Abstract
The localization of certain mRNAs to dendrites and their local translation in synaptic regions are proposed to be involved in certain aspects of synaptic plasticity. A cis-acting element within the 3′ untranslated region (3′ UTR) of the targeted mRNAs, which is bound by a trans-acting RNA-binding protein, controls the dendritic mRNA localization. Here, we identified hematopoietic zinc finger (Hzf) as a trans-acting factor that regulates the dendritic mRNA localization of the type 1 inositol 1,4,5-trisphosphate receptor (IP3RI), a dendritically localized mRNA in cerebellar Purkinje cells, via binding to the 3′ UTR. In Hzf-deficient mice, the dendritic localization of IP3RI mRNA and brain-derived neurotrophic factor-induced IP3RI protein synthesis in the cerebellum were impaired. These findings suggest that Hzf is an RNA-binding protein that controls the dendritic mRNA localization and activity-dependent translation of IP3RI, and may be involved in some aspects of synaptic plasticity.
Keywords: 3′ UTR, IP3RI, RNA-binding protein, BDNF
Interest in mRNAs that localize to the dendrites of neuronal cells is increasing. Dendritic mRNAs are thought to be locally translated in response to synaptic activities (1-3), and local protein synthesis is thought to modify the molecular composition and structure of specific populations of synapses, thereby establishing certain aspects of synaptic plasticity. Indeed, protein synthesis in the postsynapse has been shown to be required for long-term potentiation (LTP) and long-term depression (LTD) in the hippocampus (4, 5).
Most previous studies have focused on the role of the 3′ UTR in dendritic mRNA localization in the hippocampus. The 3′ UTRs of α-CaMKII and MAP2 mRNA have been shown to contain cis-elements that are required for the dendritic localization of these transcripts (6-10). Furthermore, the localization of β-actin mRNA in developing neurites and growth cones has been reported to require an interaction between a sequence known as a “zipcode” in the 3′ UTR and zipcode-binding protein 1 (ZBP1), a neuronal RNA-binding protein (11-13). Taken together, these findings indicate that RNA-binding proteins that specifically recognize sequences in 3′ UTRs are likely to play an important role in dendritic mRNA targeting and/or local translation in neuronal cells. Therefore, we investigated the role of other neuronal RNA-binding proteins in dendritic mRNA localization/translation.
IP3RI mRNA is a dendritically localized mRNA (14). IP3RI is an intracellular Ca2+-release channel that is predominantly expressed in cerebellar Purkinje cells (15) and is required for the induction of cerebellar LTD (16), which is thought to be the cellular basis for motor learning and motor coordination in the cerebellum (17). In this study, we demonstrated that the 3′ UTR of IP3RI mRNA is required as a cis-element for its dendritic localization and then identified Hzf as a trans-acting factor. Hzf protein, which was highly expressed in cerebellar Purkinje cells, associated with IP3RI mRNA by binding to the 3′ UTR. Moreover, dendritic IP3RI mRNA in Purkinje cells and BDNF-induced protein synthesis were both reduced in Hzf-deficient mice. These findings suggest that the Hzf protein is required as a trans-acting factor for the dendritic targeting of IP3RI mRNA and activity-dependent protein synthesis in cerebellar Purkinje cells and that such posttranscriptional regulation of neuronal processes may contribute to synaptic plasticity and motor learning in the cerebellum.
Materials and Methods
Studies of Primary Cell Cultures. To make the EGFP-fusion constructs, various forms of IP3RI mRNA were inserted into a pEGFP-C1 vector (Clontech). For the hippocampal cell cultures, cells dissected from the hippocampi of postnatal day 0 (P0) mouse pups were cultured in Neurobasal medium (Sigma) containing 2% B27 (Invitrogen) for 6 days and then transfected with lipofectamine 2000 (Invitrogen). After 2 days, the cells were analyzed by in situ hybridization. For quantitative analysis, we counted a neuron in which dendritic signals were clearly detected at a distance of more than the diameter of one cell body from the proximal end as a “positively localized cell” based on a definition given by Blichenberg et al. (7, 8).
RNA-Binding Assays. For the UV cross-linking assay, a 32P-labeled probe (1 × 104 cpm) was incubated at room temperature with 0.5 μg of Hzf recombinant protein in 10 μl of a solution containing 50 mM Tris·HCl (pH 7.5), 60 mM KCl, 20% glycerol, and 5× Denhardt's solution, for 30 min. In addition to full-length IP3RI 3′ UTR, the various fragments of the 3′ UTR described in Fig. 3A, constructed by PCR with a 5′primer containing the T3 promoter sequence, were used in this assay. Irradiation was carried out on ice for 5 min, and unbound RNAs were then degraded by incubation with 1 mg/ml RNase A. The UV cross-linked RNA-protein complexes were separated by SDS/12% PAGE. To quantify the RNA-binding abilities of Hzf, a gel retardation assay was performed. Briefly, the 32P-labeled probe (1 × 104 cpm) was incubated at room temperature for 30 min with various amounts of the His-Hzf protein. After the incubation, the mixtures were immediately separated by 6% PAGE.
