Abstract
Aphids maintain mutualistic symbioses involving consortia of coinherited organisms. All possess a primary endosymbiont, Buchnera, which compensates for dietary deficiencies; many also contain secondary symbionts, such as Hamiltonella defensa, which confers defense against natural enemies. Genome sequences of uncultivable secondary symbionts have been refractory to analysis due to the difficulties of isolating adequate DNA samples. By amplifying DNA from hemolymph of infected pea aphids, we obtained a set of genomic sequences of H. defensa and an associated bacteriophage. H. defensa harbors two type III secretion systems, related to those that mediate host cell entry by enteric pathogens. The phage, called APSE-2, is a close relative of the previously sequenced APSE-1 but contains intact homologs of the gene encoding cytolethal distending toxin (cdtB), which interrupts the eukaryotic cell cycle and which is known from a variety of mammalian pathogens. The cdtB homolog is highly expressed, and its genomic position corresponds to that of a homolog of stx (encoding Shiga-toxin) within APSE-1. APSE-2 genomes were consistently abundant in infected pea aphids, and related phages were found in all tested isolates of H. defensa, from numerous insect species. Based on their ubiquity and abundance, these phages appear to be an obligate component of the H. defensa life cycle. We propose that, in these mutualistic symbionts, phage-borne toxin genes provide defense to the aphid host and are a basis for the observed protection against eukaryotic parasites.
Keywords: Acrythosiphon pisum, Hamiltonella defensa, lamboid phage, aphid, Enterobacteriaceae
Aphids possess several types of symbiotic bacteria, which are now known to have a variety of effects on host growth and survival. Pea aphids (Acyrthosiphon pisum) always contain the obligate primary symbiont, Buchnera aphidicola, which provisions nutrients lacking in the diet. They can additionally harbor any of several facultative “secondary” symbionts. One of these is “Candidatus Hamiltonella defensa” (previously known as “T type” or “PABS”, ref. 1), a member of the Enterobacteriaceae that is found in aphids as well as other insect families (1-5). In pea aphids, the effects of attack by parasitoid wasps are ameliorated by infection with H. defensa: the wasp larva dies prematurely, allowing the aphid host to develop to the adult stage and to reproduce (6). Microscopy has revealed that H. defensa can live within the bacteriocytes that normally house Buchnera, within some other cell types, and extracellularly in the insect hemocoel (1, 2). In a European pea aphid strain, secondary symbionts were found to harbor an active bacteriophage, designated APSE-1; its genome sequence indicated a relationship to P22, the lambdoid phage of Salmonella enterica (7, 8). The secondary symbiont serving as host to APSE-1 was not identified initially, but, based on a survey of aphid species harboring different symbionts (2), APSE-1-like phage appear to be consistently and exclusively associated with H. defensa.
Many bacteria that live as symbionts within insects are closely related to well-studied mammalian pathogens. This close relationship is especially apparent in the Enterobacteriaceae, which includes H. defensa and other secondary symbionts of insects (2, 4, 9-12), the symbiotic Photorhabdus and Xenorhabdus species, as well as the human pathogens S. enterica, Shigella flexneri, and Yersinia pestis. Recent genomic and experimental studies have revealed that these symbionts deploy systems for infecting host cells similar to those used by mammalian pathogens (e.g., refs. 13 and 14). In the tsetse fly symbiont, Sodalis glossinidius, for example, two type III secretion systems (TTSS) are present, and at least one is required for invasion of host cells (15, 16), as is true of many pathogens, including S. enterica and Y. pestis. A TTSS also appears to underlie infection of host cells by the symbionts of Sitophilus weevils (17) and by Photorhabdus species infecting nematodes (18). Such pathogenicity determinants are often transferred horizontally among enteric bacteria via bacteriophage vectors; these transfers have a role in transforming a nonpathogenic to a pathogenic strain (19-21). Because both pathogenesis and symbiosis share the requirement of bacterial establishment within host cells or tissues, additional mechanisms, already characterized in pathogens, may be incorporated into the symbiotic lifestyle or may underlie variation among symbionts in their effects on hosts.
