Abstract
We applied genome-wide gene expression analysis to the evolutionary processes of adaptive speciation of the Israeli blind subterranean mole rats of the Spalax ehrenbergi superspecies. The four Israeli allospecies climatically and adaptively radiated into the cooler, mesic northern domain (N) and warmer, xeric southern domain (S). The kidney and brain mRNAs of two N and two S animals were examined through cross-species hybridizations with two types of Affymetrix arrays (mouse and rat) and muscle mRNA of six N and six S animals with spotted cDNA mouse arrays. The initial microarray analysis was hypothesis-free, i.e., conducted without reference to the origin of animals. Principal component analysis revealed that 20-30% of the expression signal variability could be explained by the differentiation of N-S species. Similar N-S effects were obtained for all tissues and types of arrays: two Affymetrix microarrays using probe oligomer signals and the spotted array. Likewise, ANOVA and t test statistics demonstrated significant N-S ecogeographic divergence and region-tissue specificity in gene expression. Analysis of differential gene expression between species corroborates previous results deduced by allozymes and DNA molecular polymorphisms. Functional categories show significant N-S ecologic putative adaptive divergent up-regulation of genes highlighting a higher metabolism in N, and potential adaptive brain activity and kidney urine cycle pathways in S. The present results confirm ecologic-genomic separation of blind mole rats into N and S. Gene expression regulation appears to be central to the evolution of blind mole rats.
Keywords: adaptive speciation, genome-wide expression, microarray, ecological stress, cross-species hybridization
Evolutionary change is primarily driven by ecological stress (1-3). Climatic stresses are instrumental in advancing the twin evolutionary processes of adaptation and speciation. Darwin recognized that “climate plays an important part in determining the average number of a species, and periodical seasons of extreme cold or drought seem to be the most effective of all checks” (4).
Evolutionary Ecological Genomics
The genomic revolution has linked molecular genomics with organismic evolution (3). The ongoing problems associated with speciation and adaptation (3) could fruitfully be explored by genome-wide gene expression analysis (5). How many genes, which ones, and, most importantly, what regulatory variations across the genome are possibly adaptive, associated with ecological stress, and affect adaptation and speciation? What genes and/or regulatory elements are associated with ecological speciation? Ecological and functional genomics (3) could elucidate some of the major yet unresolved evolutionary problems.
The Evolutionary Global Experiment of Subterranean Mammals
Subterranean mammals provide a long-term natural evolutionary experiment of mammals adapted to life underground. Subteranean mammals display mosaic evolution of regression, progression, and global convergence across all organizational levels from molecular to organismal (6). Their underground evolution originated in Middle Eocene to early Oligocene (45-35 million years ago) and continued stepwise in the Cenozoic period after climatic cooling and drying. The subterranean ecotope is characterized by simple, climatically stable, and specialized profiles but is subjected to severe stresses of darkness, energetics, infection, and hypoxia-hypercapnia (6).
Spalacids and Spalax ehrenbergi Superspecies in Israel
The Spalacidae are Eurasian, primarily East Mediterranean, subterranean rodents representing extreme adaptations to life underground (figure 7.4b of ref. 6). Spalacids originated some 25 million years ago in southwest Asia Minor based on fossils and 40 million years ago based on molecules (6). The Spalax ehrenbergi superspecies adaptively radiated in the Near East and split into at least 12 allospecies. Four species evolved in Israel during the Pleistocene, adapting to four climatic regimes (7). The two northern Israeli species, Spalax galili (2n = 52) and Spalax golani (2n = 54), range in cool humid climate, whereas the central species, Spalax carmeli (2n = 58), ranges in warm humid climate, and the southern species, Spalax judaei (2n = 60), ranges in warm dry climate (figures 5, 11, and 12 in ref. 7). Generally, the complex extends along a southern gradient of increasing aridity with underground temperature increasing and humidity decreasing southwards, although mildly and without extremes as above ground. This ecological speciation into a trend of four species subdivided into older northern (S. galili and S. golani) and younger southern (S. carmeli and S. judaei) species has been documented by diverse studies of noncoding and coding genomic regions. These regions include chromosomes (7), allozymes (8, 9), mtDNA (10), random amplified polymorphic DNAs, minisatellites and chiasma frequency (11, 12), microsatellites (13), DNA hybridization (14), mitochondrial cytochrome b (15), the mitochondrial control region (16), erythropoietin (17), neuroglobin and cytoglobin (our unpublished observations), and the Clock and MOP3 genes (18). Here, we show that previous genetic and phenetic patterns are corroborated by genome-wide gene-expression analysis based on an average of 15,000 genes, highlighting the remarkable importance of at least partly adaptive gene-expression regulation in Spalax evolution.
Materials and Methods
Animals Analyzed. We analyzed 16 animals, eight animals representing the cool mesic northern domain (N) [S. galili (2n = 52) from Dalton, Upper Galilee (seven animals), and S. golani (2n = 54) from Quneitra, Golan Heights (one animal)], and eight animals representing the warm xeric southern domain (S) [S. carmeli (2n = 58) from Mukhraka, Mt. Carmel (one animal), and S. judaei (2n = 60) from Lahav, northern Negev (seven animals)]. For the Affymetrix experiments, two N (one S. galili and one S. golani) and two S (one S. carmeli and one S. judaei) were used, with each animal on an individual chip. For the cDNA experiment, six N (six S. galili only) and six S (six S. judaei only) were used.
