Abstract
Telomeres protect chromosome ends from end-to-end fusion and degradation. Loss of telomere function causes cell-cycle arrest or cell death. Autosomal dominant dyskeratosis congenita (AD DC), a rare inherited bone marrow failure syndrome, is caused by mutations in TERC, the RNA component of telomerase. Here, we studied the telomere dynamics over three generations in a 32-member extended family with AD DC due to a TERC gene deletion. Our analysis shows that peripheral blood cells from family members haploinsufficient for TERC have very short telomeres. Telomeres are equally short in all individuals carrying the TERC gene deletion irrespective of their age. Chromosome-specific telomere analysis distinguishing the parental origin of telomeres showed that in gene deletion carriers, paternal and maternal telomeres are similarly short and similar in length to those of the affected parent. In children of affected parents who have normal TERC genes, parental telomeres are again similar in length, but two generations appear to be necessary to fully restore normal telomere length. These results are consistent with a model in which telomerase preferentially acts on the shortest telomeres. When TERC is limiting, this preference leads to the accelerated shortening of longer telomeres. The limited amount of active telomerase in TERC RNA haploinsufficiency may not be able to maintain the minimal length of the increasing number of short telomeres. Thus, the number of cells with excessively short telomeres and the degree of residual telomerase activity may determine the onset of disease in patients with AD DC.
Keywords: dyskeratosis congenita, anticipation, quantitative-FISH
Dyskeratosis congenita (DC) is a rare inherited bone marrow failure syndrome. Diagnostic features of DC include nail dystrophy, abnormal skin pigmentation, and mucosal leukoplakia (1, 2). DC has multiple patterns of inheritance (3). X-linked DC is caused by mutations in DKC1 encoding dyskerin (4), and individuals with autosomal dominant (AD) DC are heterozygous for mutations in the RNA component (TERC) of telomerase (5). The underlying molecular defect(s) for individuals with autosomal recessive disease remain to be identified. All patients with DC have very short telomeres (6), suggesting that dysfunctional telomere maintenance is the common pathway affected in this disease. In AD DC, haploinsufficiency for the TERC RNA is thought to be the disease mechanism (reviewed in ref. 7). Dyskerin, on the other hand, is a pseudouridine synthase, involved in the modification of rRNA molecules. Recently, however, it has emerged that dyskerin is also a component of the telomerase ribonucleoprotein complex, essential for the accumulation and stabilization of the TERC RNA in the nucleus (8).
Most of what we know about how alterations in TERC RNA influence telomere length in mammalian cells comes from studies in mice (reviewed in ref. 9). However, there are important differences in telomere maintenance between mouse and human. Laboratory mice, in comparison with humans, have very long telomeres (10). In addition, telomerase is constitutively active in most murine tissues (reviewed in ref. 11), whereas in humans telomerase activity is greatly diminished in most somatic cells with the exception of stem cells, their immediate progeny, and activated lymphocytes and monocytes (12-15).
Little is known about the dynamics of telomere shortening in individuals with DC. A comparison of telomere length in individuals with DC (X-linked and AD) at different ages by using terminal restriction fragment length (TRFL) analysis of the telomeres on long arm of chromosome 7 showed that the telomeres are already short at an early age (6). A second study, investigating patients with AD DC by using the same approach found that relative telomere shortening, in comparison with age-matched controls, was more pronounced in the affected child than in the affected parent. These investigations suggested that patients with DC are born with short telomeres, and that in AD DC, progressive telomere shortening may account for the earlier onset and the more severe disease manifestation observed in succeeding generations (16). Here, we further investigate the inheritance of telomere length in a large family with AD DC due to a TERC gene deletion by studying 32 family members over three generations. Our analysis shows that all family members carrying the TERC gene deletion have telomeres of similar length that, in comparison with age-matched controls, are very short. Chromosome-specific telomere analysis by using polymorphic subtelomeric probes that distinguish the parental origin of telomeres shows that in TERC gene deletion carriers, telomeres inherited from the affected and unaffected parent are similarly short and comparable in length to those of the affected parent. In contrast, in children who have inherited two normal TERC genes, telomeres derived from the affected and unaffected parent are again similar but are almost normal in length, comparable to those in the unaffected parent. These results demonstrate that in humans, as in mice, telomerase preferentially acts on the shortest telomeres.
Materials and Methods
Patients. Blood samples were obtained from multiple members of a family diagnosed with AD DC (6). The Washington University Institutional Review Board approved this study, and all subjects gave informed consent. The family pedigree is shown in Fig. 1. All family members were examined for physical signs of DC at the time of the blood draw. Peripheral blood counts were obtained at the time of examination.
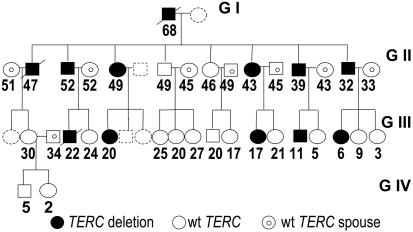
Fig. 1.