The major obstacle to the genetic and genomic characterization of most insect symbionts has been the inability to culture these organisms, thereby preventing many experimental analyses and thwarting attempts to isolate large amounts of pure DNA for shotgun sequencing. Primary symbionts are typically extremely numerous or packed within discrete organs, facilitating the recovery and sequencing of their genomic DNA without massive host contamination (e.g., refs. 22-26). However, some symbionts, including H. defensa and other insect secondary symbionts within Enterobacteriaceae, do not occupy specialized organs and reside at low titers within hosts. In such cases, DNA purification, if possible, is labor-intensive (e.g., ref. 27). As a result, we know almost nothing about the genomic content of these organisms or the mechanisms underlying their effects on hosts.
Here we report findings based on a method for whole genome amplification (WGA) of low copy number DNA template from H. defensa of pea aphids. Sequencing clones derived by this method demonstrated that the symbiont contains a diverse set of genes associated with pathogenicity in mammalian hosts, and at least two phage closely related to APSE-1 but harboring different “cargo” genes, including cdtB (cytolethal distending factor), a toxin-encoding gene previously known only from several invasive mammalian pathogens. We postulate that these phage and their toxin-encoding genes are an integral part of the aphid-bacterium mutualism.
Materials and Methods
DNA Sample. Because secondary symbionts, but not Buchnera, occur in appreciable numbers in the aphid hemocoel (1, 28), we considered hemolymph to be the optimal source for symbiont DNA without accompanying host or Buchnera DNA. Our source was an isofemale line of A. pisum infected four years earlier with H. defensa; this line, called “5AT,” is the line in which H. defensa was previously shown to provide the aphid hosts with defense against parasitoid wasps (6). Adult wingless females were sampled from the 5AT stock colony maintained continuously at 20°C, 16 h light/8 h dark. To collect hemolymph enriched in H. defensa, females were pricked in the abdomen, and if a clear droplet was exuded, it was collected with a microcapillary tube. (Cloudiness in the exuded droplet indicated that aphid cells, such as bacteriocytes housing the primary symbiont B. aphidicola, had been ruptured; such samples were discarded.) Hemolymph was pooled from four aphids, yielding a total volume of ≈0.1 μl, and used as template for WGA using the GenomiPhi kit (Amersham Pharmacia). Amplified genomic DNA was hydrosheared, endrepaired, size-selected, and cloned by using the TOPO TA (Invitrogen) cloning kit, according to the supplier's protocol.
Insert Amplification. Cloned inserts were PCR-amplified by using the M13 forward and reverse primers. Reactions conditions were 95°C (3 min), 34 cycles of 94°C (1 min), 55°C (1 min), 72°C (1 min), then 72°C (6 min). PCR products were sized and visualized on agarose gels, and those yielding a single band >1 kb in length were sequenced bidirectionally with the M13 primers.
Sequence Analyses. Sequences were initially screened and binned as phage, H. defensa, Buchnera, or host insect based on blastx similarity as follows. Any sequences with APSE-1 as the first hit were classified as phage. Because the Buchnera genome for this host species is fully sequenced (22), sequences from Buchnera were unambiguous as near perfect matches (>99% identity) with the sequenced genome. Hits to other bacteria were initially assigned as potential H. defensa sequences. ORFs with closest hits to Drosophila melanogaster or other animals, as well as noncoding sequences, were considered to be likely aphid nuclear sequences. (We expect most aphid sequences to correspond to fast-evolving intergenic regions lacking detectable homology to sequences in the database; no genome sequence for aphids or any closely related insect is available.) Sequences that did not meet criteria for any bin were temporarily placed in every bin to determine whether they formed contigs with sequences for which assignments were clearcut. Paired-end reads were placed in the bin with their mate. Also, assembly was attempted combining sequences binned as phage and as H. defensa, to uncover phage genes within the bacterial chromosome. Sequences ultimately designated as H. defensa met the following criteria: presence of ORFs with base composition of 30-45% G+C, top hit to Enterobacteriaceae, near-perfect match to one of the previously sequenced genes including the rRNA genes, paired-end mate of a sequence meeting any of the above criteria, or strongly supported assembly with a sequence meeting any of the above.