All were adult males of ≈150 g; they were captured in the field in July 2002 and housed in individual cages, each species in a separate room of the Institute of Evolution, University of Haifa. All animals were housed according to regulations conforming to animal handling protocols approved by the University of Haifa under controlled conditions of 22-24°C with seasonal light/dark hours on a diet of carrots. Animals were killed three to four months after capture (October 2002 and December 2004). Brain, kidney, and muscle tissues were removed from each animal, immersed in liquid nitrogen, and stored in -80°C until RNA processing. Brain and kidney mRNA of two animals from N and two animals from S were hybridized with Affymetrix rat and mouse oligonucleotide arrays. In addition, muscle mRNA of six animals from N and six animals from S were hybridized with a spotted mouse microarray.
Affymetrix Microarray Experiments. RNA from two Northern and two Southern individuals were analyzed with Affymetrix mouse MOE430A and rat RAE230A oligonucleotide arrays according to the manufacturer's instructions. Total RNA from each sample was used to prepare biotinylated target RNA by following the manufacturer's recommendations. Quality and quantity of starting RNA was confirmed by using agarose gel and OD measurement, respectively. After scanning, array images were assessed by eye to confirm scanner alignment and the absence of significant bubbles or scratches on the chip surface. 3′/5′ ratios for GAPDH and β-actin were confirmed to be within acceptable limits, and BioB spike controls were present on all chips, with BioC, BioD, and CreX also present in increasing intensity. When scaled to a target intensity of 150 (Affymetrix mas 5.0 array analysis software), scaling factors for all arrays were within acceptable limits, as were background, Q values, and mean intensities. Details of quality control measures are available from the National Center for Biotechnology Information or the Chaim Sheba Medical Center.
cDNA Microarray Experiments. Muscle tissue from 12 individual animals, six S [S. judaei (2n = 60) from Lahav] and six N [S. galili (2n = 52) from Dalton], were directly compared by using mouse cDNA microarrays representing 15,247 clones from the National Institute on Aging 15K libraries (19, 20) as well as positive and negative controls. We added to the cDNA arrays 32 Spalax sequences for the following genes: Epo, Epo-R, VEGF, VEGF-R(FLK), VEGF-R(FLT), HIF1A, HuR, Ngb, CYTG, Mdm2-promoter, Apaf1-promoter, p53, CARP, HPSE, LDHa, GT1, CLOCK, MOP3, Per1, Per2, Per3, Cry1, Cry2, Tim, OPN4, ADCYAP1, MAOA, LIS, DCX, CRYAB, and FGF-R3. Description of the National Institute on Aging 15K microarray printing parameters and format are available from the authors upon request. For all comparisons, 10 μg of Spalax muscle total RNA was reverse-transcribed according to indirect aminoallyl incorporation protocol and labeled with Alexa Fluor 555 or Alexa Fluor 647 dyes (Invitrogen). Slides were hybridized overnight at 42°C, washed, and scanned with an Axon GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA). Detailed protocols describing labeling and hybridization are available from the authors upon request. All slide images were analyzed by using genepix pro 6.0 software. Analyzed slide images were manually edited, and aberrant spots were flagged for exclusion in downstream analysis.
Quality Control and Normalization of Microarray Data. The initial Affymetrix hybridization probe expression signals (CEL files) were preprocessed separately for each set of hybridizations per array type (mouse or rat). Because all hybridizations were cross species and, presumably, some mouse or rat oligomer probes could be incompatible for hybridization with Spalax mRNA, we applied probe-based detection (21, 22) of N- and S-discriminating genes rather than a gene-based analysis. This approach is also expected to be generally more accurate because only a subset of each probe set may be suitable for analysis because of putative gene splice variants in the mRNA sample and possible slide artifacts (23).
The quality control analysis of the Affymetrix slides was performed by using techniques described in ref. 23. The major criterion for detection of slide artifacts was the deviation from uniformity of the distribution for log-transformed perfect match (PM) probe signals of high and low intensity over the microarray geometry. As a result of the quality control analysis, some condensed areas of high and low probe signals (“patterns”) were detected on slides, and sometimes their artifactual nature was very obvious (Supporting Materials and Methods and Fig. 3, which are published as supporting information on the PNAS web site).
Statistically significant (23) local artifactual patterns were cleaned. Corresponding probe pairs were removed from all mice and rat microarray hybridization sets. In total, 15% of the probe pairs were excluded from the subsequent analysis. Exclusion of local regions across the chip did not significantly affect the results because probe pairs of each probe set are uniformly distributed over the microarray geometry.
Considering that in cross-species hybridizations PM and mismatch (MM) probes may potentially differ in sequence from hybridized mRNA (21, 22), both probe signals were included in the analysis. However, we relied primarily on the PM signals, assuming they had higher sequence similarity. Log-transformed probe signals of each sample were quantile-normalized (24). Normalization was performed separately for PM and MM signals. The goal of the normalization was to equalize distributions of probe log-signals for all samples within the set.