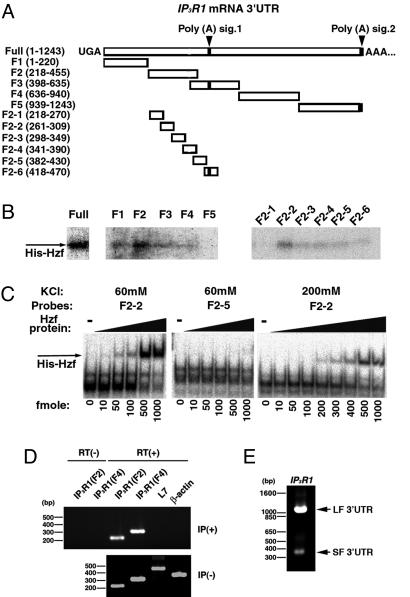
Fig. 3.
Analysis of RNA-binding activity of Hzf with IP3RI mRNA in the cerebellum. (A) Various bound fragments of IP3RI 3′ UTR are schematically represented. The black boxes show the polyadenylation signals (AAUAAA). (B) Identification of the cis-element in the 3′ UTR of IP3RI mRNA required for binding to recombinant Hzf (His-Hzf) protein by UV cross-linking analysis. (C) Quantification of the RNA-binding activity of Hzf to representative elements of the 3′ UTR of IP3RI (F2-2 and F2-5) using an RNA gel retardation assay (60 mM KCl, 200 mM KCl). (D) RT-PCR followed by immunoprecipitation with anti-Hzf antibody in adult mouse cerebellum (Upper). RT-PCR of cerebellar RNAs without immunoprecipitation (Lower). (E) 3′ RACE-PCR of IP3RI after immunoprecipitation from the cerebellum (second round PCR with nested primers). Two main bands (the upper band is LF 3′ UTR, the bottom is SF 3′ UTR) were detected.
Northern Blot Analysis. Total RNAs extracted with TRIzol Re-agent (GIBCO/BRL) were separated by electrophoresis in a 1.0% formamide agarose gel and blotted onto hybridization transfer membranes (GeneScreen Plus, NEN). A random-primed probe was generated by using a Random Primed DNA labeling kit (Roche Molecular Biochemicals). Hybridization was performed overnight at 42°C in a solution containing 5× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA) (SSPE), 50% formamide, 5× Denhardt's solution, 0.5% SDS, 0.1 mg/ml salmon sperm DNA, and 1-2 × 106 cpm/ml 32P-labeled probe.
In Situ Hybridization and Immunohistochemistry. For in situ hybridization, serial sections of frozen brains (12 μm) were fixed for 15 min with 4% PFA fixative, then rinsed in PBS. A digoxigenin (DIG)-labeled RNA probe was prepared by in vitro transcription using T3 and T7 RNA polymerases (Promega) and DIG-labeled UTP (Roche Molecular Biochemicals). The sections were hybridized overnight at 55°C with 1-2 μg/ml of an antisense or sense RNA probe in 50% formamide, 5× SSPE, 250 μg/ml yeast tRNA, and 0.25% SDS. The specimens were washed in 50% formamide, 5× SSC (30 min at 55°C), 2× SSC (45 min at 55°C), 20 μg/ml RNase A, 2× SSC (30 min at 37°C), and 0.1× SSC (twice for 20 min at room temperature). After blocking in 0.5% BSA in TBST (60 min), the slides were incubated with an alkaline phosphatase-conjugated anti-DIG antibody (Roche Molecular Biochemicals) in the above buffer (overnight at 4°C), washed in TBST (three times for 30 min), and NTMT (100 mM NaCl/50 mM MgCl2/100 mM Tris·HCl, pH 9.5/0.1% Tween-20) for 5 min. Signals were detected in NTMT containing 35 μg/ml nitrobluetetrazolium salt, and 17.5 μg/ml 5-bromo-4-chloro-3-indolylphosphate (BCIP), according to the DIG RNA Detection kit (Roche Molecular Biochemicals).
For immunohistochemistry, the dewaxed and dehydrated paraffin sections were washed in PBS and permeabilized with 0.3% Triton-X/PBS. Immunostaining was carried out by using the following primary antibodies: anti-Hzf (1:100) and anti-IP3RI monoclonal antibody 18A10 (1:200). The sections were visualized by Cy3- or FITC-labeled secondary antibodies, or by the TSA Fluorescien System (PerkinElmer·), followed by reaction with biotin-conjugated secondary antibodies (1:500, Jackson ImmunoResearch).