Sequence Assembly. Assemblies were generated by using the Institute for Genomic Research (TIGR) assembler version 2.0 (29). Manual editing was performed in consed version 14 (30). Resulting contigs were searched for ORFs (31). Gene names and functions were assigned based on sequence identity of the translated predicted ORFs as determined by blastp against the nonredundant (nr) database during March 2005. Assemblies for H. defensa were regarded as tentative and were deposited as individual sequences rather than contigs (GenBank accession numbers DQ163092 and DQ163903), except for assemblies for the type three secretions systems, which were deposited as contigs (accession numbers DQ092620-DQ092623). Phage assemblies were verified by PCR, and deposited in GenBank as contigs (accession numbers DQ092612-DQ092619).
Quantitative PCR for Gene Copy Number and Transcription Level of cdtB. We used three samples of aphid hemolymph to estimate the number of bacterial chromosomes that were used as template in the WGA. Copy number of a single copy bacterial gene (dnaK) was estimated with real-time quantitative PCR (qPCR) to index the numbers of bacterial chromosomes in each sample. Based on the initial sequences derived from the WGA experiments, we designed H. defensa-specific qPCR primers to dnaK (T70F2: 5′-GGT TCA GAA AAA AGT GGC AG-3′ and T70R2: 5′-CGA GCG AAA GAG GAG TGA C-3′). Reactions were carried out in a Roche Lightcycler using a touchdown PCR procedure as follows: 95°C (10 min), 40 cycles with denaturing temperature of 95°C (5 s), annealing temperature decreasing from 68°C to 55°C (15 s), and extension temperature of 72°C (5 s).
In addition, we used DNA samples isolated from entire, individual insects to estimate the ratio of phage genes to one another and to the single copy dnaK. Primers were designed for sequences corresponding to the homolog of P2 of APSE-1 (APSEP2F1: 5′-GTC CAG GCA TTA TTA GCG C-3′ and APSEP2R1: 5′-CAA TTT TTC TAA GGC AAC CAT G-3′), the homolog of P28 of APSE-1 (APSEP28F: 5′-TGA TAA AAG CGG ATA ATG CC-3′ and APSEP28R: 5′-GCG TTT GTC ATA CTG AAA AGG-3′), and a homolog of cdtB (ApTcdtB443F: 5′-ATA TTT TTT TTA CCG CCC CG-3′ and ApTcdtB560R: 5′-CCA GCT TCA TTT CTA CCA CCT C-3′). Reactions were performed as for dnaK.
Reverse transcriptase qPCR (RT-qPCR) was used to assess transcript levels of cdtB and two phage genes, P2 and P28 (as designated in ref. 7) as well as dnaK, a highly expressed bacterial gene. The primers were the same as those listed above, and RT-qPCR protocols followed those of Moran et al. (32).
Amplification and Cloning of cdtB-Containing Variable Region of the Phage Genome. Because assemblies from the WGA sequences indicated two versions of some sections of the phage chromosome, we used PCR to amplify a ≈6.5-kb region, which was subsequently cloned and sequenced to verify assemblies. Primers were APSE3.9F (5′-CTT CGT GCT CAG ATG AGG ATG-3′) and 8.1R (5′-TTT CAG GCG GCT GTT CTC AAC-3′) corresponding to positions near 3900 and 8100 on the APSE-1 chromosome (7). The PCR protocol was similar to the touchdown procedure described above except with denaturation at 94°C (1 min), annealing temperature dropping from 65°C to 55°C in 10 successive cycles (1 min), and extension at 72°C (2.25 min), then 28 additional cycles with a 55°C annealing temperature.
