOS Protein Expression by the Intronic 27-nt MicroRNA. We further examined the eNOS protein levels in these small RNA-treated endothelial cells. As shown in Fig. 4A, exonic and intronic 19-nt siRNAs and the intronic 27-nt microRNA caused a dose-dependent decrease in the eNOS protein levels. The extent of repression on eNOS protein expression induced by the exonic 19-nt siRNA was higher than that by the intronic 19-nt siRNA or the 27-nt microRNA (Fig. 4A). The inhibition on the eNOS protein levels by the 27-nt intronic microRNA started early at 24 h and reached maximum at 48 h after transfection (Fig. 4B). On the other hand, a 19-nt siRNA or 27-nt microRNA against luciferase gene had no effects on the eNOS protein levels. These findings suggest that the inhibition at eNOS mRNA levels is also reflected by the reduced eNOS protein levels.
Fig. 4.
Effects of small RNA on eNOS protein expression. HAECs were cultured on six-well plates and grown up to 90% confluence in EBM-2 media. Cells were then transfected with indicated RNA duplexes for 48 h. Protein samples were collected and analyzed by performing Western blotting. (A) Dose-dependent effects of the exonic and intronic RNA duplexes on eNOS protein expression. (B) Time-course changes of the 27-nt intronic microRNA duplex on eNOS protein expression as compared with the control by the luciferase 27-nt RNA duplex.
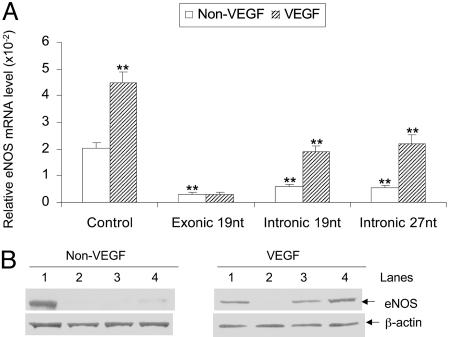
VEGF Restores the eNOS Suppression by the Exogenous Small RNA. It is generally accepted that the classical gene silencing by exonic siRNAs is irreversible due to the targeted mRNA degradation in cytoplasm. Our data suggest that the mechanisms involved in the eNOS gene silencing by intronic siRNA or microRNA might be different. It appeared that the inhibition by the intronic small RNAs might be at the transcriptional level. We then explored whether the silenced eNOS expression induced by intronic microRNA was reversible. Previous studies have shown that VEGF can increase eNOS mRNA level through enhancing the eNOS gene transcription in cultured human vascular endothelial cells (28). We examined whether VEGF could restore the intronic 27-nt microRNA induced transcription inhibition in the eNOS gene. We added 10 ng/ml of VEGF to the HAECs 12 h after the transfection with the different small RNAs. The levels of eNOS mRNA and protein were analyzed at 24 and 48 h of incubation. Cells were transfected with the 19-nt siRNA homologous to the luciferase gene and used as the control. Without VEGF treatment (showed as “non-VEGF” in Fig. 5A), the mRNA levels were significantly decreased in cells transfected with exonic 19-nt by 85% (0.306 ± 0.061 × 10-2 vs. 2.033 ± 0.215 × 10-2, P < 0.01), intronic 19-nt by 70% (0.610 ± 0.053 × 10-2 vs. 2.033 ± 0.215 × 10-2 P < 0.01), and intronic 27-nt by 73% (0.542 ± 0.081 × 10-2 vs. 2.033 ± 0.215 × 10-2, P < 0.01), respectively. VEGF treatment (showed as “VEGF” in Fig. 5A) restored the levels of the eNOS mRNA to the baseline level in HAECs transfected with intronic 19-nt siRNA or 27-nt microRNA. In the cell transfected with the intronic 19-nt and 27-nt RNA duplex, VEGF increased eNOS mRNA levels by 3.1-fold (1.903 ± 0.230 × 10-2 vs. 0.610 ± 0.053 × 10-2, P < 0.01) and 4.1-fold (2.218 ± 0.322 × 10-2 vs. 0.542 ± 0.081 × 10-2, P < 0.01) as compared with the non-VEGF treatment, respectively. On the other hand, VEGF treatment did not appear to rescue the eNOS mRNA suppression by the exonic 19-nt siRNA (0.312 ± 0.052 × 10-2 vs. 0.306 ± 0.061 × 10-2, P > 0.05). The changes in eNOS mRNA levels were also reflected by the eNOS protein levels as shown in Fig. 5B. Using 10 mM mevastatin, which appears to stabilize eNOS mRNA, we also observed a small but statistically significant increase in the eNOS mRNA in cells treated with the exogenous 27-nt microRNA (0.542 ± 0.081 × 10-2 vs. 0.918 ± 0.143 × 10-2, P < 0.05), although the rescuing effect was far less than that of the VEGF. Western blot showed the same effect on protein levels (data not shown). Our results suggest that the inhibiting effects by the intronic mRNA may be mainly at the transcriptional level rather than by affecting the mRNA stability or translational efficiency.