Immunoprecipitation and RT-PCR. Adult mouse cerebella were homogenized in NET-Triton (50 mM Tris·HCl, pH 7.5/20 mM MgCl2/1% Triton-X). Nuclei were removed by centrifugation, and the supernatant was precleared with protein A-Sepharose (Amersham Pharmacia) for 1 h at 4°C, followed by incubation with anti-Hzf antibody (1:200) and protein A-Sepharose. After five washes with cold NET-Triton, samples were resuspended in DNase buffer (50 mM Tris·HCl, pH 7.5/6 mM MgCl2) containing 20 units of DNase I and 20 units of RNasin (Promega). For RT-PCR, cDNAs were synthesized by the Superscript Preamplification System (Gibco/BRL), and amplified by PCR. 3′ RACE was performed by using a Marathon cDNA Amplification kit (Clontech).
Metabolic Pulse Labeling. From 3- to 4-week-old mice, sagittal cerebellar slices were prepared with a microslicer (400 μm in thickness; VT1000S, Leica) and placed on nylon membranes (0.4 μm pore size, Millicell sterilized culture plate insert, Millipore) immersed in 50% MEM, 20% horse serum, and 25% Hank's solution supplemented with 2% B27. The slices were perfused in serum and supplement-free MEM for 30 min before pulse labeling. Pulse labeling was conducted in 50% methionine-free MEM, 50% Hank's solution supplemented with 0.1 mCi/ml [35S]methionine and cystein (Promix [35S] in vitro cell labeling mix, Amersham Pharmacia; 1 Ci = 37 GBq), and 100 nM recombinant human BDNF (R & D Systems) for 120 min, the homogenates were immunoprecipitated with anti-IP3RI polyclonal antibody (1:200, Chemicon) and anti-α-tubulin monoclonal antibody, and loaded onto SDS/6% or 12% PAGE. The levels of [35S]methionine incorporation were measured with a bioimage analyzer (image gauge version 3.45, Fuji).
Results
The 3′ UTR of IP3RI mRNA Serves as a Cis-Element Localizing Transcript. First, to determine whether IP3RI mRNA contains a dendritic targeting element in its 3′ UTR (similar to other transcripts identified in neurons, like α-CaMKII, MAP2, and β-actin mRNAs), we transfected chimeric gene expression vectors in which the EGFP coding sequence was fused to the 3′ UTRs of IP3RI mRNA or the coding regions at the C terminus (Fig. 1A) into cultured hippocampal neurons, and the distribution of EGFP mRNA was analyzed by in situ hybridization using an antisense probe targeting the EGFP coding sequence. As illustrated in Fig. 1A, two variants of the IP3RI 3′ UTR, LF and SF, with alternative polyadenylation signals were identified (15). SF 3′ UTR is a short splicing form lacking ≈700 bp at the 3′ end. Chimeric EGFP transcripts carrying the entire 3′ UTR (EGFP-LF 3′ UTR) were significantly localized to the dendritic processes (34.3 ± 7.3%, P < 0.01; Fig. 1B and Fig. 6, which is published as supporting information on the PNAS web site), compared with EGFP mRNA (9.3 ± 3.3%, Figs. 1B and 6 A and B). The EGFP-SF 3′ UTR mRNA was also distributed to the dendrites (22.2 ± 3.3%, P < 0.05; Figs. 1B and 6B) but with a lower efficiency compared with the EGFP-LF 3′ UTR mRNA. The low percentage of cells showing dendritic labeling may be partly caused by the presence of a heterogeneous cell population in cultures. In contrast, the EGFP-chimerical constructs encoding fragment 1 (F1) and F2 coding mRNAs (F1, 5452-8248 bp; F2, 6912-8248 bp) were not localized to the dendrites (9.6 ± 6.1%, 12.0 ± 0.9%, respectively). These results suggested that 3′ UTR of IP3RI mRNA is required for its dendritic targeting and that the entire 3′ UTR is required to maximize the efficiency IP3RI mRNA trafficking to the dendrites of hippocampal neurons.
Fig. 1.
Dendritic targeting capacity of the IP3RI 3′ UTR in cultured hippocampal neurons. (A) Schematic representation of the vectors expressing fusion transcripts of EGFP. The white boxes in the 3′ UTR indicate the positions of the polyadenylation signals (AAUAAA). (B) Ratio of cells showing dendritic localization to total neurons expressing the reporter transcripts (n > 3, total 227-624 neurons per construct). Statistical significance was assessed by using a Student's t test. Values are the mean ± SEM. *, P < 0.05; **, P < 0.01.
Identification of a Trans-Acting Factor Binding to the 3′ UTR of IP3RI mRNA. Based on the finding demonstrated in Fig. 1, we screened a mouse cerebellum cDNA expression library for trans-acting factor(s) using the entire 3′ UTR sequence as a probe (see Supporting Text, which is published as supporting information on the PNAS web site). One putative positive clone was isolated from the 6 × 105 clones that were screened. An NCBI database search using blast revealed this gene to be hematopoietic zinc finger (Hzf), which encodes a protein containing three C2H2-type zinc finger motifs (see Fig. 7A, which is published as supporting information on the PNAS web site). Hzf was recently shown to be expressed predominantly in megakaryocytes and CFU-GEMMs during hematopoietic development (18). However, its expression and function within the nervous system have not been characterized.