Four-generation pedigree of the AD DC family carrying the TERC gene deletion. Filled circles and squares indicate females and males carrying the TERC gene deletion, open circles and squares indicate females and males who have two normal copies of the TERC gene but have a parent (G + 1) or grandparent (G + 2) carrying a TERC gene deletion, and circles and squares containing a smaller circle indicate spouses carrying two normal copies of TERC. Broken shapes indicate family members from whom no sample was obtained. Family members deceased before our investigation are indicated. The age at the time of investigation is shown.
TERC Gene Mutation Analysis. PCR analysis on peripheral blood was performed as described in ref. 6 and confirmed the deletion mutation of TERC for nine family members. Direct sequencing analysis of a 1,034-bp region including the 451 bp of the mature TERC RNA and 312 bp of the promoter region confirmed a wild-type sequence for the second TERC allele in the affected individuals and normal TERC RNA sequences in nonaffected family members.
TRFL Analysis. To determine the average telomere restriction fragment length (TRFL), 1-3 μg of genomic DNA digested with restriction enzymes RsaI and HinfI was electrophoresed on a 0.5%/0.6× TBE gel along with a Lambda HindIII marker. After gel drying, denaturing, and neutralization, gels were hybridized to a γ-32P end-labeled telomeric probe. The average TRFL was determined by using the program imagequant (BD Biosciences). Mean TRF length was calculated according to the method of Wynn et al. (17).
FISH Analysis by Flow Cytometry (Flow-FISH). The telomere length of peripheral blood mononuclear cells (PBMC) isolated by Ficoll/Hypaque gradient centrifugation was measured as described in ref. 18 by using FITC-conjugated (C3TA2)3 peptide nucleic acid (PNA) (Applied Biosystems) probe and flow cytometry. Relative telomere length (RTL) was determined by comparing isolated PBMC with a control cell line [GM03671C (Coriell Institute for Medical Research, Camden, NJ)], a tetraploid cell line that served as an internal control having a telomere length of 100%. Telomere lengths of 86 healthy individuals (range 3-94 years old) were used to determine median age-dependent telomere length.
Quantitative (Q)-FISH Analyses of Metaphase Chromosomes. Quantitative analyses of telomeres by FISH were performed as described in ref. 19. Briefly, fresh heparinized blood samples (1 ml) from healthy and affected individuals were diluted in RPMI/20% FBS and stimulated with phytohemagglutinin-C (30 μg/ml) for 72 h. After a 1-h incubation with colcemid (0.1 μl/ml), cells were incubated with KCl (0.075 M) at 37°C for 25 min and then fixed with ethanol/acetic acid (3:1 vol/vol). Cell suspensions were dropped onto clean slides and used the next day for the hybridization procedure with either FITC- or Cy3-labeled (CCCTAA)3 PNA probes. After hybridization, metaphases were counterstained with DAPI and visualized under an Axioplan II imaging UV microscope (Zeiss) equipped with a charge-coupled device camera (Sensys, Photometrics, Tucson, AZ) and piloted by using the program smartcapture (Digital Scientific, Cambridge, U.K.). Twenty to 30 metaphases were captured for each preparation (bin 4, 16 bit, gain 8, fixed exposure; red, 0.5 sec; green, 2 sec). Pseudocolor images were used to automatically segment telomere-specific signals, which were then individually measured on the original (black and white) image as described in ref. 19. A mean pixel value (fluorescence intensity) for either individual telomeres or metaphases was thus obtained. To measure allele-specific telomeres, after metaphase capturing, the slides were hybridized again with subtelomeric probes (20), as described in ref. 19. This procedure revealed polymorphisms that enabled us to distinguish between homologous chromosomes, to follow the segregation of chromosome extremities in the family, and to measure the fluorescence intensity of specific telomeres, either in absolute units or relative to the other telomeres in the cell. A mean value was obtained from at least 20 metaphases.
Results
Disease Anticipation Caused by TERC RNA Haploinsufficiency. Genetic anticipation is when an inherited disease manifests at increasingly younger ages and/or with increased severity with each succeeding generation. Thus, if the offspring of patients develop the disease, they will tend to do so at an earlier age and display more severe clinical manifestations than their parents. A new mechanism of anticipation was recently proposed in patients with AD DC in whom disease anticipation is associated with progressive shortening of telomeres with subsequent generations (16). According to this model, the disease is caused by the inheritance of a mutated TERC gene and the inheritance of short telomeres. To further investigate the inheritance of acquired changes in telomere length, we studied clinical features and telomere length in an extended family with AD DC due to a large TERC gene deletion. The 821-bp TERC gene deletion in this family removes the last 74 bp of the TERC RNA, including the conserved H box and ACA sequences, essential for the stability of the TERC RNA (5). Thus, in this family, AD DC is caused by TERC RNA haploinsufficiency. All TERC gene deletion carriers had at the time of the blood draw at least one clinical feature characteristic of DC including abnormal skin pigmentation, fingernail changes, and leukoplakia. Eight have or have had on a previous occasion abnormal peripheral blood values including macrocytic anemia, thrombocytopenia, or leukopenia. Table 1, which is published as supporting information on the PNAS web site, summarizes peripheral blood values and physical findings in TERC gene deletion carriers. Three deletion carriers died before this investigation (see Fig. 1). All three had phenotypic features of DC including abnormal blood values, dystrophic fingernails, and abnormal skin pigmentation. The TERC gene deletion carriers in generation GI and in GII died at the age of 68 and 47 because of lung fibrosis and liver cirrhosis, respectively. The affected individual in GIII died at the age of 22 due to pulmonary hemorrhage after allogenic bone marrow transplant for myelodysplastic syndrome with monosomy 7. Sperm preservation before bone marrow transplant revealed azospermia. Physical examination and peripheral blood cell counts were normal in family members with two intact copies of the TERC gene (data not shown). Because of ascertainment bias and a limited follow-up since diagnosis, it is difficult to determine whether the disease occurred earlier or is more severe in subsequent generations. However, the findings that deaths due to disease occurred at a younger age in subsequent generations and that azospermia was found in one GIII family member carrying the TERC gene deletion at the age of 22 but not in affected individuals of GII are consistent with disease anticipation in this family.