Fig. 5.
VEGF restored the down-regulated eNOS mRNA and protein levels by the intronic small RNA. HAECs were transfected with indicated RNA duplex. After 12-h transfection, 10 ng/ml of VEGF was added into the media. (A) Total RNA was collected after 24-h transfection for the analysis of mRNA levels by the real-time PCR. Columns represent mean + SEM of three independent experiments (*, P < 0.05 vs. non-VEGF; **, P < 0.01 vs. control by Student's t tests). (B) Protein samples were collected after 48-h transfection and measured by Western blotting. Lanes 1, 2, 3, and 4 represent control (19-nt siRNA targeting luciferase gene), 19-nt exonic siRNA, 19-nt intronic siRNA, and 27-nt intronic microRNA at the dose of 50 nmol/liter, respectively.
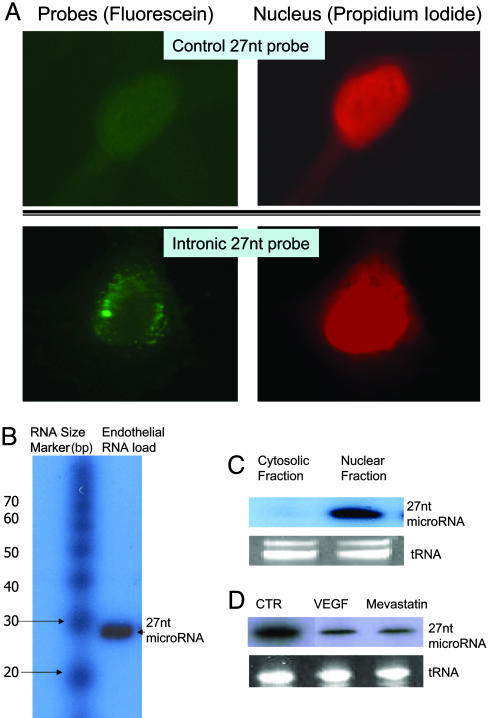
Evidence for the Intron 4-Derived 27-nt RNA Existing in Cultured HAECs. Based on the observations that exogenous 27-nt RNA duplex resulted in a repressed eNOS transcription in cultured HAECs, we then sought to investigate whether the intron 4-derived 27-nt microRNA indeed existed biologically in vivo. We used in situ hybridization technique in which the DIG-labeled anti-sense 27-nt oligo-probe (homologous to the sequence of 27-nt repeats in eNOS intron 4) was applied to the cultured HAECs, and detected with anti-DIG fluorescein conjugate. As shown in Fig. 6A by the fluorescein labeling (green staining), the 27-nt RNA distributed within the nucleus, and the perinuclear area of the HAECs in a punctate form. No fluorescein signal was detected when the same sized control probe was used. Furthermore, using the same probe, we showed that the presence of the 27-nt microRNA is likely to be derived from the 27-nt repeats in eNOS intron 4 by Northern blotting (Fig. 6B), and most of the 27-nt microRNA was located within the nucleus rather than the cytosol (Fig. 6C). Using the control 27-nt RNA probe derived from the eNOS exon 5 sequence revealed no band <200-nt in our RNA preparation. Furthermore, treating the endothelial cells with VEGF (10 ng/ml) or mevastatin (10 μM), we observed decreased levels of the 27-nt microRNA (Fig. 6D). Our data show that the intron-derived 27-nt RNA sequence exists biologically within the cultured endothelial cells. Together with the results of the exogenously transfected 27-nt small RNA, our findings suggest the possibility of the 27-nt microRNA acting as an endogenous molecule for the feedback regulation of the eNOS expression in the endothelial cells.
Fig. 6.