Similarity of Hzf mRNA and IP3RI mRNA Distributions in Adult Mouse Brain. To determine the expression pattern of the Hzf gene, we conducted Northern blot analyses and in situ hybridization in the adult mouse. A major 2.5-kb transcript was distinctly detected in samples from the brain and testis (Fig. 2A). In situ hybridization analysis showed that Hzf mRNA was expressed in the olfactory bulb, cerebral cortex, hippocampus, and cerebellum (Fig. 2B). Within the adult cerebellum, the most prominent expression site was observed in Purkinje cells (Fig. 2 C and D), which are also the site of the strongest IP3RI expression (14, 15). In addition to Purkinje cells, Hzf mRNA was observed in the satellite or basket cells of the molecular layer of the cerebellum. These results indicated that IP3RI and Hzf transcripts have similar expression profiles in the adult mouse brain.
Fig. 2.
Expression profiles of Hzf mRNA and subcellular distribution of Hzf protein in mice. (A) Northern blot analysis of Hzf mRNA in adult mice. (B-D) In situ hybridization analysis of Hzf mRNA expression in mouse brain. (B) Adult sagittal section. Ob, olfactory bulb; Ctx, cerebral cortex; CPu, caudate putamen; Hip, hippocampus; Th, thalamus; Cb, cerebellum. (C) High-magnification view of the main Hzf-expressing regions in the cerebellum. Mo, molecular layer; Pu, Purkinje cell layer; GrC granule cell layer. (D) High-magnification view of Purkinje cells in the cerebellum. Arrowheads point to the cell bodies of the Purkinje cells. (E) Double-immunohistochemistry of Hzf (FITC, green) and IP3RI (Cy3, red) in the adult cerebellum. Arrowheads show the double-stained dendrites of Purkinje cells. (Scale bars: 1 mm in B, 100 μmin C,50 μm in D, and 20 μm in E.)
Localization of Hzf Protein to the Dendrites of Cerebellar Purkinje Cells. To investigate the subcellular localization of the Hzf protein, we generated an anti-Hzf polyclonal antibody against the N-terminal side (Fig. 7 A and B, see Supporting Text). A Western blot assay of total brain, cerebellum, and testis extracts revealed three main bands at ≈43, 46, and 48 kDa (Fig. 8A, which is published as supporting information on the PNAS web site). The staining pattern in the adult mouse brain sections using this antibody corresponded closely to the distribution of Hzf mRNA (Fig. 8B). Double-immunostaining in the cerebellum with the anti-Hzf antibody and an anti-IP3RI monoclonal antibody (18A10) that selectively stains Purkinje cells within the cerebellum (19) revealed that Hzf protein was detected in both the dendrites and the cell bodies of Purkinje cells (Fig. 2E). Dendritic localization of the Hzf protein was also observed in the hippocampal neurons of the CA1 and CA2 regions (Fig. 8C).
Binding of Hzf Protein to the 3′ UTR of IP3RI mRNA in Vitro. To verify that the Hzf protein binds directly to the 3′ UTR of IP3RI mRNA, various recombinant Hzf proteins illustrated in Fig. 8A were expressed in bacterial cells (Fig. 7C, see Supporting Text); their RNA-binding activities were then examined by using a UV cross-linking assay with the radiolabeled full-length 3′ UTR of IP3RI mRNA. His-Hzf and GST-ΔZn1 bound to the 3′ UTR of IP3RI mRNA, whereas GST-ΔZn2,3 and GST-ΔZn3 did not (Fig. 7D). These results indicated that the third zinc-finger, in particular, is essential for the RNA-binding activity. To determine which regions in the IP3RI 3′ UTR were involved in the Hzf-binding, we performed UV cross-linking assays using the various IP3RI 3′ UTR RNA fragments shown in Fig. 3A as probes. Hzf most strongly interacted with the sequence of F2 (218-455 bp), which is just upstream of the first polyadenylation signal sequence (Fig. 3B Left). UV cross-linking and RNA gel retardation assays using various F2 RNA fragments revealed that Hzf bound to F2-2 (261-309 bp) with the highest affinity (Fig. 3 B Right and C). The Kd was ≈30 nM (in the presence of 200 mM KCl).
In Vivo RNA-binding Ability of Hzf Protein. To determine whether the Hzf protein is associated with IP3RI mRNA in vivo, extracts of adult mouse cerebellum were immunoprecipitated with the anti-Hzf polyclonal antibody (Fig. 8D). RNAs contained in the immunoprecipitates were reverse-transcribed and amplified by PCR using specific primers for the IP3RI 3′ UTR (F2 and F4), β-actin, and pcp-2 (L7). Pcp-2 is a representative mRNA known to be localized in the dendrites of Purkinje cells (20). Whereas neither β-actin nor pcp-2 was amplified from the immunoprecipitates, IP3RI transcripts were highly detected in an reverse transcription-dependent manner (Fig. 3D). We also performed 3′RACE with poly(A) RNAs from the immunoprecipitates to detect the different 3′ UTR lengths of the two variants, LF and SF. Both variants were amplified in this experiment (Fig. 3E). Furthermore, double immunocytochemistry followed by fluorescence in situ hybridization of the EGFP-IP3RI 3′ UTR mRNA showed the colocalization of this mRNA with endogenous Hzf protein in MAP2-positive hippocampal dendrites (Fig. 7E). Consistent with the results of the in vitro RNA-binding experiments demonstrated in Fig. 3 A-C, these results indicated that the Hzf protein selectively associates with both forms of IP3RI mRNA in vivo, although we cannot exclude the possibility that other mRNAs may also be bound by Hzf.