TERC Gene Deletion Carriers Have Very Short Telomeres. We used three different methods to determine the dynamics of telomere length; all samples were analyzed by TRFL analysis of DNA isolated from peripheral blood cells and by flow-FISH of PBMC. To determine telomere-length distribution and the parental origin of telomeres, Q-FISH was performed on phytohemagglutinin-stimulated PBMC from selected family members. First, we studied the telomere length in the nine family members with the TERC gene deletion. The average TRFL measured in peripheral blood cells from TERC gene deletion carriers was dramatically shorter (6.68 kb, range 5.53-8.45, SD 1.13), despite their younger age (average age 27.2, range 6-52, n = 9), than that from unrelated family members (spouses, average telomere length 9.15 kb, range 8.56-10.77, SD 1.22, P = 0.0009, average age 43.5, range 33-52, n = 7; see also Fig. 2). Similarly, when measured by flow-FISH the average relative telomere length in PBMC from TERC gene deletion carriers was far below the first percentile of age-matched controls and significantly shorter (7.8, range 7.0-8.4, SD 0.5) compared with unrelated family members (13.2, range 11.2-15.1, SD 1.3, P = 6.6 × 10-9), which had telomere length similar to age-matched control individuals (13.4, SD 2.1, P = 0.8; see also Fig. 3). The results obtained by TRFL and flow-FISH showed a strong correlation (r2 = 0.82, P < 0.0001).
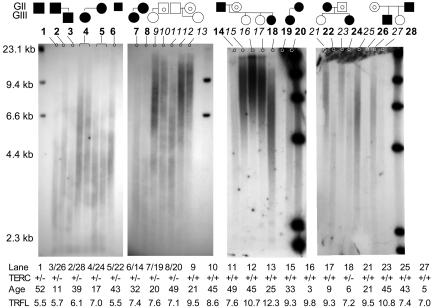
Fig. 2.
TRFL analysis of DNA isolated from PBMC from family members carrying the TERC gene deletion. TRFL of DNA isolated from peripheral blood leukocytes was measured by in-gel hybridization with radioactive labeled telomeric probe. DNA was digested with restriction enzymes RsaI and HinfI before electrophoresis. The family relationships are schematically indicated above. Filled circles and squares indicate females and males carrying the TERC gene deletion, open circle and squares indicate females and males who have two normal copies of the TERC gene but have a parent (G + 1) or grandparent (G + 2) carrying a TERC gene deletion, and circles and squares containing a smaller circle indicate spouses carrying two normal copies of TERC. The table below indicates the genotype the age and the average telomere length measured. Some of the family members are represented more than once with DNA isolated from independent samples. Note that the TRFL is shorter when the affected parent is the father than when the affected parent is the mother.
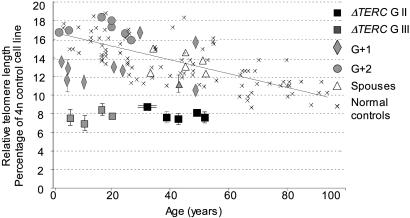
Fig. 3.
Telomeres in PBMC from TERC gene deletion carriers are very short. Relative telomere length in comparison with a control cell line was measured by using flow-FISH and a fluorochrome-labeled telomeric PNA probe. The relative telomere lengths in PBMC isolated from TERC gene deletion carriers in generation GII are shown as filled squares; the relative telomere lengths of TERC gene deletion carriers in generation GIII are shown in gray squares. The telomere lengths of family members with two normal TERC gene copies are shown as gray diamonds (G + 1) and gray circles (G + 2), those of spouses are shown as open triangles, and normal healthy controls are indicated by “×.” Relative telomere length is on the y axis, and the age in years is on the x axis. The means and SDs of several family members in whom the telomere lengths have been determined multiple times are indicated.