Four of the resulting cloned products were completely sequenced by primer-walking beginning with the M13 primers. The resulting fragments were assembled in sequencher version 4.2. As a control, we also amplified, cloned, and sequenced the corresponding fragments from the aphid stock that served as the original source of the H. defensa isolate (A. pisum line 8-2-B).
Distribution of Phage Genes in Different Isolates of H. defensa. To determine the distribution of phages and of toxin-encoding genes among H. defensa isolates, we screened DNA samples from insects previously shown to contain H. defensa (2, 4). These included other A. pisum collections, other aphid species, and one psyllid and one whitefly species (Table 1). PCR primers were designed to detect close homologs of several regions of the APSE genome, including conserved regions common to all isolates, as well as the stx homolog from APSE-1 (P7 in ref. 7) and the cdtB sequences present in our assemblies. Primers for phage genes were APSEP2F1 and APSEP2R1, APSE3.9F and APSE8.1R (all listed above), and APSE3.6F (5′-GGA GCA AAA AAA CAT GAG CAG-3′) and APSE4.2R (5′-CTC CGG GTC CAT GTC TAA TCG-3′). In addition, primers for P7 were APSEP7F (5′-CAA CTT TAC TCT TTT CGT GCG-3′) and APSEP7R (5′-TGT TAC CTT TTC ACC TAC GGC-3′); and for cdtB, cdtB F1 (5′-CCA ATA TCC TAC TCT CAG AGC-3′), cdtB R1 (5′-GAA GGA TTC ATA CTA AGC TGC-3′), and cdtB R2 (5′-GCT GAC ATG GTG ATC ACT ATG AG-3′). For the Shiga-toxin (stx) homolog, primers were APSE P7F (5′-CAA CTT TAC TCT TTT CGT GCG-3′) and APSE P7R (5′-TGT TAC CTT TTC ACC TAC AGC-3′).
Table 1. General features of H. defensa and APSE-2 predicted genes.
| Feature | Number |
|---|---|
| H. defensa | |
| Total number of predicted CDS | 473 |
| E. coli homologs with known function | 249 |
| Non-E. coli homologs with known function | 88 |
| Hypothetical or putative function | 135 |
| TTSS genes | 21 |
| Toxin genes (e.g., leukotoxin A, RTX) | 6 |
| APSE-2 | |
| Total number of predicted CDS | 57 |
| APSE-1 homologs | 48 |
| Homologs to other genes in other phage | 6 |
| Putative ORFs | 3 |
Three percent of clones were assigned to Buchnera, and 26% were excluded because of sequence quality or host contamination.
Electrophoretic Determination of H. defensa Chromosome Size. We used samples of two lines of the same aphid clone, one infected with H. defensa (5AT) and one lacking secondary symbionts (5A). The latter served as a control for discrimination between DNA fragments corresponding to H. defensa and its phage (present only in the infected line) and those from the aphid, Buchnera, or other organisms (such as insect viruses) present in both lines. For each sample, 1-2 g of mixed-age insects were crushed in 15 ml of PBS and passed through a 100-μm filter. Debris on the filter was collected in 15 ml of PBS and passed again through a 100-μm filter. The combined sample was centrifuged at 170 × g for 25 min at 4°C, the supernatant collected and passed through a 20-μm and then an 11-μm filter. Cells were collected by centrifugation at 1,500 × g at 4°C. The pellet was resuspended in 600 μl of PBS containing 1% proteinase K, and mixed with 1.5 ml of 1% agarose in TE to form plugs. The agarose plugs containing filter-purified cells were then subjected to a series of six washes, each at 55°C for 30 min: twice with lysis buffer with proteinase K, once in water, and three times in TE. Plugs were stored at 4°C in TE. Digestion with AscI and NotI was carried out as in Moran et al. (33). These enzymes were selected because neither cuts the sequenced Buchnera genome (22), and because they were expected to cut the H. defensa genome several times based on the base composition (39% G+C) of sequences assigned to H. defensa. Pulsed-field gel electrophoresis (PFGE) proceeded as described (33) with runs of different pulse times to resolve bands of different size ranges. Conditions for discriminating smaller fragments were 200 V, 5-40 s for 22 h; conditions for discriminating larger fragments were 200 V, 70 s for 18 h followed by 160 V, 120 s, for 20 h.
