Abstract
Small ubiquitin-like modifier (SUMO) modification is emerging as an important control in transcription regulation. Here, we show that CREB-binding protein (CBP), a versatile transcriptional coactivator for numerous transcription factors in response to diverse signaling events, can be modified by SUMO-1 at lysine residues 999, 1034, and 1057 both in vitro and in vivo. Mutation of the SUMO acceptor lysine residues either individually or in combination enhanced CBP transcriptional activity, and expression of a SUMO protease SENP2 potentiated the transcriptional activity of CBP wild-type but not its sumoylation mutant, indicating that SUMO modification negatively regulates CBP transcriptional activity. Furthermore, we demonstrated an interaction of SUMO-1-modified CBP with the transcriptional corepressor Daxx and an essential role of Daxx in mediating SUMO-dependent transcriptional regulation of CBP through histone deacetylase 2 recruitment. Together, our findings indicate that SUMO modification and subsequent recruitment of Daxx represent a previously undescribed mechanism in modulating CBP transcriptional potential.
Keywords: protein-protein interaction, posttranslational modification, transcriptional repression, SENP2, histone deacetylase 2
Sumoylation, the covalent attachment of the small ubiquitin-like modifier (SUMO) peptide to lysine residues of targeted substrate, has recently emerged as an important mechanism in transcriptional control (1-3). With an increasing number of sumoylated transcription factors and cofactors being identified, SUMO modification, in most cases, appears to repress the activity of targeted transcriptional activators through altering their subcompartmentalization and/or molecular interaction properties. For example, sumoylation silences the transcriptional activity of Sp3 by translocating it to nuclear domain 10, also named promyelocytic leukemia protein oncogenic domains (4). In addition to the regulation of the nucleo-cytoplasmic shuttling (5), Elk-1 sumoylation further recruits the histone deacetylase (HDAC) 2 to Elk-1-regulated promoters, thereby repressing their transcription (6). Sumoylation of transcriptional coactivator p300 also mediates the recruitment of HDAC6, leading to SUMO-dependent transcriptional repression (7).
The CREB-binding protein (CBP), a paralogue of p300, functions as a transcriptional coactivator in multiple, signal-dependent transcription events (for reviews, see refs. 8-11). The coactivator activity of CBP appears to be exerted through linking different sequence-specific transcription factors to the general transcriptional machinery and/or through its acetyltransferase activity that can acetylate histones and/or transcription factors, thereby activating transcription. Recent studies revealed that the activity of CBP can be dynamically regulated by posttranslational modifications such as phosphorylation (12-15) and methylation (16, 17). Whether CBP can be also regulated by SUMO modification remains unknown.
Daxx, initially identified as a cytoplasmic signaling molecule linking Fas receptor to Jun N-terminal kinase signaling (18), has recently been reported to function as a transcriptional repressor in the nuclear compartments. Daxx was found to interact with and suppress several transcription factor-mediated reporter activities, including ETS1 (19), Pax3 (20, 21), glucocorticoid receptor (22, 23), p53 family proteins (24, 25), mineralocorticoid receptor (26), androgen receptor (27), and Smad4 (28). Because Daxx was shown to associate with multiple proteins that are critical for transcriptional repression, such as HDAC1 (29), HDAC2 (30), and ATRX, a protein binding to heterochromatin protein 1 and functioning as part of a chromatin-remodeling complex (31-33), Daxx-mediated repressive effects appear to involve HDACs and chromatin silencing factors.
In the present study, we show that CBP can be covalently modified by SUMO-1 both in vitro and in vivo. This covalent modification takes place at lysine residues 999, 1034, and 1057 of CBP. We further demonstrate that SUMO modification negatively modulates CBP transcriptional activity by recruiting Daxx, facilitating the HDAC2 to associate with CBP. Our results provide a molecular mechanism of sumoylation in repressing CBP transcriptional activity.
Materials and Methods
Cell Culture, Transfection, Reporter Gene Assays, and Quantification of IRF1 Expression Level. COS-1, HeLa, and 293 cells are from the American Type Culture Collection. Daxx (+/+) and Daxx (-/-) mouse embryonic cells described in ref. 33 were cultured in DMEM supplemented with 10% FBS. Transfections were performed by using LipofectAmine (Invitrogen) according to the manufacturer's instructions. For reporter gene assays, cells were transfected in 24-well plates with 500 ng of DNA, including the indicated reporter constructs, expression vectors, and 100 ng of pRL-TK plasmid as an indicator for normalization of transfection efficiency. The luciferase activities (firefly luciferase for the reporter and Renilla luciferase for the indicator) were measured by using the Dual-Luciferase Assay System (Promega). For IFN-γ-induced IRF1 expression, 293 cells transfected with Flag-tagged CBP wild-type or K999/1034/1057R triple lysine mutant (3KR) were cultured for 24 h and subsequently treated with or without IFN-γ (5 ng/ml) for 6 h. Total cellular RNAs from these cells were extracted by using the TRIzol reagent (Invitrogen) and then reverse transcribed by using ThermoScript RT-PCR system (Invitrogen). The resulting RT reaction product was analyzed for the level of IRF1 and GAPDH RNA by semiquantitative and real-time PCR analysis as described in Supporting Text, which is published as supporting information on the PNAS web site.