The six spotted array hybridizations were normalized by nonlinear lowess regression (25). The slide artifacts for spotted array hybridizations were checked with the procedure described in ref. 23.
ANOVA Model for Log-Signal Gene Expression Values. We used a general additive ANOVA model for the influence of effects of two factors (ecogeography and tissue) and their interaction on expression log-signal of a probe (see Supporting Materials and Methods). The effects of the model were estimated through general linear model regression (26). The significance of the estimated effect for individual probe profile (the vector of probe signals across all samples) was detected through t-distributed statistic (26). The ANOVA estimation was used to detect genes with significant geography-tissue interaction effect.
The Statistical Significance of Gene Regulation. For the Affymetrix array experiments, the statistical significance of ANOVA effects for a given gene were evaluated by Wald statistics (27). First, for each probe profile the t test statistic (see Eq. 2 in Supporting Materials and Methods) of the effect estimation was calculated as a measure of normalized deviation (including direction) of the factor combination-specific log-signal from the probe log-signal average. The null hypothesis assumes that a significant individual effect (t test) would appear randomly for probe profiles across the entire set of probes of the set of hybridizations. For a specific gene, the null hypothesis assumes that all probes within the gene probe set demonstrate effects of factors by chance (see The Independence of Probe Signals Within Probe Set in Supporting Materials and Methods; see also Figs. 4-6, which are published as supporting informtion on the PNAS web site). Afterward, two Wald statistics (the sum of squares of t statistics) were calculated for every gene effect (N-S effect, tissue effect, and their interaction): one for all probes with positive effects and another one for all negative effect probes. Under the null hypothesis, these are χ2-distributed (27). The P value of the ratio of these two χ2 statistics, scaled by corresponding degrees of freedom (F test), reflects the significance of the effect for a gene in the corresponding direction.
The same Wald approach was applied for detection of N-S differentiating genes per tissue. Considering that fold changes detected by the Affymetrix experiments were rather small (≈1.5) even for the best discriminating probes and because the t test significance of the effect for an individual probe was based on only four animals, we used the above mentioned probe set “voting” approach to determine whether an individual gene was differentially expressed in N and in S and whether there is a specificity of expression under some geography-tissue combination. The N-S discriminating power of a gene was estimated through voting of its probe set t-test values either “for” or “against” regulation in a given direction. Two Wald statistics were calculated for each gene: with N up-regulated probes of the gene probe set and with S up-regulated probes of the same gene. If one Wald statistic was significant and the other was not, we considered that gene to be significantly up-regulated in the direction of the significant Wald statistic. To flag a gene as significantly differentially expressed, a single Wald test P value (calculated according to all 11 probes of the probe set) needed to pass the 10% false discovery rate (28) significance level.
For spotted cDNA arrays, genes with significant N-S differences of expression signals (N-S effect) were detected with a t test taking into consideration three direct and three dye-swapped hybridizations. Because all 12 RNA samples represent individual animals (six N and six S), all direct vs. all swapped hybridizations were paired to estimate the N-S effect that is not confounded by dye. The nine estimations of N-S effect are slightly interdependent. Indeed, the estimation of every one of nine N-S effects is based on a pair of direct and dye-swapped hybridizations. Each pair has six related pairs: three pairs with the same first hybridization and three pairs with the same second hybridization. This weak interdependence of nine N-S difference estimations decreases the effective number of independent members for the χ2 statistic of the corresponding t test. The Satterthwaite's formula was used to calculate the effective number of degrees of freedom for the variance estimation according to interpenetrating subsamples (29).
Results
After quality control filtering of the Affymetrix signals, we analyzed a data set that contained 430,509 probe pairs representing 17,941 genes and 308,042 probes representing 14,708 genes on the mouse and rat GeneChips, respectively. For the mouse cDNA, 16,416 spots were analyzed, including control spots.
The process of data analysis was carried out by using a multistep approach. First, a hypothesis-free principal component (PC) analysis (PCA) was done to detect what factors explain the major trends in the overall variability. Next, we investigated the N vs. S differences as these stem from the PCA analysis of two types of Affymetrix microarray data (PM and MM probe oligomer signals separately for the mouse and rat) and from spotted array data (Fig. 1a). Finally, we characterized the effect of N vs. S species for all three tissues at the individual probe level (Affymetrix hybridizations) or at the gene level (spotted arrays). The last step of our analysis was a “supervised approach,” for which we distinguished genes with significant N-S expression divergence within each individual tissue by estimating differences on spotted arrays and the correlated differential behavior of probe sets for Affymetrix arrays. Also, genes with geography-tissue specificity in expression were detected by ANOVA.
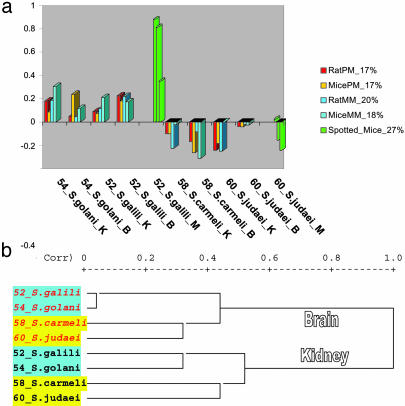
Fig. 1.