The Inheritance of Short Telomeres in TERC Gene Deletion Carriers. As shown in Fig. 3, TERC gene deletion carriers already have very short telomeres at a very young age, whereas telomere lengths in normal controls gradually shorten with age. To further investigate the dynamics of telomere length caused by TERC RNA haploinsufficiency, we compared the absolute telomere lengths in peripheral blood cells from the affected parent and child in each nuclear family within this large pedigree. By flow-FISH, the average telomere length in three of the four children, despite their younger age, was less than in the affected parent. However, this difference did not reach statistical significance (average telomere length: ΔTERC GIII, 7.7, SD 0.6; ΔTERC GII, 8.0, SD 0.6; P = 0.6.) One daughter's mononuclear cells had longer telomeres than the mononuclear cells of her affected mother. TRFL analysis of DNA isolated from peripheral blood of the affected children and their parents showed that two children had shorter TRFLs, whereas in the other two affected children, the TRFLs were slightly longer than in the affected parent (see Fig. 2). These findings were confirmed in two independent blood samples. It is interesting that in both of the cases with longer TRFLs in the child (as well as the one measured by flow-FISH), the affected parent was the mother (see discussion below).
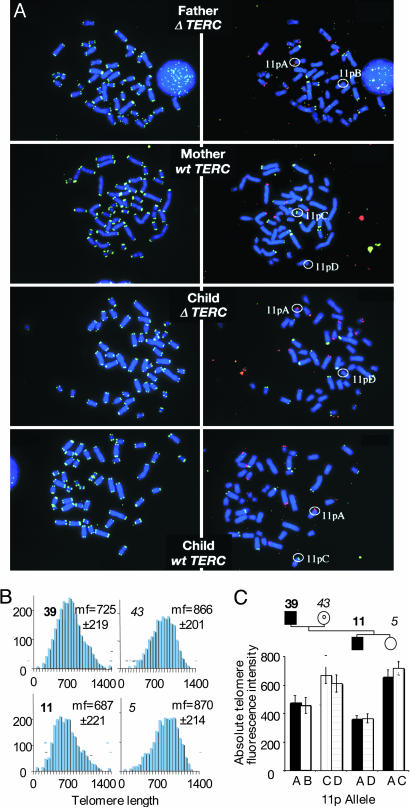
Accelerated Shortening of Long Telomeres in Generations Haploinsufficient for TERC RNA. According to the model of anticipation by telomere shortening, an affected child should inherit a set of short telomeres from his affected parent and a set of normal length telomeres from his unaffected parent. Flow-FISH does not provide information on the length distribution of telomeres in individual cells. TRFL analysis, because of the reduced number of telomeric repeats in the affected individual, shows a telomere signal that is very weak, just above background, with no evidence of a bimodal telomere length distribution (see also Fig. 2). To further investigate the length of paternally and maternally derived telomeres, we compared the intensity of telomere signals within a single metaphase using Q-FISH. Remarkably, in both nuclear families a uniform distribution of telomeres was found in the affected child and in the affected parent, with the child's telomeres being shorter than the parent's (Fig. 4B and data not shown), consistent with the findings by TRFL and flow-FISH. Interestingly, although rare relatively long and relatively very short telomeres were observed in the TERC gene deletion carriers, we did not observe a general broadening of telomere-length distribution, suggesting that the telomeres on chromosomes derived from either parent are equally short (Fig. 4B). To investigate this further, we used subtelomeric polymorphic probes that can distinguish between maternally and paternally inherited telomeres. One nuclear family was informative for the polymorphic probe on chromosome 11p and, in part, for the subtelomeric probe on chromosomes 7p and 1p. Fig. 4A Left shows one representative metaphase from each nuclear family member hybridized to the FITC-conjugated telomeric PNA probe. Fig. 4A Right shows the same metaphases hybridized to the subtelomeric probes distinguishing the parental 11p, 7p, and 1p telomeres. Fig. 4C shows the absolute telomere length of the telomeres on chromosome 11p. Both the paternal 11pA and maternal telomere 11pD telomere are equally short in the affected child; the paternal 11pA in the affected child is shorter than in the affected father. The maternal 11pD telomere is very much shorter than in the nonaffected mother but not significantly different from the length of the paternally derived telomere 11pA (Fig. 4C). Thus, chromosome-specific telomere analysis shows that in the affected child the telomeres inherited from the affected parent are further shortened to some extent but that those inherited from the unaffected parent are shortened extensively to reach a similar length.
Fig. 4.
Q-FISH and identification of chromosome-specific telomere signals. (A) Metaphase spread after hybridization with telomeric PNA (Left) followed by a second hybridization step with polymorphic subtelomeric probes, f7501 and ICRF10 (Right). Chromosomes were counterstained with DAPI. In this particular nuclear family the second step allows distinction between the parental telomeres for chromosome 11p. (B) Quantification of telomere fluorescence in the affected father (age 39), the unaffected mother (age 43), the affected child (age 11), and the nonaffected sibling (age 5). Mean fluorescence (mf) and SD in arbitrary units are shown. Although mean absolute fluorescence intensities are significantly different between affected and healthy individuals, the distribution of relative fluorescence intensities within cells are not (not shown). (C) Absolute fluorescence intensity of the telomere on chromosome 11p. The affected child (filled square) has 11pA (filled bar) from the affected father (filled square) and 11pD (vertically striped bar) from the healthy mother (open circle), whereas the healthy sibling (open circle) has inherited 11pA from her affected father and 11pC (horizontally striped bar) from her unaffected mother. Note that in the affected child, both 11p alleles are shorter than in the parents (P < 0.05 for allele A and P < 0.005 for allele D), whereas in his sibling with two normal TERC genes, allele A is significantly longer (P < 0.05). SEM is indicated.