Immunoprecipitation and Western Analyses. Transfected 293, COS-1, and HeLa cells were lysed directly in a modified RIPA buffer containing 50 mM Tris (pH 7.8), 0.15 M NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.5% Nonidet P-40, and a 0.1% sodium deoxycholate/protease inhibitor mixture (Complete, Roche Molecular Biochemicals) with or without 10 mM N-ethylmaleimide (NEM). Lysates were further subjected to immunoprecipitation and Western analyses as described in ref. 22. For endogenous Daxx-CBP coimmunoprecipitation, 5 × 107 HeLa cells were harvested for nuclear extract preparation as described by Dignam (34) with some modification (Supporting Text).
Yeast Two-Hybrid and β-Gal Assays. Yeast two-hybrid assays were performed as described in ref. 22. L40 yeast transformants with indicated bait and prey constructs were selected on medium lacking histidine, leucine, and tryptophan for 4 days. His+ colonies were further analyzed for β-gal activity as described in ref. 22. The β-gal activities were determined from three separate liquid yeast cultures according to the instructions of the Galacto-Light Plus kit (Tropix).
In Vitro Sumoylation and Protein Interaction Assays. In vitro sumoylation assays were performed as described in ref. 27 with immunoprecipitated CBP or Flag-tagged CBP5 proteins bound to beads or GST-CBP5 recombinant protein. The resulting samples were washed extensively with PBS for Western analyses. For Daxx binding studies, half of the resulting sample was examined for sumoylation by Western analysis. Another half was further incubated with in vitro synthesized [35S]methionine-labeled Daxx. After 2 h of binding at 37°C, beads of samples were washed with PBS, and bound proteins were fractionated by SDS/PAGE and analyzed by autoradiography.
Chromatin Immunoprecipitation (ChIP) Analysis. The ChIP experiments were performed essentially as described in ref. 35. Briefly, ≈5 × 106 293 cells were transfected with 5 μg of p5XGal-Luc and 5 μg of Gal4-DBD, Gal4-CBP WT, or 3KR expression construct along with or without pSUPER or pSUPER-Daxx, cultured for 48 h, and then subjected to ChIP procedure (Supporting Text) by using 5 μg of anti-Daxx, anti-Gal4, anti-HDAC1, anti-HDAC2, anti-HDAC3 antibody, or no antibody (as input chromatin control).
Plasmids and Antibodies. For details of plasmid constructs and antibodies used in this study can be found in Supporting Text.
Results and Discussion
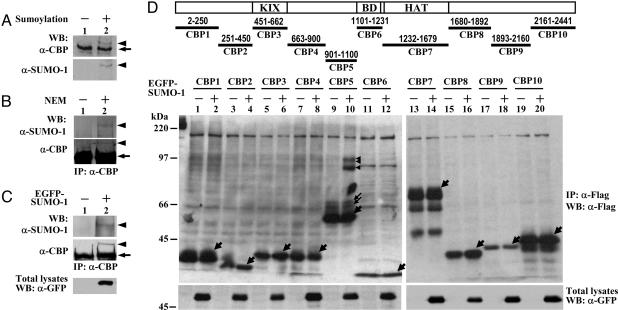
Sumoylation of CBP. To test whether CBP could be covalently modified by SUMO, CBP immunoprecipitated from COS-1 cells was subjected to in vitro sumoylation assays and then analyzed by immunoblotting with anti-CBP or anti-SUMO-1 antibodies. As shown in Fig. 1A, anti-CBP antibodies detected a slower migrating form of CBP in the sumoylation sample (Upper, lane 2, arrowhead), and this slower-migrating CBP was also immunoreactive to anti-SUMO-1 antibody (Lower). These results indicate that CBP can be modified by SUMO-1 in vitro.
Fig. 1.
CBP can be covalently modified by SUMO-1 in vitro and in vivo. (A) Western blot analysis of in vitro sumoylation of CBP proteins immunoprecipitated from COS-1 cells. (B and C) Western blot analyses of immunoprecipitated endogenous CBP from HeLa cells with or without 10 mM NEM (B) or with EGFP-SUMO-1 transfection (C). (D) Schematic presentation of different CBP fragments analyzed by in vivo sumoylation assays. COS-1 cells transfected with various Flag-tagged CBP fragments along with EGFP-SUMO-1 as indicated were precipitated and immunoblotted with anti-Flag antibody. The arrowhead and arrow indicate SUMO-1-modified CBP or CBP fragments and unmodified CBP or CBP fragments, respectively.
To examine whether CBP is sumoylated in cells, endogenous CBP was immunoprecipitated from HeLa cells in the presence or absence of NEM, a cysteine protease inhibitor usually used to preserve the sumoylation of cellular proteins, and then blotted with anti-CBP or anti-SUMO-1 antibody. A slow migrating band immunoreactive to anti-SUMO-1 and anti-CBP antibodies was observed in the sample treated with NEM (Fig. 1B, lane 2). Furthermore, expression of EGFP-SUMO-1 proteins in HeLa cells also yielded a slower migrating band of CBP, which can be recognized by anti-SUMO-1 and anti-CBP antibodies (Fig. 1C, lane 2). Likewise, SUMO modification of ectopically expressed Flag-tagged mouse CBP was also observed in 293 cells transfected with EGFP-SUMO-1 (Fig. 6A, lane 2, which is published as supporting information on the PNAS web site). Together, these results suggest that CBP can be covalently modified by SUMO-1 in cells.