PCA and cluster analysis of probe oligomer (Affymetrix arrays) and gene signals (spotted array). Profiles of the second PC of PM and MM log signals for mouse and rat Affymetrix microarray experiments and mouse spotted-array experiment. (a) PC2 reflects the general signal difference between N and S animals for two types of Affymetrix microarrays [PM (red bars) and MM (blue bars) signals] and for the spotted array experiment (green bars). This PC2 component explains 20-30% of total signal variability. (b) The dendrogram of correlation distances between Affymetrix hybridizations (average linkage) after the cleaning of artifacts. All nonpatterned probe oligomers with average signals of >300 according to all eight hybridizations were taken. Clear N-S (S. galili plus S. golani vs. S. carmeli plus S. judaei) separation is seen for both tissues as in all other evolutionary discrimination of Spalax species based on diverse molecular markers and morphological criteria (figures 16, 17, 19, 20-22, 23 a and b, 31, and 33 in ref. 7).
PCA. The general picture of PM and MM probe log signals according to the PCA (30) demonstrated similar patterns for the variability of Affymetrix array signals across the two series of hybridizations. This similarity supports the hypothesis of small differences between PM and MM probes in cross-species hybridizations.
The first PC of all Affymetrix hybridizations (mouse and rat arrays) reflected the difference between tissues. PC1 of spotted hybridizations is related to the variability of individual slides. In all cases, PC1 covers 40-50% of total signal variability.
Notably, PC2 reflected an important pattern of signal variability for all sets of hybridizations (Affymetrix and spotted arrays) and accounts for gene expression divergence between N and S species (Fig. 1a). This PC explains 20-30% of the total signal variability.
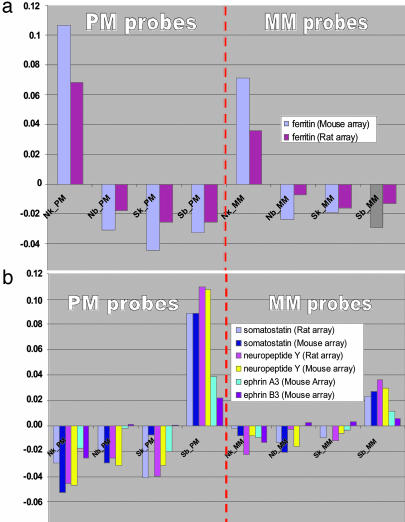
Genes with Ecogeography-Tissue Specificity in Their Expression. The ANOVA analysis detected ≈100 genes with ecogeography-tissue specificity in their expression (Data Set 1, which is published as supporting information on the PNAS web site). We selected several of the most representative genes that demonstrate their specificity for mouse and rat array hybridizations (according to PM and MM signals) (Fig. 2).
Fig. 2.
The estimated expression profiles of several proteins that have ecogeography-tissue specificity (see the description of proteins in the text). The log values of averaged gene expression for four factor combinations were calculated through estimated parameters of the ANOVA model according to all PM and MM probes. The same type of gene expression regulation was detected by mouse and rat array hybridizations. (a) N kidney up-regulated ferritin. The strong N up-regulation of this gene for muscle was detected by spotted array hybridizations. (b) S brain up-regulated genes: somatostatin, neuropeptide Y, and ephrin.
S brain up-regulated neuropeptide Y is involved in the regulation of circadian rhythms, sexual functioning, anxiety and stress response, peripheral vascular resistance, and contractility of the heart. Neuropeptide Y is also involved in the regulation of feeding behavior, including food intake as well as metabolic and lipogenic rates (31). Injection of neuropeptide Y into cerebral ventricles or directly into the hypothalamus of rats potently stimulates food intake and decreases energy expenditure. S brain up-regulated somatostatin hormone inhibits the release of growth hormone; thus, it could be responsible for the smaller size of S animals. S brain up-regulated ephrin is crucial for axonal pathfinding in vertebrates and invertebrates. Ephrin signaling is important for synaptic remodeling in the adult brain (32); therefore, it could be related to the better topographic orientation in S, where territories are larger. Profilin, a low-molecular-mass actin-binding protein is up-regulated in N animals, both in kidney and brain. Proifilin is an actin monomer-sequestering protein (33). Profilin polymerizes actin onto the barbed ends of actin filaments that participate in muscle contraction. The up-regulation of profilin results in the perfection of muscles in N animals. The N kidney, up-regulated ferritin, an iron-storing protein, was also detected by cDNA hybridizations. The up-regulation of ferritin in N saturates the blood with O2, thus contributing to higher hypoxia tolerance in N. N brain up-regulated cytoplasmic dynein light-chain Tctex-1 plays a key role in multiple steps of hippocampal neuron development, including initial neurite sprouting, axon specification, and, later, dendritic elaboration (34); therefore, it may have an important role in cell-cell signaling during nervous system formation. N brain up-regulated amyloid precursor-like protein 1 accumulates in neuritic plaques in Alzheimer's disease, and it may contribute to neurodegeneration.