Restoration of Telomere Length in Individuals with Two Normal TERC Alleles. According to the model of anticipation by telomere shortening, short telomeres would be passed on to the next generation regardless of the inheritance of the TERC gene deletion. We therefore studied telomere length in children of affected parents that did not inherit the TERC gene deletion (G + 1) and compared it (i) with the telomere length of their siblings and parents who carry the TERC gene deletion, (ii) with the telomere length of the non affected parent (spouse), and (iii) with the telomere length of age-matched controls. As shown in Fig. 2, in individuals who have a normal TERC genotype but a heterozygous parent, telomere length is significantly longer compared with the telomere length of their affected siblings or affected parent and similar to the telomere length of the unaffected parent. In comparison with age-matched controls, however, telomere length in the G + 1 generation varies greatly, ranging from below the 1st to the 90th percentile. In the second generation with a normal TERC genotype but a heterozygous grandparent, telomere length was at or above the 50th percentile in all children investigated. Q-FISH analysis of chromosome-specific telomeres in the G + 1 generation revealed a uniform distribution of telomere length (Fig. 4 B and C and data not shown).
Discussion
DC is the first human disease whose pathogenesis has been directly linked to an impairment of telomere maintenance (5, 21, 22). Telomeres are complex nucleoprotein structures at the ends of eukaryotic chromosomes that protect the chromosomes from end-to-end fusion, degradation, and inappropriate recombination. Telomeric DNA consists of tandemly repeated G-rich sequences that are synthesized by the telomerase ribonucleoprotein complex whose integral RNA component, the telomerase RNA or TERC RNA, contains the sequences that act as a template for the synthesis of these repeats (23-25). The telomeric repeats serve as a scaffold for many of the proteins involved in telomere maintenance (26). Thus, a minimal number of telomeric repeats is required to ensure proper telomere function. When telomeres become critically short, cell-cycle arrest or cell death occurs (27). Telomere length is highly variable between different species. Within an individual species the number of telomeric repeats is usually maintained within a well defined range. In humans, telomeres shorten with age (28, 29). In peripheral blood cells, rapid telomere shortening occurs during the first year of life, followed by a more gradual decline over time (30, 31). Although considerable variation is present among individuals of the same age, reports based on twin studies showed that telomere length is familial (32). The relative lengths of individual telomeres are defined in the zygote and strictly maintained during life (33).
Patients with DC have very short telomeres (6). Patients with AD DC are heterozygous for mutations in the TERC gene. The absence of a dominant negative effect in in vitro analysis of telomerase activity indicates that haploinsufficiency is the mechanism for telomere shortening in TERC gene mutation carriers. Interestingly, the extent to which the telomerase activity is impaired by the TERC gene mutations varies, suggesting that the balance of telomere maintenance is very subtle because even a mild impairment may lead to telomere shortening and disease (reviewed in ref. 34). Here, we study the telomere length dynamics in a large family with AD DC haploinsufficient for the TERC RNA due to a large TERC gene deletion. The investigation of telomere length within a single family has the advantage that the molecular lesion responsible for telomere shortening is uniform and that the contribution of other genetic components influencing telomere length is similar. Our investigations show that in all TERC gene deletion carriers, the telomeres measured in peripheral blood cells are excessively short, far shorter than any telomeres measured in healthy controls (Figs. 2 and 3). The average telomere length in all TERC gene deletion carriers appeared to be similar. In contrast to normal controls, there was no apparent age-dependent decrease in telomere size, even when TERC gene deletion carriers in GII and GIII were analyzed separately. The similar absolute average telomere length in the affected members of this family may be explained in two ways. (i) GIII family members have inherited preshortened telomeres, resulting in a similar telomere length in GIII and GII family members despite their age difference. Unfortunately, we do not have a long-term follow-up of telomere length in this family, nor do we know for how many generations the TERC gene deletion has been inherited in this family. (ii) An alternative explanation for the lack of an age-dependent decrease in telomere length in TERC gene mutation carriers, which we actually favor, might be that, in this family, circulating PBMC have reached a minimal telomere length and that further shortening of telomeres would prevent the cells from contributing to the peripheral blood cell pool. Indeed, the peripheral blood cell counts were lower in TERC gene deletion carriers compared with the unaffected family members, and seven out of nine deletion carriers had abnormal blood values (see Table 1), suggesting a reduced contribution of the bone marrow progenitor cells to the peripheral blood pool and a stressed hematopoiesis. Naturally, these two explanations are not mutually exclusive. The analysis of two generations of mutation carriers did not reveal whether this minimal telomere length is due to consecutive shortening of telomeres over several generations or whether telomeres are already critically short in the first generation carrying the TERC gene deletion. However, the clinical assessment is consistent with disease anticipation, and that in this family, with increasing generations the critical telomere length is reached at an earlier age due to the inheritance of preshortened telomeres. The analysis of telomere length in sperm would be interesting and strengthen the hypothesis of disease anticipation caused by the inheritance of short telomeres. The findings in our family suggest that at the time of disease manifestation, the PBMC show minimal telomere length necessary for cell survival and that the onset of disease and disease severity is dictated by the number of cells, most likely progenitor cells, with telomeres below the minimal length incompatible with cell proliferation or survival.