To map the sumoylation site(s) within CBP, 10 fragments encompassing the entire mouse CBP (Fig. 1D Upper) were constructed into a mammalian expression vector with a SV40 nuclear localization signal fused to the amino terminus to ensure the localization of each fragment in the nucleus and two copies of Flag epitope tag fused to the carboxyl terminus for protein detection. These CBP fragments were able to be expressed in COS-1 cells, although to a different extent (Fig. 1D Center). It is noteworthy that CBP5 containing amino acid residues 901-1100 rendered a pattern of one major band (thick arrow) and two minor bands (thin arrow). The nature of these two minor species is currently unknown but is likely due to posttranslational modification(s). When coexpressed with EGFP-SUMO-1, only CBP5 fragment conferred a pattern of three slower-migrating bands, whereas no change was observed in any other CBP fragments, as evidenced by Western blot analysis with anti-Flag (Fig. 1D, arrowheads) or anti-SUMO-1 antibody (Fig. 6B). These results have further been confirmed by in vitro sumoylation assays (Fig. 6C). Our findings suggest that the CBP901-1100 fragment contains the sumoylation site(s).
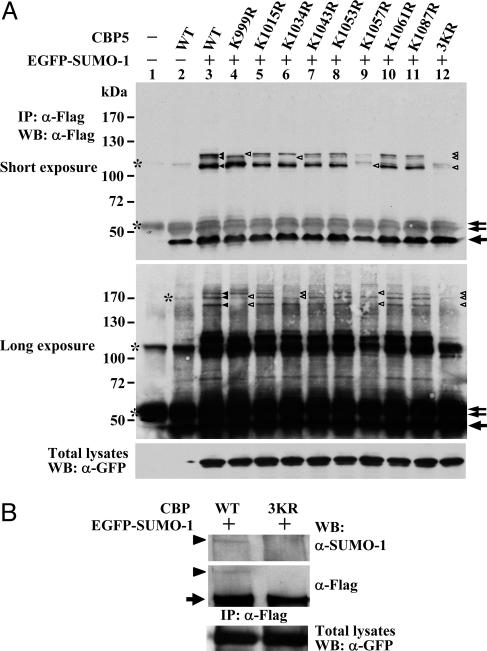
SUMO-1 Modification of CBP Occurs at Lys-999, 1034, and 1057. To further map the sumoylation site(s) within CBP, we mutated eight lysines, which resemble the consensus sumoylation motif “ΨKXE” (where Ψ is a large hydrophobic residue, K is the lysine to which SUMO-1 is conjugated, X is any amino acid, and E is glutamic acid) to arginine residue within CBP901-1100 fragment (Fig. 7, which is published as supporting information on the PNAS web site) and conducted in vivo sumoylation assays. The pattern of shifted bands caused by sumoylation of the K1015R, K1043R, K1053R, K1061R, and K1087R mutants is very similar, if not identical, to that of wild-type CBP901-1100 (Fig. 2A, lanes 3, 5, 7, 8, 10, and 11). By contrast, the K999R, K1034R, and K1057R mutants abrogated the formation of shifted bands corresponding to a single EGFP-SUMO-1 molecule conjugated to a specific lysine residue (Fig. 2 A Top, open arrowheads) and double EGFP-SUMO-1 modification at different lysine residues when immunoblotting was exposed longer (Center, open arrowheads). We have enlarged the regions of sumoylated bands to indicate these slowly migrating bands resulted from specific sumoylated lysine residues within CBP5 (Fig. 8). Notably, the smaller amount of EGFP-SUMO-1 conjugation on K999 or K1034 in K1057R mutant (lane 9) was due to the experimental variation in K1057R construct transfection, which yielded a lower expression level of K1057R mutant protein, because we did observe the extent of sumoylation at K999 or K1034 of K1057R mutant comparable to that of wild-type in a separate experiment (Fig. 9, which is published as supporting information on the PNAS web site). Furthermore, the 3KR completely abolished all SUMO modified forms of CBP5 (Fig. 2 A, lane 12).
Fig. 2.
K999, K1034, and K1057 are the SUMO acceptor sites in CBP. COS-1 cells transfected with expression construct of Flag-tagged CBP5 WT and various KR mutants (A) or Flag-tagged CBP WT and 3KR mutants (B) along with or without EGFP-SUMO-1 as indicated were lysed in RIPA buffer with 10 mM NEM followed by immunoprecipitation and Western analysis with indicated antibodies. A scheme indicating the migrating position of the single and double EGFP-SUMO-1-conjugated CBP5 fragments are shown in Fig. 8, which is published as supporting information on the PNAS web site. The arrowhead depicts the SUMO-1-modified CBP fragments (A) or full-length CBP (B). The arrow and asterisk indicate CBP fragments and nonspecific bands, respectively. The open arrowhead indicates the position of sumoylated CBP species is missing because of mutation.
It should be noted that we did not detect a band representing the triple EGFP-SUMO-1 modification of CBP901-1100 in cells but did observe such triple sumoylated band by in vitro sumoylation assays with recombinant GST-CBP5 protein (Fig. 6C). Apparently, the efficiency of the concomitantly SUMO-1-modified CBP901-1100 in vivo is lower than in vitro. Moreover, the nature causing the aberrant migration of individual sumoylated band is unknown. It likely results from the distinct branching position of SUMO-1 modification at different lysine residues and the charge surrounding the sumoylation site, as suggested by a similar observation in the study of basic Kruppel-like factor sumoylation (36).