To determine whether phages existed as autonomous elements, as temperate prophage in the H. defensa chromosome, or both, we searched by Southern blotting for large DNA fragments containing known phage genes. DNA from the PFGE gels was transferred to nylon membranes, which were probed with fragments corresponding to cdtB and P2.
Results
Sequences Assigned to H. defensa. A total of 3,650 sequences were obtained by sequencing both ends of 1,825 clones recovered from the WGA of DNA present in aphid hemolymph. Most clones could be assigned to an APSE-1-like phage (hereby designated as APSE-2) or to H. defensa, based on criteria outlined in Materials and Methods. Assemblies of the H. defensa sequences yielded 416 contigs containing 473 ORFs with orthologs in numerous related Gammaproteobacteria (Table 1 and Data Set, which is published as supporting information on the PNAS web site). These sequences were consistently A+T-biased (39% G+C), and several showed exact or nearly exact (>99%) identity to the few available sequences for H. defensa [16S rDNA, 23S rDNA, and gyrB (1, 2)]. None matched (>95% amino acid identity) any other sequence in GenBank, including our vector organism Escherichia coli; furthermore, all four of the rDNA clones sampled corresponded to H. defensa, indicating that contamination from other bacteria was absent or rare. H. defensa sequences contained homologs of genes for metabolic, replicative, and structural pathways that are characteristic of bacteria.
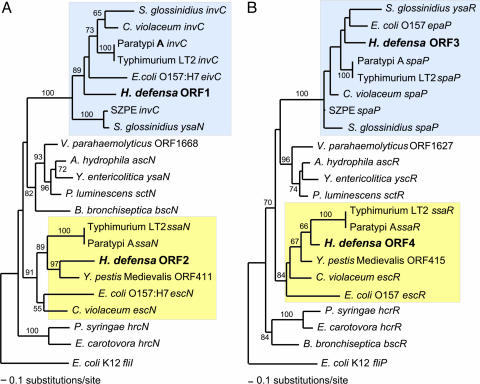
Of the sequenced bacterial ORFs, 22 were homologous to TTSS genes. For several of the TTSS gene families, two distinct homologs are present in H. defensa and these formed two sets of contigs corresponding to portions of two TTSS-encoding regions. Phylogenetic analysis of these sequences revealed that they belong to subfamilies corresponding to the two TTSS encoded in the SPI-1 and SPI-2 islands of S. enterica and as present in other Proteobacteria (Fig. 1). Other genes in H. defensa that have likely roles in its relationship to its host are fur, which encodes the ferric uptake regulation protein involved in regulating lysis in lambda-like phage containing stx. Also, the genome contains multiple genes in the pore-forming RTX toxin pathway (rtx) and leukotoxin (ltx) genes.
Fig. 1.
Phylogenetic analyses of homologs of genes encoding components of type III secretion systems (TTSS), including those newly discovered in H. defensa. (A) invC/ssaN. (B) spaP/ssaR; H. defensa possesses genes for two TTSS, corresponding to those specified within the SPI-1 and SPI-2 pathogenicity islands of Salmonella enterica (highlighted in blue and yellow, respectively). Identical results were obtained for homologs to spaQ/ssaS and to spaR/ssaT. Analyses were based on nucleotide alignments fit to alignments of inferred amino acid sequences by using CLUSTALW (63), followed by identification of initial maximum likelihood (ML) model of evolution (using AIC as implemented in MODELTEST version 3.16; ref. 64). ML heuristic searches were performed by using PAUP* version 4b10 (65) under the TBR branch-swapping option. Bootstrap support was estimated in parallel by using 100 searches and starting from 10 random trees.