Analysis of telomeres by using TRFL revealed that in two heterozygous children the telomeres were shorter, whereas in another two they were longer, than those of their affected parent (see Fig. 3). TRFL, in contrast to flow-FISH, analyzes telomeres in DNA isolated from mononuclear and polymorphonuclear cells. This may explain why by flow-FISH only in one child, but by the TRFL analysis in two children, the telomeres were longer than in their affected parents. In individuals with bone marrow failure, telomere shortening is usually more pronounced in the polymorphonuclear cells compared with the mononuclear cells, which are mainly lymphocytes. Interestingly, in both cases the transmitting parents were the mothers who had similar TRFL to their affected brothers but had shorter TRFL than their children. This finding raises the interesting possibility that affected fathers transmit shorter telomeres than affected mothers. Telomerase is highly expressed in the oogonia and in the spermatogonia. In humans, mature sperm have longer telomeres than most somatic tissues (28, 35). Mitosis of spermatogonia occurs throughout adult life, whereas it ceases at 25 weeks of gestation in the oogonia, thus providing a possible reason why haploinsufficiency may affect telomeres in the sperm more than in the oogonia. However, to prove this point, more large-family studies would have to be conducted.
Q-FISH analysis confirmed the presence of short telomeres in deletion carriers. The mean metaphase intensities observed were consistent with flow-FISH and TRFL analyses. In the TRFL analysis the hybridization signal from the very short telomeres is weak and difficult to distinguish from the background caused by the hybridization to repeats in subtelomeric regions, resulting in a rather broad hybridization signal. However, when using Q-FISH analysis, which allows a very accurate comparison of telomere lengths within an individual metaphase, the relative telomere length distribution within cells was close to normal rather than bimodal, and no broadening of the frequency distribution of telomere length was observed (data not shown). Interestingly, we did not observe a narrowing of the telomere-length distribution either, as one would expect if telomerase only acted on the shortest telomere. It would appear that a normal relative length distribution is maintained in cells even when telomeres are becoming critically short. Q-FISH analysis in TERC gene deletion carriers in combination with probes distinguishing the paternal from the maternal telomere showed equally short lengths of telomeres derived from the affected and nonaffected parent. This finding implies that the diminished telomerase activity preferentially adds telomeric repeats onto the shortest class of chromosomes. On the other hand, in the children who inherited two normal copies of TERC the telomeres derived from the affected parent were similar in length to those derived from the unaffected parent, suggesting that restored telomerase activity reestablishes the normal telomere length. However, in six out of eight such children the average telomere length in PBMC was shorter than in age-matched controls. After two generations, inheriting two normal TERC genes (G + 2) telomere length was equal to or higher than in age-matched controls, indicating that two generations are needed to restore the original equilibrium of telomere maintenance. No clinical manifestations were observed in G + 1 individuals even when telomeres where shorter than in age-matched controls, suggesting that normal telomerase activity precludes the manifestation of DC or, if our interpretation is correct, prevents significant cell-cycle arrest or cell death in progenitor cells due to short telomeres. The unimodal distribution of telomere length in these children indicates that with normal levels of telomerase activity telomeric repeats once more are preferentially added to the shortest telomeres.
The priority of telomerase to maintain the shortest telomeres has elegantly been shown in mice with short telomeres due to telomerase deficiency. Mice with loss of telomerase function (telomerase-null mice) due to either a of Terc gene deletion (Terc RNA -/- mice) or a null mutation in the Tert gene, encoding the catalytic component of telomerase (Tert -/- mice), show heritable progressive telomere shortening resulting in a “short telomere phenotype” resembling the clinical features seen in patients with DC (22). Three to six generations of inbreeding is needed to produce a “short telomere phenotype” (22, 36-39). The reintroduction of one normal copy of the deleted gene did not have a major effect on the average telomere length in mice but reduced the numbers of very short telomeres (40-42), indicating that with limiting levels telomerase acts preferentially on the shortest telomeres. In mice, as in humans, telomerase haploinsufficiency is associated with telomere shortening, but, in contrast to humans, mice, even after 10 generations, do not develop a “short telomere phenotype” characterized by an increased rate of apoptosis and increased frequency of chromosomal fusions (38, 43). Interestingly, in the patients with TERC haploinsufficiency studied here, metaphase analysis of peripheral mononuclear cells failed to show end-to-end fusions of chromosomes or telomere free ends (data not shown), suggesting that in phytohemagglutinin-responsive peripheral blood lymphocytes, residual telomerase activity maintains the integrity of telomeres.