Presence of endogenous 27-nt microRNA in cultured HAECs. (A) In situ hybridization was performed in cultured HAECs. HAECs were grown up to 50% confluence. After fixation and dehydration, cells were incubated in hybridization buffer containing the DIG-labeled 27-nt oligo-probe at 42°C for 16 h. The anti-DIG-fluorescein (dilution of 1:500 in blocking solution) was then applied to the cells at room temperature for 45 min. After counterstaining the nuclei with propidium iodide, samples were visualized under fluorescence microscopy. The fluorescein (green) showed that the 27-nt RNA was distributed within the nucleus, and in the perinuclear area of the HAECs. The staining appears to be in a punctate form. No obvious fluorescein signal was detectable in HAECs when control probe (27-nt single strand DNA complementary to luciferase gene) was used. (B) Northern blotting for the 27-nt microRNA from the endothelial small RNA extracts. The sizes of small RNA markers (Ambion) are labeled on the left. The band at the position approximate to 27-nt was hybridized to the [γ-32P]-labeled 27-nt oligo-probe is indicated on the right. (C) Northern blotting shows that the 27-nt microRNA is mainly located in the nuclear fraction. The cytosolic fraction contains none or only trace of the 27-nt microRNA. (D) A representative Northern blot of three separate experiments for the total cellular 27-nt microRNA from the endothelial cells treated by 10 ng/ml VEGF or 10 μM mevastatin for 24 h during culture. Both VEGF and mevastatin reduced 27-nt microRNA significantly (48.2 ± 11.1% and 39.6 ± 7.9% of the levels in the control cells, respectively).
Discussion
We have designed the current study to understand how the 27-nt repeat variant in the eNOS intron 4 affects the eNOS expression. Based on all possible functional mechanisms that a 27-nt repeat sequence can have, we hypothesized that the eNOS intron 427-nt repeats could be the source for endogenous microRNA, possibly derived during eNOS pre-mRNA processing to function as a negative feedback regulator on eNOS expression. Theoretically, the more the eNOS is expressed, the more 27-nt microRNA will be produced. Hence, the overexpressed eNOS will be controlled to a reasonable level. This negative feedback regulatory loop could be an important physiological regulator of the adequate eNOS expression. When the eNOS expression is too high, more free radicals, e.g., superoxide or peroxynitrite, derived from the eNOS associated reaction could be harmful. Indeed, our experiments show the biological existence of the 27-nt microRNA with the sequence complementary to the sense strand of the 27-nt repeats in eNOS intron 4. Using Northern blotting analysis (Fig. 6 B and C), we demonstrate that the 27-nt microRNA is mainly located within the nucleus indicating the possible functional sites at the eNOS transcription or pre-mRNA processing. The positive signals in immunofluorescence or Northern blotting assay are less likely to be caused by nonspecific reaction because the luciferase gene based DNA probe produced no positive signal.
To investigate this hypothesis further, we have taken the siRNA approach by designing the 27-nt microRNA and applied to the endothelial cells as exogenous microRNA to examine its effect on the eNOS gene expression. Our study has shown that an intron-based microRNA or siRNA can gene-specifically suppress the eNOS expression. Classically, exon-based siRNA inhibits gene expression by increasing the mRNA degradation (29-31), which requires perfect sequence match. Alternatively, some microRNA could exert translational inhibition via imperfect sequence match at 3′ UTR. Our data show that the intron sequence-based microRNA can directly affect gene transcription efficiency. The inhibition of the 27-nt microRNA on the transcription efficiency in the nuclear run-on transcription assay and the predominant nuclear presence of the endogenous 27-nt microRNA support this mechanism. The 27-nt microRNA mediated gene suppression appears to be sequence specific because the inhibitory effect is seriously compromised when three bases in the 27-nt microRNA were mutated and the same sized 27-nt microRNA matched to luciferase gene had no effect. Notably, the synthesized exonic 27-nt microRNA also showed the ability to inhibit the eNOS gene transcription efficiency in the nuclear run-on assay at the high dose (500 nM). The mechanism(s) of these effects on the eNOS gene transcriptional efficiency is not defined. We propose that the 27-nt microRNA could directly target the complementary genomic sequence to inhibit the transcriptional factor binding or modify nucleotide methylation status or the histone acetylation status, or affect pre-mRNA splicing; all of these pathways have been proposed as possible mechanisms (13, 22). This negative regulatory mechanism based on intron-derived microRNA could be potentially applied to many other genes that contain the similarly sized repeat sequences in noncoding regions.