N-S Differentiating Genes. Data Sets 2 (Affymetrix rat array) and 3 (Affymetrix mouse array), which are published as supporting information on the PNAS web site, contain significant N and S up-regulated genes for the kidney and brain, respectively.
The spotted array genes with significant N-S divergence of expression signals (N-S effect) were subjected to a t test (see Materials and Methods). Data Set 4, which is published as supporting information on the PNAS web site, contains significant N and S up-regulated genes for muscle.
Gene Ontology (GO) Analysis. The GO analysis for differentially expressed genes in the Affymetrix experiments was carried out by using the rat and mouse array data. A total of six sets of N-S differentially expressed genes were analyzed according to the different tissue × species factor combinations.
We identified GO categories overrepresented with N-S differentially expressed genes, which were identified with the National Institutes of Health david (Database for Annotation, Visualization, and Integrated Discovery) server (35). The significance of overrepresentation was based on all rat and mouse LocusLink genes, respectively [Data Set 4 (for spotted mouse array) and Data Sets 5 (for Affymetrix rat array) and 6 (for Affymetrix mouse array), which are published as supporting information on the PNAS web site].
GO analysis demonstrates that the main distinguishing feature for animals of N (of all three tissues) was the increase in metabolic processes involving practically all biochemical subclasses. For S, the most distinguishing features were amino acid and amine metabolism, and induction of apoptosis in muscles (Data Sets 4-6).
N- and S-up-regulated genes in kidney and brain, common for Affymetrix rat and mouse arrays, are detailed in Table 1, which is published as supporting information on the PNAS web site. The following gene groups exemplify the combined N up-regulation of brain and kidney: protein metabolism; nucleobase, nucleoside, nucleotide and nucleic acid metabolism, cell growth and maintenance, and carbohydrate metabolism. N-brain-specific up-regulation includes O2 and electron transport, ion channel activity, ion transport, hemoglobin complex, O2 transport, O2 transporter activity, mitochondrial electron transport, and oxidoreductase activity. N kidney up-regulation includes oxidoreductase activity and phosphorous metabolism. S up-regulation (both kidney and brain) includes urea cycle, aminopeptidase activity; arginase activity, arginine catabolism, arginine metabolism, Mn ion binding, and axon guidance.
Ecological Phylogeny of the S. ehrenbergi Superspecies in Israel. The phylogenetic analysis based on gene expression in tissues reflects the ecological separation of the Israeli N and S species (S. galili/S. golani and S. carmeli/S. judaei) (Fig. 1b). The pattern precisely fits the PCA of gene expression data and corroborates all earlier protein and DNA phylogenetic trees of the S. ehrenbergi superspecies in Israel, into a N older pair of species vs. a S younger species pair (figures 16, 17, 33, and 45 in ref. 7). Hence, the N-S ecological divergence underlies the phylogeny of S. ehrenbergi superspecies in Israel displaying a climatically southward-oriented trend of speciation from N to S environments (7).
Discussion
Climatic Adaptive Complexes and Molecular Divergence. Multiple systems (genetic, ecological, biochemical, morphological, physiological, and behavioral) characterize each of the Israeli Spalax species adapting it to its unique climate as well as the prime ecological separation between mesic N and xeric S (7). Competitive exclusion between neighboring species, apparently determined by climatic selection, leads to parapatric distribution. The four species of the Spalax chrenlergi superspecies in Israel represent four allospecies with unique ecophysiological, ethological, and genetic syndromes and partial premating and postmating reproductive isolation, despite narrow zones of natural hybridization between species (7). Chromosome, protein, and DNA diversities support species divergence without introgression across hybrid zones (7). DNA-DNA hybridization (14) indicated that the percent of nucleotide substitution in single-copy nuclear DNA among the species ranged from 0% to 5%. This finding suggests that speciation had occurred with relatively minor genomic changes, although the present microarray evidence indicates dramatic regulatory divergence between the older N species pair [S. galili (2n = 52) and S. golani (2n = 54)] and the younger S species pair [S. carmeli (2n = 58) and S. judaei (2n = 60)] (Figs. 1 and 2; see also Table 2, Fig. 7, and Data Sets 2-4, which are published as supporting information on the PNAS web site). The youngest species pair differs by 0.2-0.6% base pair MM (i.e., one to three base-pair differences in a 500-bp fragment) (14).
The interspecific values of percent nucleotide differences permit the recognition of two well separated speciation events in the S. ehrenbergi superspecies: the older event (Lower Pleistocene) having isolated the N species pair [S. golani (2n = 54) and S. galili (2n = 52)] before the divergence of the S species pair [S. carmeli (2n = 58) and S. judaei (2n = 60)], based on climatic and ecological N-S contrasts. Genomic-wide gene expression remarkably corroborates this speciation junction of the two species pairs by muscle, kidney, and brain expression patterns (Fig. 1b) based on 16 specimens, three tissues, two Affymetrix microarrays, and one spotted cDNA mouse array for 15,000 genes. This unique separation is based on the differential regulation of the genome because the gene content appears similar in the four allospecies (7).