To further confirm these three lysines are SUMO acceptor sites in the context of full-length CBP, in vivo sumoylation assay was conducted by using full-length CBP with 3KR mutation. Consistent with the above results, full-length CBP-3KR failed to be sumoylated as compared to CBP-WT (Fig. 2B). These findings indicate that K999, K1034, and K1057 are the SUMO-1 acceptor sites within CBP.
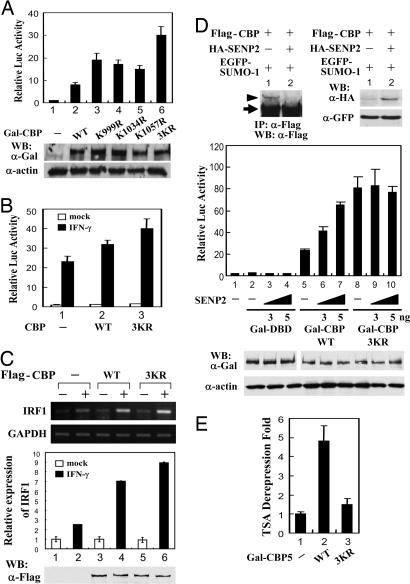
Sumoylation Suppresses the Transcriptional Activity of CBP. We next examined whether sumoylation regulates the transactivation activity of CBP. Because CBP does not bind to DNA on its own, the intrinsic transactivation activity of CBP was assessed by Gal-CBP fusion protein in which CBP was linked to a heterologous DNA-binding domain of Gal4. The transcriptional potential of Gal-CBP WT and various sumoylation mutants was analyzed in COS-1 cells with cotransfection of the p4XGal-TK-Luc reporter construct, containing four copies of Gal4 binding sites in the thymidine kinase promoter fused to the luciferase reporter gene. As expected, Gal-CBP WT conferred the transcriptional activation on this reporter (Fig. 3A, lane 2). Interestingly, individual sumoylation site mutant K999R, K1034R, or K1057R caused a marked increment in the transcriptional potential of CBP. This effect was even more significant when using the 3KR mutant lacking all of the CBP sumoylation (lane 6). The enhancement of CBP-3KR transactivation capacity was apparently not due to the protein expression level because the steady-state level of this mutant was very similar, if not identical, to that of CBP-WT, as assessed by immunoblotting (Fig. 3A Lower).
Fig. 3.
Sumoylation negatively modulates CBP transcriptional activity. (A Upper) COS-1 cells transfected with 200 ng of p4XGal-TK-Luc reporter construct along with 200 ng of Gal-CBP WT or individual or 3KR sumoylation mutant as indicated were incubated 24 h and then harvested for reporter gene assays. The relative luciferase activity is represented as mean ± SD (i.e., firefly luciferase light units/Renilla luciferase light units). (A Lower) The expression levels of Gal-CBP WT and sumoylation mutants are represented in three independent experiments. (B) COS-1 cells transfected with 300 ng of p3xLy6E-Luc along with Flag-CBP WT or 3KR expression vector for 24 h were treated with or without IFN-γ (5 ng/ml) for an additional 6 h then harvested for reporter gene analyses. (C) Semiquantitative PCR and real-time PCR analyses of endogenous IRF1 RNA from Flag-CBP-transfected 293 cells with or without IFN-γ treatment as described in Materials and Methods are shown. The expression levels of transfected Flag-CBP WT and 3KR are indicated by immunoblotting analysis. (D) COS-1 cells transfected with Flag-CBP and EGFP-SUMO-1 along with or without 100 ng of pCMV-HA-SENP2 were subjected to immunoprecipitation and Western analysis with indicated antibodies (Top). COS-1 cells transfected with increasing amount of HA-SENP2 along with p4XGal-TK-Luc reporter construct and Gal-DBD, Gal-CBP WT, or 3KR mutant as indicated were subjected to reporter gene assays. Relative luciferase activity of each sample was determined as described above. The expression levels of transfected Gal-DBD, Gal-CBP WT, and 3KR are indicated by immunoblotting analysis. (E) COS-1 cells transfected with 4XGal-TK-Luc reporter construct along with Gal-DBD (indicated as “-”), Gal-CBP5 WT, or 3KR were treated with 100 nM TSA for 24 h and then subjected to reporter gene assays. The data are presented as fold derepression of reporter activity, which is calculated from the activity of each TSA-treated vs. -untreated sample after normalization to the TSA-mediated derepression of the Gal4 binding domain alone (taken as 1). The results of relative luciferase activities are shown in Fig. 10.
Besides a Gal4-fusion heterologous system, the transcriptional coactivator activity of CBP WT and 3KR for IFN-γ-induced Stat1-mediated reporter activity and endogenous gene expression was also examined. As expected, CBP-3KR gave a greater transactivation activity than CBP-WT in IFN-γ-induced Stat1-mediated reporter activity in COS-1 cells (Fig. 3B). Likewise, the induction level of endogenous IRF1 gene expression by IFN-γ is higher in CBP-3KR-transfected cells than in CBP-WT-transfected cells as evidenced by semiquantitative PCR and real-time PCR analyses (Fig. 3C).
Furthermore, cotransfection of SENP2, a SUMO-specific protease that can reverse CBP sumoylation (Fig. 3D Top), increased the transcriptional activity of CBP WT but not 3KR mutant (Fig. 3D Center). Together, these findings suggest that SUMO modification negatively modulates CBP transcriptional activity.