Disrupted Dendritic Localization of IP3RI mRNA in Cerebellar Purkinje Cells of Hzf-Deficient Mice. Next, we used loss-of-function studies to investigate the in vivo function of Hzf. Hzf-mutant mice exhibit a pronounced tendency to rebleed and have low levels of α-granule substances in both their megakaryocytes and their platelets (21). To confirm the role of Hzf as a trans-acting factor for the dendritic mRNA localization of IP3RI, we investigated the subcellular localization and the levels of IP3RI transcripts in these mutant mice. We first confirmed that endogenous Hzf protein was completely absent in Hzf-mutant mice (Fig. 9 A and B, which is published as supporting information on the PNAS web site). The cerebellum of Hzf-mutant mice was normal in size, foliation, and trilaminar structure (data not shown).
However, in situ hybridization showed that the level of IP3RI mRNA in the molecular layer was remarkably reduced in the cerebellum, whereas IP3RI mRNA appeared to be normally expressed in other regions, including the cell bodies of Purkinje cells (Fig. 4 A and B), indicating that the dendritic IP3RI mRNA of Purkinje cells was specifically decreased in the Hzf-mutant mice. A quantitative analysis revealed that the level of IP3RI mRNA in the molecular layer of the mutant mice was significantly reduced (22.6 ± 6.8%, P < 0.01; Fig. 4C Center). In contrast, no significant difference in the amount of IP3RI mRNA in the Purkinje cell layer was seen between the wild-type and mutant mice (Fig. 4C Left). In the mutant mice, the mRNA level in the molecular layers normalized to that in the Purkinje cell layer was also <30% of that in wild-type mice (28.9 ± 9.5%, P < 0.01; Fig. 4C Right). In addition, a Northern blot analysis using total RNAs extracted from the cerebellum showed that the lack of Hzf protein did not significantly affect the overall levels of IP3RI transcripts (Fig. 9 B and C).
Fig. 4.
Impaired dendritic localization of IP3RI mRNA in Hzf-mutant mice. (A) In situ hybridization analysis of IP3RI transcripts in the cerebella of Hzf-mutant mice. Dendritic localization within the Purkinje cells was reduced in the Hzf-mutant mice. ML, molecular layer; PCL, Purkinje cell layer. (B) Scan analysis of representative in situ hybridization signal for IP3RI mRNAs in wild-type and mutant Purkinje cells. PCL, Purkinje cell layer; ML, molecular layer. (C) Quantitation of IP3RI mRNAs in the Purkinje cell layer (PCL, Top) and molecular layer (ML, Middle) of wild-type and Hzf-mutant mice (n = 3 mice per genotype, each >20 cells per slice). Furthermore, the mRNA level in the molecular layer was normalized to that in the Purkinje cell layer positioned just below the molecular layer (Bottom). Values are the mean ± SEM. **, P < 0.01. (D) Dendritic localization of transfected 3′ UTR of IP3RI mRNA was reduced in cultured hippocampal neurons from Hzf-mutant mice (n = 4, total 247-675 neurons per construct); the reduced dendritic mRNA localizations were rescued by cotransfection with a CMV-Hzf construct (n = 3, total 103-466 neurons per construct). Values are the mean ± SEM. *, P < 0.05. Statistical significance was assessed by using a Student t test in C and D. (Scale bars: 25 μm in A and B.)
To confirm that Hzf functions as a trans-acting factor for the dendritic mRNA localization of IP3RI, we analyzed the distribution of exogenously expressed EGFP mRNA fused to the 3′ UTR of IP3RI in cultured hippocampal neurons from Hzf-mutant mice by using the protocol described in Fig. 1. The localization efficiency of the EGFP-IP3RI 3′ UTR mRNA in the mutant mice was significantly lower than that in the wild-type mice (P < 0.05; Fig. 4D, open bar). We then examined whether these low efficiencies observed in the Hzf-mutant neurons would recover under rescue conditions produced by the cotransfection of Hzf expression vectors driven by the CMV promoter. When cultured hippocampal neurons from Hzf-mutant mice were cotransfected with the Hzf expression vector, EGFP-IP3RI 3′ UTR mRNA became strongly localized to the dendrites, compared with its localization in untransfected Hzf-mutant neurons (P < 0.02; Fig. 4D, shaded bar). Taken together, these observations suggested that Hzf plays an important role in the dendritic targeting of IP3RI mRNA via binding to the 3′ UTR.