Ecological Radiation of the S. ehrenbergi Superspecies. The climatic radiation of Spalax has multiple correlates: genetic (allozyme DNA markers, random amplified polymorphic DNAs, variable number tandem repeats, simple sequence repeats, and mtDNA), morphological (size, shape, skull, molar, incisor, penial, and pelage variations), physiological (basal and energy metabolism, urinary concentration ability, water turnover rate, caecum anatomy, thermoregulation, nonshivering thermogenesis, respiratory adaptations, hematocrit, and hypoxia survival), population structure (differential species survivorship in the laboratory), behavioral patterns (ethological isolation, odor variation, habitat selection, swimming ability, geographic dialects, seismic communication, magnetic orientation, activity pattern, exploratory behavior, rest-activity patterns, and aggression), and parasites (gamaside mites and fleas, helminthes, and coccidians) (7). Fig. 4 and Data Sets 2-4 display the up-regulated kidney, brain, and muscle genes associated with energetics, respiratory adaptations, and behavioral patterns. Some major physiological and behavioral correlates related to microarray results are represented in Figs. 1 and 2, Tables 1 and 2, and Data Sets 2-4.
Spalax Population Structure and Geographic Variation Southwards in Israel. Population density of Spalax in Israel declines from the mesic N to the xeric S toward the northern Negev desert, following the decline in productivity (figure 47 in ref. 7). The differential survivorship of the four species under standardized laboratory conditions (figure 48 in ref. 7) also reflects the differential (N-S) climatic fitness of the species (pp. 92-95 in ref. 7). Thus, the demographic decline southward displays increasing climatic stress toward the desert, revealed by the N-S divergent genome-wide gene expression and diverse physiological (energetics and respiration) and behavioral phenotypic divergence.
Energetics. Basal metabolic rates (p. 69 and table 10 in ref. 7) decline southward from N S. galili to S S. judaei (1.03→0.62 cm3 of O2·g-1·h-1). The lower basal metabolic rates value of S S. judaei (7) is adaptive by reducing water expenditure and overheating. Similar geographical variation of energy metabolism was found in gross energy intake, both southward and eastward (p. 69 and table 11 in ref. 7): water turnover (figure 34 in ref. 7) correlated with energy turnover and water intake by food and urinary concentration ability in xeric S. judaei under protein and salt loads (table 12a in ref. 7). All of these energetics variations relate to structural and functional adaptations of kidney water conservation to increasing aridity (figure 35 of ref. 7). Likewise, adaptive thermoregulatory and nonshivering thermogenesis are climatically correlated with increasing temperatures southward (pp. 69-81 in ref. 7). Because productivity declines toward the desert, all of the aforementioned evidence suggests strong climatic selection associated with energetic stresses.
Remarkably, genes up-regulated in N reflect the putative adaptation of higher metabolism (Table 2, Fig. 7, and Data Sets 2-4) (7). For example, kidney and/or brain metabolic up-regulated genes in N include glucose-6-phosphatase, catalytic ATP synthase, H+ transporting, mitochondrial F0 complex subunit b, NADH dehydrogenase (ubiquinone), and flavoprotein 2, as shown on the rat and mouse arrays.
Respiratory Adaptations. Hypoxic-hypercapnic stresses increase northward in mole rats and were maximized in N species rather than in S species. O2 and CO2 measurements in Spalax burrows show 7% O2 and 6% CO2 in heavy soils after flooding (36), and estimates may even become more extreme under heavy flooding. Several respiratory components vary geographically according to the variation in hypoxia-hypercapnia stress. These components include O2 and CO2 pressure in subcutaneous gas pockets (figure 40 in ref. 7), breathing and heart frequencies decreasing southward (pp. 83-85 in ref. 7), hematocrit and hemoglobin concentrations increasing northward (figure 42 a and b in ref. 7), interspecies higher hypoxic survival in S. galili (≈2% O2) as compared with S. judaei (≈3% O2) (table 18 and figure 43 in ref. 7), correlated with additional traits and substantiating respiratory adaptations in the more flooded N (pp. 83-86 in ref. 7). Even the molecular evolution of cytochrome b (figure 44 in ref. 7) and several globins, e.g., haptoglobin (figure 46 in ref. 7), neuroglobin, and cytoglobin (ref. 36 and our unpublished results) reflect N-S stress gradient correlations. Notably, a complex of hypoxia-tolerant genes in rat and mouse arrays were up-regulated in N, including hemoglobin β-chain complex, hemoglobin α-adult-chain-1, cardiotrophin, cardiac muscle slow twitch 2, hypoxia-inducible factor 1α, myoglobin (37), muscle protein, and p53 (38), which control cellular stress responses, including hypoxia (17), and substantially contribute to the Spalax putative adaptive strategy of hypoxia tolerance.