Currently, there are several ways of producing siRNAs or microRNAs. In addition to chemical synthesis, in vitro transcription, or RNase III/Dicer digestion of long dsRNAs, siRNAs or microRNAs can be expressed in vivo from plasmids, PCR cassettes, or viral vectors that contain a CMV or polymerase III (pol III) transcription unit (32). In this study, we used the chemically synthesized RNA duplex as the tool. There are several reasons for choosing the size of 27-nt. First, the 27-nt microRNA is homologous to the 27-nt repeats in the eNOS intron 4, which is polymorphic and related to cardiovascular risk. Second, in the eNOS genomic sequence, there always exist “AG” and “GT” bases at 5′ and 3′ end regions of exons. Coincidentally, the “AG” and “GA” bases are also present at the 5′ and 3′ ends of the 27-nt repeats in eNOS intron 4. These similar “splice sites” may result in the formation of intact microRNAs from the 27-nt repeats because of the pre-mRNA splicing. The amount of these repeat-derived 27-nt microRNA would vary from individual to individual depending on the number of repeats, therefore accounting for between-individual differences in eNOS expression and cardiovascular risk. Many microRNAs at the size of 27-nt have been identified and have the functions of regulating gene expression during development (32). In our study, we also compared the effects of the 27-nt with the 19-nt small RNA because 19-nt has been classically used as siRNA for gene silencing. Our experiments have shown little size-dependent difference.
As expected, the eNOS exon sequence based siRNA may mainly repress the eNOS expression by increasing mRNA degradation. This observation is supported by the findings that the exon 19-nt siRNA did not change the transcription efficiency significantly at low doses in the nuclear run-on assay (Fig. 2). The treatment by VEGF also did not rescue the suppressing effect of the exonic siRNA (Fig. 5A). On the other hand, the intron-based small RNAs may mainly repress the expression through the transcription efficiency in the nucleus. This possibility is supported by the effective inhibition in the nuclear run-on assay at low doses (Fig. 2), and the effect was reversible by the eNOS transcription enhancer VEGF (28, 33, 34) (Fig. 5A). Although these small RNAs may operate through different pathways, they all result in a decreased eNOS mRNA and protein levels when the intact cells are transfected with these different types of small RNAs (Figs. 3 and 4).
If the 27-nt microRNA is derived from the eNOS pre-mRNA processing, we should expect an increased endogenous 27-nt microRNA when eNOS transcription is enhanced by VEGF. Conversely, our results showed a reduced level of endogenous 27-nt microRNA (Fig. 6D). Although more studies are needed to dissect the mechanisms, our findings suggest that the regulation of the intron 4-derived 27-nt microRNA might be independent of the eNOS transcription. Alternatively, VEGF may regulate eNOS expression by directly inhibiting the eNOS specific 27-nt microRNA production.
In summary, we have demonstrated the biological existence of the 27-nt microRNA derived from the eNOS intron 4 27-nt repeats. The transfection of the exogenous 27-nt microRNA can significantly reduce the eNOS expression, which is more likely to have occurred at the eNOS gene transcription efficiency. The 27-nt microRNA-mediated eNOS regulation is gene-specific and is potentially reversible by VEGF. Our findings provide evidence for a previously undescribed negative feedback regulatory mechanism in eNOS expression; a molecular model could be applicable to other genes sharing a similar genomic structure.
Supplementary Material
Acknowledgments
We thank Ms. Irene Harrison for editorial assistance. The work is supported by National Institutes of Health Grant R01-HL066053. X.L.W. is an American Heart Association Established Investigator.
Author contributions: M.-X.Z., H.O., Jian Wang, J.C., and X.L.W. designed research; M.-X.Z., H.O., Y.H.S., and Jing Wang performed research; M.-X.Z., H.O., Y.H.S., Jing Wang, Jian Wang, J.C., and X.L.W. analyzed data; and H.O., Jian Wang, J.C., and X.L.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: eNOS, endothelial nitric oxide synthase; HAEC, human aortic endothelial cell; siRNA, small interfering RNA.