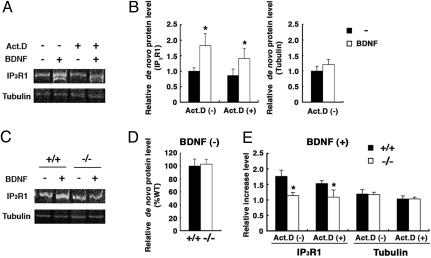
Posttranscriptional de Novo IP3RI Protein Synthesis Promoted by BDNF. We finally examined whether IP3RI protein expression was disturbed in the Hzf-mutant mice in a manner similar to its altered mRNA expression profile. Large RNA granules containing dendritic mRNAs docked near postsynaptic sites have been suggested to serve as local storage sites, where the mRNA is in a state of translational arrest (22). Therefore, the majority of dendritic IP3RI mRNAs might reside in dendritic processes in a state that enables them to be translated in an activity-dependent manner. To address this possibility, we investigated activity-dependent IP3RI protein synthesis using metabolic pulse labeling in cerebellar slices stimulated with BDNF, which induces the local translation of dendritic mRNA (3). IP3RI and α-tubulin proteins were immunoprecipitated from [35S]methionine-labeled slices. As expected, the incorporation of [35S]methionine into IP3RI increased significantly with BDNF treatment in the absence and presence of actinomycin D, an inhibitor of RNA polymerase II (1.83 ± 0.38, 1.64 ± 0.37, P < 0.03, respectively; Fig. 5 A and B Left), whereas that into α-tubulin was not significantly affected (Fig. 5 A and B Right). These results suggested that stored IP3RI mRNA was actively translated in the cerebellum. Surprisingly, the BDNF-dependent protein synthesis of IP3RI was significantly reduced in the Hzf-mutant slices, compared with that in the wild-type slices (+/+, 1.53 ± 0.09; -/-, 1.09 ± 0.23, P < 0.03, in the presence of actinomycin D; Fig. 5 C and E). In contrast, the level of de novo α-tubulin protein was not significantly affected (Fig. 5 C and E). In addition, the basal level of de novo IP3RI protein synthesis without BDNF treatment was similar in the wild-type and mutant slices (Fig. 5D). These results suggested that Hzf may be involved in the activity-dependent protein synthesis of IP3RI, in addition to its role in the dendritic targeting of IP3RI mRNA in Purkinje cells in vivo.
Fig. 5.
Disruption of BDNF-induced IP3RI protein synthesis in Hzf-mutant mice. (A) De novo protein synthesis of IP3RI and α-tubulin in the cerebellum with BDNF (100 nM) treatment in the absence or presence of actinomycin D (40 μM) (n = 6, eight in each case). (B) The relative amounts of IP3RI (Left) and α-tubulin (Right) protein synthesis in the cerebellar slices increased with BDNF treatment in the absence or presence of actinomycin D. (C) BDNF-induced IP3RI protein synthesis in the presence of actinomycin D was impaired in Hzf-mutant slices (n = 11, nine per genotype). (D) The amounts of constitutive IP3RI protein synthesized in the cerebellum are comparable in wild-type and mutant slices (n = 4 per genotype). (E) Comparison of de novo protein synthesis, induced by BDNF treatment, between wild-type and Hzf-mutant slices (n = 10, six per genotype in the absence of actinomycin D, n = 11, nine per genotype in the presence of actinomycin D). Statistical significance was assessed by using a paired t test in B and a Student's t test in D and E. Values are the mean ± SEM. *, P < 0.05.
Discussion
Molecular Mechanism of Dendritic Targeting of IP3RI mRNA Regulated by Hzf. The results of the in situ hybridization studies using cultured hippocampal neurons expressing chimerical transcripts suggested that the 3′ UTR of IP3RI mRNA plays an important role in the dendritic targeting of IP3RI mRNA. We also identified the RNA-binding protein Hzf as one of the trans-acting factors. Hzf is strongly expressed in the brain, especially in the olfactory bulb, cortex, hippocampus, and cerebellum (similar to the expression profile of IP3RI), and is coimmunoprecipitated with IP3RI mRNA from cerebellar lysate. Furthermore, the dendritic localization of IP3RI mRNA is significantly reduced in Hzf-mutant mice, but the overexpression of exogenous Hzf restores this phenotype. These biochemical and loss-of-function studies demonstrated that Hzf regulates the dendritic targeting of IP3RI mRNA by binding directly to the 3′ UTR of IP3RI mRNA in neurons. In particular, the 49-nt RNA fragment located upstream of the first polyadenylation signal (261-309 in 3′ UTR) exhibited the highest affinity for Hzf protein in in vitro RNA-binding assays. Thus, this region may be at least one of the Hzf-binding sites.