We next examined whether the transcriptional activity of CBP attenuated by SUMO modification is through a HDAC-dependent manner. COS-1 cells transfected with the expression construct of Gal-CBP901-1100 WT or 3KR mutant along with p4XGal-TK-Luc reporter construct were treated with the deacetylase inhibitor trichostatin A (TSA). As shown in Fig. 3E, the CBP901-1100 WT fragment is more sensitive to TSA treatment than 3KR mutant in derepressing the reporter activity, indicating an involvement of HDAC in controlling the transcriptional activity of sumoylated CBP (see also Fig. 10, which is published as supporting information on the PNAS web site).
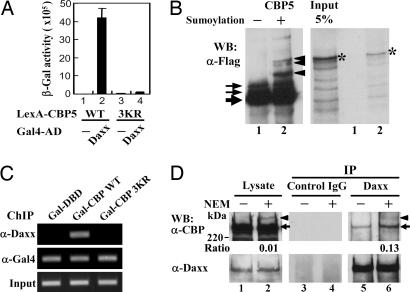
Daxx Interacts with Sumoylated CBP. Previous reports indicate that Daxx associates with HDACs (29, 30) and functions as a transcriptional corepressor. More recently, we have demonstrated that Daxx regulates the transcriptional activities of androgen receptor (27) and Smad4 (28) via a sumoylation-dependent manner. These findings, along with the recent report that CBP associates with Daxx by coimmunoprecipitation assays (37), raise the possibility that Daxx is involved in the interaction with sumoylated CBP, leading to a reduction of CBP transcriptional activity. To test this possibility, we first carried out the interaction studies between Daxx and CBP. Yeast two-hybrid assays indicated that Daxx binds to the CBP901-1100 fragment but not to any other regions of CBP (Table 1, which is published as supporting information on the PNAS web site) and the CBP-Daxx interaction depends on the sumoylation sites of CBP, as evidenced by the quantitative β-gal assays (Fig. 4A), suggesting the SUMO modification of CBP may mediate Daxx interaction.
Fig. 4.
SUMO modification of CBP mediates Daxx interaction. (A) L40 yeast cells cotransformed with the plasmid constructs as indicated were subjected to quantitative β-gal assays. The data represent the mean ± SD of three independent experiments. (B) Flag-tagged CBP901-1100 proteins prepared from pRSV-NLS-CBP5-Flag transfected COS-1 cells were subjected to an in vitro sumoylation assay (Left) and binding studies with Daxx proteins (Right) as described in Materials and Methods. The arrow, arrowhead, and asterisk indicate the CBP5 fragments, sumoylated CBP5 species, and Daxx protein, respectively. (C) ChIP analysis of 293 cells transfected with p5XGal-E1B-Luc and constructs expressing Gal-CBP WT or 3KR as described in Materials and Methods with indicated antibodies. Immunoprecipitated DNA was amplified in PCRs by using primers encompassing the Gal-driven E1B-promoter. (D) Nuclei isolated from HeLa cells were lysed in the absence or presence of 10 mM NEM as described in Materials and Methods. Equal amount of nuclear extracts were immunoprecipitated with a control antibody (Middle) or anti-Daxx antibody (Right) followed by immunoblotting with anti-CBP or anti-Daxx antibody as indicated. The arrowhead and arrow corresponds to the SUMO-modified and unmodified CBP, respectively. The intensity of SUMO-modified and -unmodified CBP bands was quantified by densitometry, and the ratio of modified to unmodified CBP band is indicated below.
To further substantiate the SUMO-dependent interaction between CBP and Daxx, we performed in vitro binding assays. The CBP901-1100 peptide was sumoylated in vitro, then subjected to binding assays with in vitro synthesized Daxx. As expected, sumoylated CBP901-1100 protein, but not the unmodified one, pulled down Daxx (Fig. 4B Right, lane 2), suggesting a critical role of SUMO modification of CBP in Daxx interaction. Consistent with this notion, Gal-CBP WT, but not 3KR protein, is able to associate with VP16-Daxx in mammalian two-hybrid assays (Fig. 11, which is published as supporting information on the PNAS web site). Accordingly, Gal-CBP WT, but not 3KR, can recruit endogenous Daxx to the promoter region of the p5XGal-E1B-Luc reporter, consisting of five tandem Gal4 binding elements in front of the minimal adenoviral E1B promoter fused to the luciferase reporter gene, by ChIP analysis (Fig. 4C). Collectively, these results suggest that sumoylation of CBP is essential for CBP-Daxx association.
We next examined the interaction between endogenous Daxx and CBP in cells. Nuclei of HeLa cells were lysed in the presence or absence of NEM, then subjected to immunoprecipitation experiments with anti-Daxx antibody followed by immunoblotting with anti-CBP antibody. As seen in Fig. 4D, the nuclear extracts prepared from nuclei lysed with NEM yielded a slower-migrating CBP band (arrowhead, lane 2 vs. lane 1), which corresponds to CBP SUMO-1 modification as shown in Fig. 1B. The coimmunoprecipitation experiments revealed that the SUMO-modified and -unmodified bands of CBP were detected in the Daxx immunoprecipitates from nuclei lysed with NEM (Fig. 4D, lane 6), whereas a small amount of unmodified CBP was precipitated by Daxx from nuclei lysed without NEM (Fig. 4D, lane 5), indicating that NEM treatment enhances CBP-Daxx association. Furthermore, the ratio of SUMO-modified to unmodified CBP was significantly increased in the Daxx immunoprecipitates relative to that observed in input lysate (Fig. 4D, lane 6 vs. lane 2), suggesting that Daxx preferentially associate with sumoylated CBP.