References
- 1.Gilbert, W. (1978) Nature 271, 501. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, V. (2003) Cell 113, 673-676. [DOI] [PubMed] [Google Scholar]
- 3.Lee, R. C. & Ambros, V. (2001) Science 294, 862-864. [DOI] [PubMed] [Google Scholar]
- 4.Forrest, S. T., Barringhaus, K. G., Perlegas, D., Hammarskjold, M. L. & McNamara, C. A. (2004) J. Biol. Chem. 279, 32897-32903. [DOI] [PubMed] [Google Scholar]
- 5.Liu, X., Yue, P., Khuri, F. R. & Sun, S. Y. (2004) Cancer Res. 64, 5078-5083. [DOI] [PubMed] [Google Scholar]
- 6.Virts, E. L. & Raschke, W. C. (2001) J. Biol. Chem. 276, 19913-19920. [DOI] [PubMed] [Google Scholar]
- 7.Lu, S. & Cullen, B. R. (2003) RNA 9, 618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi, K., Toyota, M., Sasaki, Y., Yamashita, T., Ishida, S., Ohe-Toyota, M., Maruyama, R., Hinoda, Y., Saito, T., Imai, K., et al. (2004) Oncogene 23, 7791-7798. [DOI] [PubMed] [Google Scholar]
- 9.Reddy, V. S. & Reddy, A. S. (2004) Plant Mol. Biol. 54, 273-293. [DOI] [PubMed] [Google Scholar]
- 10.Hui, J., Stangl, K., Lane, W. S. & Bindereif, A. (2003) Nat. Struct. Biol. 10, 33-37. [DOI] [PubMed] [Google Scholar]
- 11.Wang, X. L., Sim, A. S., Badenhop, R. F., McCredie, R. M. & Wilcken, D. E. (1996) Nat. Med. 2, 41-45. [DOI] [PubMed] [Google Scholar]
- 12.Wattanapitayakul, S. K., Mihm, M. J., Young, A. P. & Bauer, J. A. (2001) Trends Pharmacol. Sci. 22, 361-368. [DOI] [PubMed] [Google Scholar]
- 13.Wang, J., Dudley, D. & Wang, X. L. (2002) Arterioscler. Thromb. Vasc. Biol. 22, e1-e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miska, E. A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M. & Horvitz, H. R. (2004) Genome Biol. 5, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon, K., Cole-Strauss, A. & Kmiec, E. B. (1996) Proc. Natl. Acad. Sci. USA 93, 2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grewal, S. I. & Moazed, D. (2003) Science 301, 798-802. [DOI] [PubMed] [Google Scholar]
- 17.Zeng, Y., Yi, R. & Cullen, B. R. (2003) Proc. Natl. Acad. Sci. USA 100, 9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambros, V., Lee, R. C., Lavanway, A., Williams, P. T. & Jewell, D. (2003) Curr. Biol. 13, 807-818. [DOI] [PubMed] [Google Scholar]
- 19.Lin, S. L., Chang, D., Wu, D. Y. & Ying, S. Y. (2003) Biochem. Biophys. Res. Commun. 310, 754-760. [DOI] [PubMed] [Google Scholar]
- 20.Ying, S. Y. & Lin, S. L. (2005) Biochem. Biophys. Res. Commun. 326, 515-520. [DOI] [PubMed] [Google Scholar]
- 21.Ying, S. Y. & Lin, S. L. (2004) Gene 342, 25-28. [DOI] [PubMed] [Google Scholar]
- 22.Morris, K. V., Chan, S. W., Jacobsen, S. E. & Looney, D. J. (2004) Science 305, 1289-1292. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki, H. & Taira, K. (2004) Nature 431, 211-217. [DOI] [PubMed] [Google Scholar]
- 24.Marsden, P. A., Heng, H. H., Scherer, S. W., Stewart, R. J., Hall, A. V., Shi, X. M., Tsui, L. C. & Schappert, K. T. (1993) J. Biol. Chem. 268, 17478-17488. [PubMed] [Google Scholar]
- 25.Fleming, I. & Busse, R. (2003) Am. J. Physiol. 284, R1-R12. [DOI] [PubMed] [Google Scholar]
- 26.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg, M. (1987) in Current Protocols in Molecular Biology, eds. Ausubel, F., Brent, R., Kingston, R., Moore, D., Smith, J., Seidman, J. & Struhl, K. (Wiley, New York), Vol. 1, pp. 4.10.1-4.10.11. [Google Scholar]
- 28.Aida, K., Shi, Q., Wang, J., VandeBerg, J. L., McDonald, T., Nathanielsz, P. & Wang, X. L. (2004) J. Steroid Biochem. Mol. Biol. 91, 219-224. [DOI] [PubMed] [Google Scholar]
- 29.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25-33. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 31.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravin, A. A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J. & Tuschl, T. (2003) Dev. Cell 5, 337-350. [DOI] [PubMed] [Google Scholar]
- 33.Shen, B. Q., Lee, D. Y. & Zioncheck, T. F. (1999) J. Biol. Chem. 274, 33057-33063. [DOI] [PubMed] [Google Scholar]
- 34.Bouloumie, A., Schini-Kerth, V. B. & Busse, R. (1999) Cardiovasc. Res. 41, 773-780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.