Behavioral Patterns. Behavior also varies geographically and adaptively between N mesic species (S. galili and S. golani) and S xeric species (particularly S. judaei) (pp. 95-130 in ref. 7). Assortative mating is much stronger in N species pair (tables 21 and 22 and figures 49 and 50 in ref. 7) based on nonvisual cues (olfaction, vocalization, seismic communication, tactility, and magnetic orientation). Geographic variation in habitat selection, swimming ability, activity patterns, exploratory behavior, circadian rhythmicity, and aggressive behavior (figures 61-63 in ref. 7) all display clinal patterns associated with increasing climatic stress toward the desert. Remarkably, aggression increases northward and “pacifism” increases southward (pp. 127-132 in ref. 7). Notably, the 5-hydroxytryptamine (serotonin) receptor expression associated with aggression levels is significantly up-regulated (our unpublished results) in S. judaei, possibly relating to its more pacifistic behavior in xeric environments.
Two antagonistic biological trends occur in S. ehrenbergi superspecies in Israel; metabolism increases northward, but relative brain size and encephalization, underlying complex behavioral patterns, increase southward and climax in S. judaei (2n = 60) (tables 3 and 4 a-c in ref. 7), (i.e., one of the S species) the smallest species in size (figure 25 in ref. 7). This extraordinary pattern is presumably adaptive, reflecting the Bergmann rule and driven by natural selection. Larger relative brain size in the southern S. judaei appears to be associated with increasing aridity stress and climatic unpredictability. The more significantly N-up-regulated kidney genes (Table 2 and Fig. 7) display the higher metabolism in N. Behavioral traits like memory and orientation tend to increase in S. Microarray results support the previously described physiological and behavioral opposite N-S trends (7).
Kidney and Brain Function Based on GO Classification. How does the genome-wide gene expression underline Spalax adaptations to underground life? The S kidney is characterized by extraordinary urinary concentration ability under protein and salt loads (pp. 71-73 in ref. 7). Only the kidney of S. judaei (2n = 60) produced hyperosmotic urine adapted to the desert succulent and halophyte plants, pushing speciation southward (pp. 71-77 and figure 35 in ref. 7). Genes of the GO categories protein signaling, acid metabolism, and amine transport characterize the presumed adaptive uniqueness of the Spalax desert kidney, associated with increasing ecological stresses of aridity and climatic unpredictability (pp. 35 and 173-175 in ref. 7). Intriguingly, the up-regulated genes in the relatively larger brain of S. judaei in S relate to the GO category phosphate metabolism and are significantly overrepresented regarding all LocusLink rat genes. These genes include those in the GO categories phosphorous metabolism, phosphorylation, protein modification, and protein amino acid phosphorylation (see Data Set 5).
Conclusions and Prospects
This study highlights the fact that allelic diversity is not the only pattern that ecologically differentiates N from S Spalax. Most remarkably, genome-wide gene expression regulation in speciation and adaptation seems to be of paramount importance in Spalax evolution. This regulatory impact resides primarily in the noncoding genome (e.g., random amplified polymorphic DNAs, amplified fragment-length polymorphisms, variable number tandem repeats, simple sequence repeats), which has been repeatedly implicated in diverse environmental, climatic, and respiratory stresses. Climatic selection seems to be instrumental in driving the noncoding and coding Spalax genome to presumably adapt to life underground and speciate in a N-S trend along a southward gradient of increasing aridity stress. Environmentally stressed regulation of genome-wide gene expression in Spalax and probably more generally in other organisms across life appears to be the hallmark of the ecological evolutionary process of speciation and adaptation.
We plan to deeply explore the nature of the divergent genes in Spalax and unravel their functional significance. Future studies also could elucidate the interaction of potential adaptive evolution of Spalax genomics, proteomics, and phenomics to local and regional stresses underground by focusing on sequencing and SNP of stress genes and their evolutionary environmental regulation (e.g., ref. 30).
Supplementary Material
Acknowledgments
We thank Eshel Ben Jacob and Yossi Hillel for reviewing the paper and Harris Lewin and Lena Feinstein for comments that helped improve the paper. This work was supported by an Arison family donation (to the Center of DNA Chips in Pediatric Oncology, The Chaim Sheba Medical Center) and by the Ancell Teicher Research Foundation for Genetics and Molecular Evolution.
Author contributions: A.A., L.T., N.A., A.B.K., M.B., G.R., and E.N. designed research; J.J.-H., A.A., L.T., S.Z., N.A., and M.B. performed research; L.I.B., J.J.-H., S.Z., A.P., A.B.K., G.R., and E.N. analyzed data; and L.I.B. and E.N. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: N, northern domain; S, southern domain; PC, principal component; PCA, PC analysis; PM, perfect match; MM, mismatch.
References
- 1.Hoffman, A. R. & Parsons, P. (1991) Environmental Stress (Oxford Univ. Press, Oxford).
- 2.Nevo, E. (1998) Heredity 81, 591-593. [Google Scholar]
- 3.Nevo, E. (2001) Proc. Natl. Acad. Sci. USA 98, 6233-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darwin, C. (1859) On the Origin of Species by Means of Natural Selection (Murray, London).
- 5.Brown, P. O. & Botstein, D. (1999) Nat. Genet. 21, 33-37. [DOI] [PubMed] [Google Scholar]
- 6.Nevo, E. (1999) Mosaic Evolution of Subterranean Mammals, Regression, Progression and Global Convergence (Oxford Univ. Press, Oxford).