Notably, the unmodified CBP precipitated down by Daxx may have resulted from some other factor(s) indirectly mediating CBP-Daxx interaction or from the rapid desumoylation of CBP after coimmunoprecipitation. Because yeast and mammalian two-hybrid analyses (Figs. 4A and 11), ChIP analysis (Fig. 4C), and GST pull-down assays (Fig. 4B) clearly indicate that CBP-Daxx interaction strictly depends on the presence of CBP SUMO acceptor residues and sumoylation event per se, respectively, the observed coprecipitation of Daxx with unmodified CBP is likely due to the latter scenario. This notion is further supported by the significant reduction of CBP-Daxx coprecipitation in the absence of NEM (Fig. 4D, lane 5 vs. lane 6).
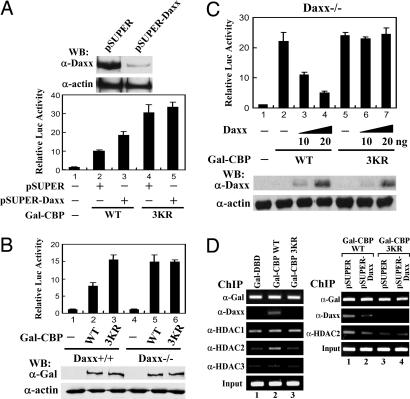
Daxx Mediates Sumoylation-Dependent Inhibition of CBP Transcriptional Activity. Having shown that Daxx binds to sumoylated CBP, we next ask whether endogenous Daxx represses CBP transcriptional activity through a SUMO-dependent mechanism. To this end, we depleted endogenous Daxx by an RNA interference approach. As shown in Fig. 5A, pSUPER-Daxx transfection resulted in a decrease of endogenous Daxx without altering the protein level of actin in 293 cells (Upper). Under such condition, the CBP WT-activated reporter gene activity was enhanced by pSUPER-Daxx transfection (Fig. 5A Lower, lane 3 vs. lane 2). By contrast, pSUPER-Daxx transfection did not alter Gal-CBP 3KR mutant activity (Fig. 5A, lane 5 vs. lane 4). These results suggest that Daxx suppresses the CBP transactivation potential through a SUMO-dependent manner. We further tested this model in the context of Daxx (-/-) cells. The CBP WT and 3KR were analyzed in the Daxx (+/+) and Daxx (-/-) mouse embryonic cells transfected with p4XGal-TK-Luc reporter construct. As shown in Fig. 5B, Gal-CBP 3KR conferred a higher level of reporter activity than the Gal-CBP WT in Daxx (+/+) cells (Fig. 5B, lane 2 vs. lane 3), consistent with the results in COS-1 cells and 293 cells (Figs. 3A and 5A). Notably, in Daxx (-/-) cells, Gal-CBP WT activated the reporter gene activity to the extent similar to that of Gal-CBP 3KR (Fig. 5B, lane 5 vs. lane 6), suggesting the essential role of Daxx in SUMO-dependent repression of CBP. Accordingly, reintroduction of Daxx into Daxx (-/-) cells resulted in the repression of CBP WT activity in a dose-dependent manner (Fig. 5C, lanes 2-4). By contrast, Daxx failed to suppress the transcriptional activity of the CBP 3KR (Fig. 5C, lanes 5-7). Together, these results strongly suggest that the sumoylation of CBP is crucial for Daxx interaction, leading to suppression of CBP transcriptional activity.
Fig. 5.
Daxx suppresses the transcriptional activity of sumoylated CBP via HDAC2 recruitment. (A) Western blot analysis of 293 cells transfected with pSUPER or pSUPER-Daxx construct (Upper). Reporter gene analysis of 293 cells transfected with 100 ng of p4XGal-TK-Luc and 50 ng of Gal-CBP WT or 3KR mutant along with 250 ng of pSUPER-Daxx or pSUPER empty vector is as indicated. The data represent mean ± SD of three independent experiments. (B and C) Daxx (+/+) and Daxx (-/-) cells separately transfected with 200 ng of p4XGal-TK-Luc and 50 ng of Gal-CBP WT or 3KR mutant and increasing amount of HA-Daxx as indicated were harvested for reporter gene assays. The data represent mean ± SD of three independent experiments. (B and C Lower) The expression levels of transfected Gal-CBP (B) or HA-Daxx (C). (D) ChIP analysis of 293 cells transfected p5XGal-E1B-Luc reporter along with Gal fusion constructs and pSUPER-Daxx or pSUPER control vector as indicated. Transfected 293 cells were cultured for 48 h and then subjected to ChIP procedures as described in Materials and Methods by using indicated antibodies. Five percent of the immunoprecipitated and purified DNA were subjected to PCR amplification. Input represents 0.5% of the chromatin used for the immunoprecipitation.