In summary, telomere analysis in our family suggests that in cells expressing telomerase, as in activated lymphocytes, the residual level of telomerase is sufficient to rescue the shortest telomeres but leads to accelerated shortening of the longer telomeres and the accumulation of cells with very short telomeres. Eventually, the limited amount of active telomerase in TERC RNA haploinsufficiency may not be able to maintain the minimal length of the increasing number of short telomeres. Thus, the number of cells with excessively short telomeres and the degree of residual telomerase activity may determine the onset and severity of disease in TERC RNA haploinsufficiency and in families and patients with AD DC.
Supplementary Material
Acknowledgments
We thank Sheila Stewart for interesting discussion and suggestions; the Hereditary Cancer Core, in particular Jennifer Ivanovich, for help; and the Procurement Core of the Alvin J. Siteman Cancer Center at Washington University (National Cancer Institute Cancer Center Support Grant P30 CA91842) for patient accruement and DNA isolation. This work was supported by National Institutes of Health Grant R01 1RO-1 CA105312, American Cancer Society Opportunity Grant ROG-03-104-01, and the Bursary Award from the Aplastic Anemia and MDS International Foundation. Work in the laboratory of A.L.-V. was supported by Association pour la Recherche sur le Cancer Grant ARC3588.
Author contributions: M.B. designed research; R.B., S.K., S.F., H.-Y.D., A.L.-V., and M.B. performed research; L.H. and A.L.-V. contributed new reagents/analytic tools; P.J.M., A.L.-V., and M.B. analyzed data; P.J.M. and M.B. wrote the paper; and F.G. accrued and examined the study participants.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AD, autosomal dominant; DC, dyskeratosis congenita; flow-FISH, FISH analysis by flow cytometry; PBMC, peripheral blood mononuclear cells; Q-FISH, quantitative FISH; TRFL, terminal restriction fragment length.
References
- 1.Cole, H., Rauschkolb, J. & Toomey, J. (1930) Arch. Dermatol. Syphiligraph. 21, 71-95. [Google Scholar]
- 2.Knight, S., Vulliamy, T., Copplestone, A., Gluckman, E., Mason, P. & Dokal, I. (1998) Br. J. Haematol. 103, 990-996. [DOI] [PubMed] [Google Scholar]
- 3.Drachtman, R. A. & Alter, B. P. (1992) Am. J. Pediatr. Hematol. Oncol. 14, 297-304. [PubMed] [Google Scholar]
- 4.Heiss, N. S., Knight, S. W., Vulliamy, T. J., Klauck, S. M., Wiemann, S., Mason, P. J., Poustka, A. & Dokal, I. (1998) Nat. Genet. 19, 32-38. [DOI] [PubMed] [Google Scholar]
- 5.Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P. J. & Dokal, I. (2001) Nature 413, 432-435. [DOI] [PubMed] [Google Scholar]
- 6.Vulliamy, T. J., Knight, S. W., Mason, P. J. & Dokal, I. (2001) Blood Cells Mol. Dis. 27, 353-357. [DOI] [PubMed] [Google Scholar]
- 7.Mason, P. J., Wilson, D. & Bessler, M. (2005) Curr. Mol. Med. 5, 159-170. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell, J. R., Cheng, J. & Collins, K. (1999) Mol. Cell. Biol. 19, 567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blasco, M. A. (2005) EMBO J. 24, 1095-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allshire, R. C., Dempster, M. & Hastie, N. D. (1989) Nucleic Acids Res. 17, 4611-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsyth, N. R., Wright, W. E. & Shay, J. W. (2002) Differentiation 69, 188-197. [DOI] [PubMed] [Google Scholar]
- 12.Feng, J., Funk, W. D., Wang, S. S., Weinrich, S. L., Avilion, A. A., Chiu, C. P., Adams, R. R., Chang, E., Allsopp, R. C., Yu, J., et al. (1995) Science 269, 1236-1241. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura, T. M., Morin, G. B., Chapman, K. B., Weinrich, S. L., Andrews, W. H., Lingner, J., Harley, C. B. & Cech, T. R. (1997) Science 277, 955-959. [DOI] [PubMed] [Google Scholar]
- 14.Kim, N. W., Piatyszek, M. A., Prowse, K. R., Harley, C. B., West, M. D., Ho, P. L., Coviello, G. M., Wright, W. E., Weinrich, S. L. & Shay, J. W. (1994) Science 266, 2011-2015. [DOI] [PubMed] [Google Scholar]
- 15.Masutomi, K., Yu, E. Y., Khurts, S., Ben-Porath, I., Currier, J. L., Metz, G. B., Brooks, M. W., Kaneko, S., Murakami, S., DeCaprio, J. A., et al. (2003) Cell 114, 241-253. [DOI] [PubMed] [Google Scholar]