- 7.Nevo, E., Ivanitskaya, E. & Beiles, A. (2001) Adaptive Radiation of Blind Subterranean Mole Rats (Backhuys, Leiden, Germany).
- 8.Nevo, E. & Shaw, C. R. (1972) Biochem. Genet. 7, 235-241. [DOI] [PubMed] [Google Scholar]
- 9.Nevo, E. Filippucci, M. G. & Beiles, A. (1994) Heredity 72, 465-487. [DOI] [PubMed] [Google Scholar]
- 10.Nevo, E., Honeycutt, R. L., Yonekawa, H., Nelson, K. & Hanzawa, N. (1993) Mol. Biol. Evol. 10, 590-604. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Shlomo, R., Fahima, T. & Nevo, E. (1996) Isr. J. Zool. 42, 317-326. [Google Scholar]
- 12.Nevo, E., Ben-Shlomo, R. Beiles, A., Ronin, Y. I., Blum, S. & Hillel, J. (1995) in Gene Families: Structure, Function, Genetics Evolution, eds. Holmes, R. S. & Lim, H. A. (World Scientific, Singapore), pp. 55-70.
- 13.Karanth, P. K., Avivi, A., Beharav, A. & Nevo, E. (2004) Biol. J. Linn. Soc. 83, 229-241. [Google Scholar]
- 14.Catzeflis, F. M., Nevo, E., Ahlquist, J. E. & Sibley, C. G. (1989) J. Mol. Evol. 29, 223-232. [DOI] [PubMed] [Google Scholar]
- 15.Nevo, E., Beiles, A. & Spradling, T. (1999) J. Mol. Evol. 49, 215-226. [DOI] [PubMed] [Google Scholar]
- 16.Reyes, A., Nevo, E. & Saccone, C. (2003) Mol. Biol. Evol. 20, 622-632. [DOI] [PubMed] [Google Scholar]
- 17.Shams, I., Avivi, A. & Nevo, E. (2004) Proc. Natl. Acad. Sci. USA 101, 9698-9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avivi, A., Albrecht, U., Oster, H., Joel, A., Beiles, A. & Nevo, E. (2001) Proc. Natl. Acad. Sci. USA 98, 13751-13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka, T. S., Jaradat, S. A., Lim, M. K., Kargul, G. J., Wang, X., Grahovac, M. J., Pantano, S., Sano, Y., Piao, Y., Nagaraja, R., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 9127-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kargul, G. J., Dudekula, D. B., Meng, Y. K., Saied, K. L., Jaradat, A., Tanaka, T. S., Carter, M. G. & Ko, M. S. H. (2001) Nat. Genet. 28, 17-18. [DOI] [PubMed] [Google Scholar]
- 21.Ji, W., Zhou, W., Gregg, K., Yu, N. & Davis, S. (2004) Nucleic Acids Res. 32, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautier, L., Cope, L. M., Bolstad, B. M. & Irizarry, R. A. (2004) Bioinformatics 20, 307-315. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky, L., Leontovich, A., Shtutman, M. & Feinstein, E. (2004) Nucleic Acids Res. 32, E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. (2003) Bioinformatics 19, 185-193. [DOI] [PubMed] [Google Scholar]
- 25.Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J. & Speed, T. P. (2002) Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draper, N. R. & Smith, H. (1981) Applied Regression Analysis (Wiley, New York).
- 27.Agresti, A. (1990) Categorical Data Analysis (Wiley, New York).
- 28.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. Ser. B 57, 289-300. [Google Scholar]
- 29.Qian, J. (1998) Proc. Survey Res. Methods Section, Am. Stat. Assoc., 704-708.
- 30.Alter, O., Brown, P. & Botstein, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billington, C. J., Briggs, J. E., Harker, S., Grace, M. & Levine, A. S. (1994) Am. J. Physiol. 266, R1765-R1770. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin, T., Hindges, R., Yates, P. A. & O'Leary, D. D. (2003) Development (Cambridge, U.K.) 130, 2407-2418. [DOI] [PubMed] [Google Scholar]
- 33.Kang, F., Purich, D. L. & Southwick, F. S. (1999) J. Biol. Chem. 274, 36963-36972. [DOI] [PubMed] [Google Scholar]
- 34.Chuang, J. Z., Yeh, T. Y., Bollati, F., Conde, C., Canavosio, F., Caceres, A. & Sung, C. H. (2005) Dev. Cell. 9, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R60. [PubMed] [Google Scholar]
- 36.Shams, I. Avivi, A. & Nevo, E. (2005). Comp. Biochem. Physiol. 142, 376-382. [DOI] [PubMed] [Google Scholar]
- 37.Gurnett, A. M., O'Connell, J. P., Harris, D. E., Lehmann, H., Joysey, K. A. & Nevo, E. (1984) Protein Chem. 3, 445-454. [Google Scholar]
- 38.Ashur-Fabian, O., Avivi, A., Trakhtenbrot, L., Adamsky, K., Cohen, M., Kajakaro, G., Joel, A., Amariglio, N., Nevo, E. & Rechavi, G. (2004) Proc. Natl. Acad. Sci. USA 101, 12236-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.