Given that sumoylation inhibits CBP activity via an HDAC-dependent mechanism (Fig. 3E) and Daxx has the capacity to associate with HDAC1 and HDAC2 (29, 30), we next examined whether sumoylation of CBP leads to the association of HDAC1 and HDAC2 through a Daxx-dependent manner, contributing to the transcription inhibition. To this end, 293 cells transfected with Gal-CBP WT or 3KR and p5XGal-E1B-Luc constructs were subjected to ChIP analyses. As shown in Fig. 5D Left, the recruitment of endogenous HDAC2 to the Gal-driven E1B promoter, like Daxx, was markedly elevated in the Gal-CBP WT, but not 3KR, transfected samples as compared to the control Gal-DBD-transfected samples. By contrast, the binding of HDAC1 and HDAC3 to this promoter region is irrelevant to CBP or CBP sumoylation, as evidenced by the extents of HDAC1 and HDAC3 recruited by Gal-CBP WT and 3KR were very similar to that of Gal-DBD. These findings suggest that sumoylation of CBP specifically causes HDAC2 association. To further demonstrate the HDAC2 recruited by sumoylated CBP is Daxx-dependent, we performed the ChIP experiments along with pSUPER-Daxx transfection. Notably, depletion of endogenous Daxx protein by pSUPER-Daxx attenuated the level of HDAC2 recruited by Gal-CBP WT to the Gal-driven E1B promoter to the extent close to that of Gal-CBP 3KR (Fig. 5D Right, lanes 2 and 3), suggesting an essential role of Daxx for HDAC2 to associate with sumoylated CBP. Collectively, these findings suggest a model that sumoylation negatively regulates CBP transcriptional potential by recruiting Daxx and HDAC2.
Although the SUMO-mediated transcriptional repression of both CBP and its paralogue p300 is via an HDAC-dependent manner (Fig. 3E and ref. 7), the present study, along with the report from Hay and coworkers (7), clearly indicate that the sumoylation-dependent repression mechanism of CBP and p300 is distinct. Sumoylation of p300 leads to recruit HDAC6 directly, whereas sumoylation of CBP recruits Daxx then HDAC2. Such distinct specificity in recruiting transcriptional corepressors likely results from the sequences surrounding the SUMO modification sites that provide a conformational preference for specific recognition by these transcriptional corepressors. Indeed, the sumoylation sites of p300 coactivator were mapped to K1020 and K1024 (7). Sequence alignment of both CBP and p300 revealed that p300 K1020 resembles CBP SUMO-conjugation site K1057, whereas other SUMO acceptor lysines in CBP and p300 are unrelated (Fig. 12, which is published as supporting information on the PNAS web site), indicating that both factors have different contexts of SUMO modification. This diverse sumoylation context may determine the distinct recruitment of HDAC6 and Daxx by p300 and CBP, respectively.
Because CBP functions as a coactivator of many transcription factors, our findings that Daxx suppresses CBP transcriptional activity may explain, in part, that cotransfection of Daxx represses various reporter activities (38). However, Emelyanov et al. (37) have recently shown that Daxx acts as a transcriptional coactivator or corepressor of Pax5 protein depending on the cellular contexts. In certain mouse B cells, Daxx apparently enhances Pax5-mediated reporter activity through the recruitment of CBP to Pax5-Daxx complexes (37). In this study, Daxx functions as a bridge factor between Pax5 and CBP, instead of acting as a transcriptional corepressor. Whether specific configuration of transcriptional complexes of transcription factor, Daxx, and CBP determines the role of Daxx in transactivation or transrepression requires further investigation.
Supplementary Material
Acknowledgments
We thank Drs. Hsing-Jien Kung, Ruey-Hwa Chen, and Li-Jung Juan for critical comments on this manuscript. This work was supported by National Science Council Grants NSC93-3112-B-001-033, NSC93-2321-B-001-022, and NSC94-2311-B-001-066 (to H.-M.S.).
Author contributions: H.-Y.K. and H.-M.S. designed research; H.-Y.K., C.-C.C., J.-C.J., and H.-M.H. performed research; D.-Y.L. G.G.M., and R.P.S.K. contributed new reagents/analytic tools; H.-Y.K. and H.-M.S. analyzed data; and H.-Y.K. and H.-M.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CBP, CREB-binding protein; ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase; NEM, N-ethylmaleimide; SUMO, small ubiquitin-like modifier; 3KR, K999/1034/1057R triple lysine mutant; TSA, trichostatin A.