An RNA-binding protein, the mammalian homologue of Staufen (mStau), has been suggested to be one of the proteins involved in mRNA transport into dendrites, because mStau localizes to the somatodendritic domain and colocalizes with a ribonucleoprotein complex in cultured hippocampal neurons (23-26). Similarly, we found that Hzf is localized to the somatodendritic region of cerebellar Purkinje cells and specific hippocampal neurons. Recently, a kinesin superfamily protein, KIF5, has been reported to transport RNA selectively to dendrites via large RNA granules; Hzf protein (ZFP385) was isolated from these KIF5-transporting RNA granules (27). Furthermore, the 3′ UTR of IP3RI mRNA is cotransported with SYNCRIP, one of the components of RNA granules, into the dendrites of cultured hippocampal neurons (28). These reports suggest that both Hzf and IP3RI mRNA are components of the neuronal RNA granules. Therefore, we speculate that Hzf recruits IP3RI mRNA into large RNA granules via binding to the 3′ UTR; these granules then remain in a translationally quiescent state while they are transported by motor proteins, such as KIFs, along the cytoskeletal structure and into the dendritic processes.
Activity-Dependent Translational Control of IP3RI by BDNF. Various dendritic mRNAs are known to be locally translated in an activity-dependent manner (29). In the present study, we biochemically demonstrated that BDNF promotes de novo IP3RI protein synthesis at the posttranscriptional level in the cerebellum and that this process was impaired in Hzf-mutant mice. BDNF leads to activity-dependent RNA translation, perhaps by activating translation factors through extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK-MAPK) (30) or a mammalian target of the rapamycin-phosphatidylinosotol 3-kinase-dependent (mTOR-PI3K) pathway (31, 32). How does the loss of Hzf lead to a reduction of the BDNF-induced translation of IP3RI? Neuronal RNA granules are thought to function as mRNA storage compartments equipped with translation machinery, in which the mRNA is under translational arrest but can be translated by reorganization of the RNA granules; stored mRNA and some translational components are then released in response to neuronal activity (22). Because Hzf might play an important role in mRNA storage by incorporating mRNA into the neuronal RNA granules, the disruption of BDNF-induced IP3RI protein synthesis in Hzf-mutant mice is likely in part due to the absolute reduction of the stored IP3RI mRNA pool within the RNA granules.
Putative Role of Hzf in Cerebellar Function. BDNF induces local protein synthesis in cultured hippocampal neurons (3) and developing axons (33). Recently, impairments in dendritic mRNA localization and activity-dependent protein synthesis have been shown to affect long-lasting forms of synaptic plasticity like latephase LTP, which is maintained for several hours (10, 30). In contrast, early-phase LTP can be established in the absence of de novo mRNA and protein synthesis (34). In addition, BDNF has been shown to play an important role in long-lasting synaptic plasticity in the hippocampus (35). Therefore, Hzf may be involved in long-lasting synaptic plasticity in the cerebellum, possibly by controlling dendritic mRNA localization and activity-dependent translation. Interestingly, Hzf mutant mice exhibit tremors, ataxic gate, and poor motor performance in behavioral analyses despite the grossly normal morphology and cytoarchitecture of the cerebellum (unpublished data). Future studies including physiological and pharmacological analyses designed to assess late-phase synaptic plasticity in the cerebellum are warranted to reveal the biological significance of dendritic mRNA localization and translation in cerebellar learning and memory.
Supplementary Material
Acknowledgments
We thank Dr. Teiichi Furuichi (RIKEN Brain Science Institute, Saitama, Japan) for providing the mouse cerebellum cDNA library and probes for the in situ hybridization of IP3RI. We appreciate all members of the Okano laboratory for invariable assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.O.), a Grant-in-aid for 21st Century COE program to Keio University, and Ontario Research and Developmental Challenge Fund (to Y.K.).
Author contributions: H.J.O. and H.O. designed research; T. Iijima and T. Imai performed research; Y.K. and A.B. contributed new reagents/analytic tools; H.J.O. and M.Y. analyzed data; and T. Iijima wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LTP, long-term potentiation; LTD, long-term depression.