- 16.Vulliamy, T., Marrone, A., Szydlo, R., Walne, A., Mason, P. J. & Dokal, I. (2004) Nat. Genet. 36, 447-449. [DOI] [PubMed] [Google Scholar]
- 17.Wynn, R. F., Cross, M. A., Hatton, C., Will, A. M., Lashford, L. S., Dexter, T. M. & Testa, N. G. (1998) Lancet 351, 178-181. [DOI] [PubMed] [Google Scholar]
- 18.Knudson, M., Kulkarni, S., Ballas, Z. K., Bessler, M. & Goldman, F. (2005) Blood 105, 682-688. [DOI] [PubMed] [Google Scholar]
- 19.Londoño-Vallejo, J. A., DerSarkissian, H., Cazes, L. & Thomas, G. (2001) Nucleic Acids Res. 29, 3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Der-Sarkissian, H., Vergnaud, G., Borde, Y. M., Thomas, G. & Londoño-Vallejo, J. A. (2002) Genome Res. 12, 1673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, J. R., Wood, E. & Collins, K. (1999) Nature 402, 551-555. [DOI] [PubMed] [Google Scholar]
- 22.Marciniak, R. A., Johnson, F. B. & Guarente, L. (2000) Trends Genet. 16, 193-195. [DOI] [PubMed] [Google Scholar]
- 23.Greider, C. W. & Blackburn, E. H. (1985) Cell 43, 405-413. [DOI] [PubMed] [Google Scholar]
- 24.Yu, G. L., Bradley, J. D., Attardi, L. D. & Blackburn, E. H. (1990) Nature 344, 126-132. [DOI] [PubMed] [Google Scholar]
- 25.Blackburn, E. H. (2000) Nature 408, 53-56. [DOI] [PubMed] [Google Scholar]
- 26.Smogorzewska, A. & de Lange, T. (2004) Annu. Rev. Biochem. 73, 177-208. [DOI] [PubMed] [Google Scholar]
- 27.Wright, W. E. & Shay, J. W. (2001) Curr. Opin. Genet. Dev. 11, 98-103. [DOI] [PubMed] [Google Scholar]
- 28.Hastie, N. D., Dempster, M., Dunlop, M. G., Thompson, A. M., Green, D. K. & Allshire, R. C. (1990) Nature 346, 866-868. [DOI] [PubMed] [Google Scholar]
- 29.Allsopp, R. C., Vaziri, H., Patterson, C., Goldstein, S., Younglai, E. V., Futcher, A. B., Greider, C. W. & Harley, C. B. (1992) Proc. Natl. Acad. Sci. USA 89, 10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rufer, N., Brummendorf, T. H., Kolvraa, S., Bischoff, C., Christensen, K., Wadsworth, L., Schulzer, M. & Lansdorp, P. M. (1999) J. Exp. Med. 190, 157-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson, J. D., Gale, R. E., Wynn, R. F., Dougal, M., Linch, D. C., Testa, N. G. & Chopra, R. (2000) Br. J. Haematol. 109, 272-279. [DOI] [PubMed] [Google Scholar]
- 32.Slagboom, P. E., Droog, S. & Boomsma, D. I. (1994) Am. J. Hum. Genet. 55, 876-882. [PMC free article] [PubMed] [Google Scholar]
- 33.Graakjaer, J., Pascoe, L., Der-Sarkissian, H., Thomas, G., Kolvraa, S., Christensen, K. & Londoño-Vallejo, J. A. (2004) Aging Cell 3, 97-102. [DOI] [PubMed] [Google Scholar]
- 34.Bessler, M., Wilson, D. B. & Mason, P. J. (2004) Rev. Curr. Opin. Pediatr. 16, 23-28. [DOI] [PubMed] [Google Scholar]
- 35.de Lange, T., Shiue, L., Myers, R. M., Cox, D. R., Naylor, S. L., Killery, A. M. & Varmus, H. E. (1990) Mol. Cell. Biol. 10, 518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A. & Greider, C. W. (1997) Cell 91, 25-34. [DOI] [PubMed] [Google Scholar]
- 37.Herrera, E., Samper, E., Martin-Caballero, J., Flores, J. M., Lee, H. W. & Blasco, M. A. (1999) EMBO J. 18, 2950-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Y., Snow, B. E., Hande, M. P., Yeung, D., Erdmann, N. J., Wakeham, A., Itie, A., Siderovski, D. P., Lansdorp, P. M., Robinson, M. O. & Harrington, L. (2000) Curr. Biol. 10, 1459-1462. [DOI] [PubMed] [Google Scholar]
- 39.Niida, H., Shinkai, Y., Hande, M. P., Matsumoto, T., Takehara, S., Tachibana, M., Oshimura, M., Lansdorp, P. M. & Furuichi, Y. (2000) Mol. Cell. Biol. 20, 4115-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemann, M. T., Strong, M. A., Hao, L. Y. & Greider, C. W. (2001) Cell 107, 67-77. [DOI] [PubMed] [Google Scholar]
- 41.Samper, E., Flores, J. M. & Blasco, M. A. (2001) EMBO Rep. 2, 800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erdmann, N., Liu, Y. & Harrington, L. (2004) Proc. Natl. Acad. Sci. USA 101, 6080-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, H. W., Blasco, M. A., Gottlieb, G. J., Horner, J. W., II, Greider, C. W. & DePinho, R. A. (1998) Nature 392, 569-574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.