References
- 1.Gill, G. (2003) Curr. Opin. Genet. Dev. 13, 108-113. [DOI] [PubMed] [Google Scholar]
- 2.Girdwood, D. W., Tatham, M. H. & Hay, R. T. (2004) Semin. Cell. Dev. Biol. 15, 201-210. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, E. S. (2004) Annu. Rev. Biochem. 73, 355-382. [DOI] [PubMed] [Google Scholar]
- 4.Ross, S., Best, J. L., Zon, L. I. & Gill, G. (2002) Mol. Cell 10, 831-842. [DOI] [PubMed] [Google Scholar]
- 5.Salinas, S., Briancon-Marjollet, A., Bossis, G., Lopez, M. A., Piechaczyk, M., Jariel-Encontre, I., Debant, A. & Hipskind, R. A. (2004) J. Cell. Biol. 165, 767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang, S. H. & Sharrocks, A. D. (2004) Mol. Cell 13, 611-617. [DOI] [PubMed] [Google Scholar]
- 7.Girdwood, D., Bumpass, D., Vaughan, O. A., Thain, A., Anderson, L. A., Snowden, A. W., Garcia-Wilson, E., Perkins, N. D. & Hay, R. T. (2003) Mol. Cell 11, 1043-1054. [DOI] [PubMed] [Google Scholar]
- 8.Goodman, R. H. & Smolik, S. (2000) Genes Dev. 14, 1553-1577. [PubMed] [Google Scholar]
- 9.Vo, N. & Goodman, R. H. (2001) J. Biol. Chem. 276, 13505-13508. [DOI] [PubMed] [Google Scholar]
- 10.McManus, K. J. & Hendzel, M. J. (2001) Biochem. Cell Biol. 79, 253-266. [PubMed] [Google Scholar]
- 11.Chan, H. M. & La Thangue, N. B. (2001) J. Cell Sci. 114, 2363-2373. [DOI] [PubMed] [Google Scholar]
- 12.Janknecht, R. & Nordheim, A. (1996) Biochem. Biophys. Res. Commun. 228, 831-837. [DOI] [PubMed] [Google Scholar]
- 13.Ait-Si-Ali, S., Ramirez, S., Barre, F. X., Dkhissi, F., Magnaghi-Jaulin, L., Girault, J. A., Robin, P., Knibiehler, M., Pritchard, L. L., Ducommun, B., et al. (1998) Nature 396, 184-186. [DOI] [PubMed] [Google Scholar]
- 14.Zanger, K., Radovick, S. & Wondisford, F. E. (2001) Mol. Cell 7, 551-558. [DOI] [PubMed] [Google Scholar]
- 15.Impey, S., Fong, A. L., Wang, Y., Cardinaux, J. R., Fass, D. M., Obrietan, K., Wayman, G. A., Storm, D. R., Soderling, T. R. & Goodman, R. H. (2002) Neuron 34, 235-244. [DOI] [PubMed] [Google Scholar]
- 16.Xu, W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B. M., Montminy, M. & Evans, R. M. (2001) Science 294, 2507-2511. [DOI] [PubMed] [Google Scholar]
- 17.Chevillard-Briet, M., Trouche, D. & Vandel, L. (2002) EMBO J. 21, 5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, X., Khosravi-Far, R., Chang, H. Y. & Baltimore, D. (1997) Cell 89, 1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, R., Pei, H., Watson, D. K. & Papas, T. S. (2000) Oncogene 19, 745-753. [DOI] [PubMed] [Google Scholar]
- 20.Hollenbach, A. D., Sublett, J. E., McPherson, C. J. & Grosveld, G. (1999) EMBO J. 18, 3702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehembre, F., Muller, S., Pandolfi, P. P. & Dejean, A. (2001) Oncogene 20, 1-9. [DOI] [PubMed] [Google Scholar]
- 22.Lin, D. Y. & Shih, H. M. (2002) J. Biol. Chem. 277, 25446-25456. [DOI] [PubMed] [Google Scholar]
- 23.Lin, D. Y., Lai, M. Z., Ann, D. K. & Shih, H. M. (2003) J. Biol. Chem. 278, 15958-15965. [DOI] [PubMed] [Google Scholar]
- 24.Kim, E. J., Park, J. S. & Um, S. J. (2003) Nucleic Acids Res. 31, 5356-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao, L. Y., Liu, J., Sidhu, G. S., Niu, Y., Liu, Y., Wang, F. & Liao, D. (2004) J. Biol. Chem. 279, 50566-50579. [DOI] [PubMed] [Google Scholar]
- 26.Obradovic, D., Tirard, M., Nemethy, Z., Hirsch, O., Gronemeyer, H. & Almeida, O. F. (2004) Mol. Pharmacol. 65, 761-769. [DOI] [PubMed] [Google Scholar]
- 27.Lin, D. Y., Fang, H. I., Ma, A. H., Huang, Y. S., Pu, Y. S., Jenster, G., Kung, H. J. & Shih, H. M. (2004) Mol. Cell. Biol. 24, 10529-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang, C. C., Lin, D. Y., Fang, H. I., Chen, R. H. & Shih, H. M. (2005) J. Biol. Chem. 280, 10164-10173. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., Leo, C., Zhu, J., Wu, X., O'Neil, J., Park, E. J. & Chen, J. D. (2000) Mol. Cell. Biol. 20, 1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollenbach, A. D., McPherson, C. J., Mientjes, E. J., Iyengar, R. & Grosveld, G. (2002) J. Cell Sci. 115, 3319-3330. [DOI] [PubMed] [Google Scholar]
- 31.Xue, Y., Gibbons, R., Yan, Z., Yang, D., McDowell, T. L., Sechi, S., Qin, J., Zhou, S., Higgs, D. & Wang, W. (2003) Proc. Natl. Acad. Sci. USA 100, 10635-10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, J., Wu, S., Liu, H., Stratt, R., Barak, O. G., Shiekhattar, R., Picketts, D. J. & Yang, X. (2004) J. Biol. Chem. 279, 20369-20377. [DOI] [PubMed] [Google Scholar]
- 33.Ishov, A. M., Vladimirova, O. V. & Maul, G. G. (2004) J. Cell Sci. 117, 3807-3820. [DOI] [PubMed] [Google Scholar]
- 34.Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinmann, A. S. & Farnham, P. J. (2002) Methods 26, 37-47. [DOI] [PubMed] [Google Scholar]
- 36.Perdomo, J., Verger, A., Turner, J. & Crossley, M. (2005) Mol. Cell. Biol. 25, 1549-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emelyanov, A. V., Kovac, C. R., Sepulveda, M. A. & Birshtein, B. K. (2002) J. Biol. Chem. 277, 11156-11164. [DOI] [PubMed] [Google Scholar]
- 38.Ecsedy, J. A., Michaelson, J. S. & Leder, P. (2003) Mol. Cell. Biol. 23, 950-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.