References
- 1.Steward, O. & Halpain, S. (1999) J. Neurosci. 19, 7834-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ouyang, Y., Rosenstein, A., Kreiman, G., Schuman, E. M. & Kennedy, M. B. (1999) J. Neurosci. 19, 7823-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aakalu, G., Smith, W. B., Nguyen, N., Jiang, C. & Schuman, E. M. (2001) Neuron 30, 489-502. [DOI] [PubMed] [Google Scholar]
- 4.Kang, H. & Schuman, E. M. (1996) Science 273, 1402-1406. [DOI] [PubMed] [Google Scholar]
- 5.Huber, K. M., Kayser, M. S. & Bear, M. F. (2000) Science 288, 1254-1257. [DOI] [PubMed] [Google Scholar]
- 6.Mayford, M., Baranes, D., Podsypanina, K. & Kandel, E. R. (1996) Proc. Natl. Acad. Sci. USA 93, 13250-13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blichenberg, A., Shuwanke, B., Rehbein, M., Garner, C. C., Richer, D. & Kindler, S. (1999) J. Neurosci. 19, 8818-8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blichenberg, A., Rehbein, M., Muller, R., Garner, C. C., Richter, D. & Kindler, S. (2001) Eur. J. Neurosci. 13, 1881-1888. [DOI] [PubMed] [Google Scholar]
- 9.Mori, Y., Imaizumi, K., Katayama, T., Yoneda, T. & Tohyama, M. (2000) Nat. Neurosci. 3, 1079-1084. [DOI] [PubMed] [Google Scholar]
- 10.Miller, S., Yasuda, M., Coats, J. K., Jones, Y., Martone, M. E. & Mayford, M. (2002) Neuron 36, 507-519. [DOI] [PubMed] [Google Scholar]
- 11.Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L. & Singer, R.H. (1997) Mol. Cell. Biol. 17, 2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, H. L., Eom, T., Oleynikov, Y., Shenoy, S. M., Liebelt, D. A., Dictenberg, J. B., Singer, R. H. & Bassell, G. J. (2001) Neuron 31, 261-275. [DOI] [PubMed] [Google Scholar]
- 13.Tiruchinapalli, D. M., Oleynikov, Y., Kelic, S., Shenoy, S. M., Hartley, A., Stanton, P. K., Singer, R. H. & Bassell, G. J. (2003) J. Neurosci. 23, 3251-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuichi, T., Simon-Chazottes, D., Fujino, I., Yamada, N., Hasegawa, M., Miyawaki, A., Yosikawa, S., Guenet, J. L. & Mikosiba, K. (1993) Recept. Channels 1, 11-24. [PubMed] [Google Scholar]
- 15.Furuichi, T., Yosikawa, S., Miyawaki, A., Wada, K., Maeda, N. & Mikoshiba, K. (1989) Nature 342, 32-38. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, T., Kato, K., Kohda, K. & Mikoshiba, K. (1998) J. Neurosci. 18, 5366-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, M. (1989) Annu. Rev. Neurosci. 12, 85-102. [DOI] [PubMed] [Google Scholar]
- 18.Hidaka, M., Caruana, G., Stanford, W. L., Sam, M., Correll, P. H. & Bernstein, A. (2000) Mech. Dev. 90, 3-15. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, N., Niinobe, M., Nakahira, K. & Mikoshiba, K. (1988) J. Neurochem. 51, 1724-1730. [DOI] [PubMed] [Google Scholar]
- 20.Bian, F., Chu, T., Schilling, K. & Oberdick, J. (1996) Mol. Cell. Neurosci. 7, 116-133. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, Y., Hart, A., Hirashima, M., Wang, C., Holmyard, D., Pittman, J., Pang, X. L., Jackson, C. W. & Bernstein, A. (2002) J. Exp. Med. 195, 941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krichevsky, A. M. & Kosik, K. S. (2001) Neuron 32, 683-696. [DOI] [PubMed] [Google Scholar]
- 23.Kiebler, M. A., Hemraj, I., Verkade, P., Kohrmann, M., Fortes, P., Marion, R. M., Ortin, J. & Dotti, C. G. (1999) J. Neurosci. 19, 288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohrmann, M., Luo, M., Kaether, C., DesGroseillers, L., Dotti, C. G. & Kiebler, M. A. (1999) Mol. Biol. Cell 10, 2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monshausen, M., Putz, U., Rehbein, M., Schweizer, M., DesGroseillers, L., Kuhl, D., Richter, D. & Kindler, S. (2001) J. Neurochem. 76, 155-165. [DOI] [PubMed] [Google Scholar]
- 26.Tang, S. J., Meulemans, D., Vazquez, L., Colaco, N. & Schuman, E. (2001) Neuron 32, 463-475. [DOI] [PubMed] [Google Scholar]
- 27.Kanai, Y., Dohmae, N. & Hirokawa, N. (2004) Neuron 43, 513-525. [DOI] [PubMed] [Google Scholar]
- 28.Bannai, H., Fukatsu, K., Mizutani, A., Natsume, T., Iemura, S., Ikegami, T., Inoue, T. & Mikoshiba, K. (2004) J. Biol. Chem. 279, 53427-53434. [DOI] [PubMed] [Google Scholar]
- 29.Steward, O. & Schuman, E. M. (2001) Annu. Rev. Neurosci. 24, 299-325. [DOI] [PubMed] [Google Scholar]
- 30.Kelleher, R. J., III, Govindarajan, A., Jung, H. Y., Kang, H. & Tonegawa, S. (2004) Cell 116, 467-479. [DOI] [PubMed] [Google Scholar]
- 31.Schratt, G. M., Nigh, E. A., Chen, W. G., Hu, L. & Greenberg, M. E. (2004) J. Neurosci. 24, 9366-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takei, N., Inamura, N., Kawamura, M., Namba, H., Hara, K., Yonezawa, K. & Nawa, H. (2004) J. Neurosci. 24, 9760-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X. & Poo, M. M. (2002) Neuron 36, 675-688. [DOI] [PubMed] [Google Scholar]
- 34.Kandel, E. R. (2001) Science 294, 1030-1038. [DOI] [PubMed] [Google Scholar]
- 35.Korte, M., Kang, H., Bonhoeffer, T. & Schuman, E. (1998) Neuropharmacology 37, 553-559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.