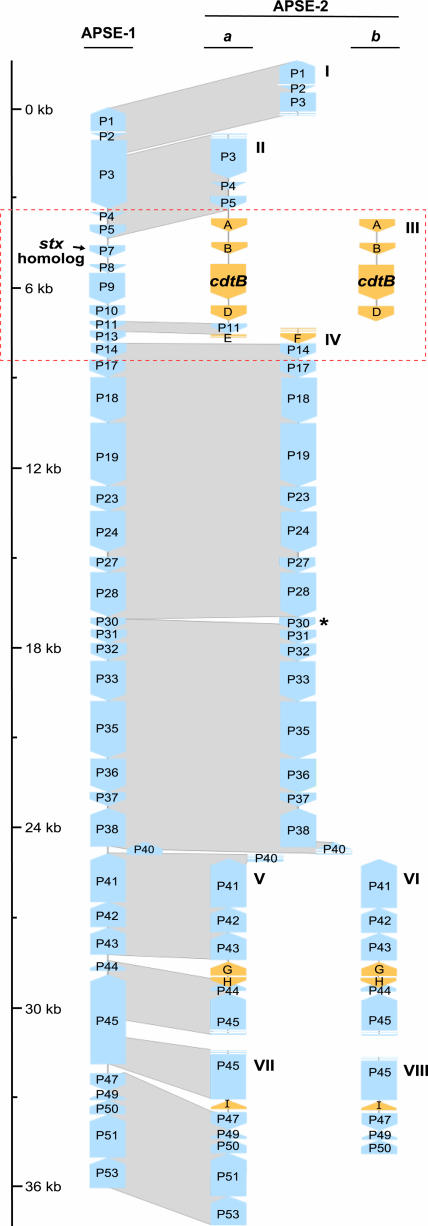
Sequences Assigned to Phage. Assembly and annotation of those sequences with similarity to APSE-1 resulted in eight large contigs representing two distinct phage haplotypes, designated APSE-2a and APSE-2b (Fig. 2). The clones yielded an average of 8.5× coverage across these phage contigs. Most genes of APSE-2 were 95-100% identical to those in APSE-1. Despite this high level of sequence similarity, the gene content of the APSE-2 haplotypes differed from that of APSE-1. First, both haplotypes contained two small insertions of genes encoding hypothetical proteins, each of which was confirmed by targeted PCR. Next, a large fragment containing several new ORFs including a homolog of cdtB, a toxin-encoding gene (Fig. 3), has replaced the region of APSE-1 that was previously found to contain several ORFs including a homolog of stx, encoding Shiga toxin (7).
Fig. 2.
Schematic of genome assemblies for APSE-2 phage of H. defensa. ORFs in blue represent those genes identified in APSE-1, numbered according to van der Wilk et al. (7), as well as corresponding homologs identified in the APSE-2 haplotypes (designated a and b). ORFs in yellow represent genes present in APSE-2 but absent from APSE-1. Phage contigs are denoted by roman numerals (I through VIII), and corresponding regions containing toxin genes (stx and cdtB) in the APSE-1 and APSE-2 genomes are highlighted. The APSE-2 homolog of P30 (marked with asterisk) contains an insertion of 58 aa.
Fig. 3.
Alignment of the CdtB homolog from H. defensa APSE-2 with those of other bacteria and with human DNase1. GenBank accession numbers are as follows: Escherichia coli MBU, AF373206; Escherichia coli O157, AJ508930; Salmonella enterica sv Typhi, NC_004631; Campylobacter jejuni, AL139074; Homo sapiens DNase1, D83195. Shading denotes positions where residues are conserved between APSE-2 CdtB and at least one other homolog. Yellow shading denotes conserved active sites of the protein, as determined for human DNase1. Note that residues at active sites are invariant among homologs. Alignment was constructed by using CLUSTALW (63).
Quantitative PCR to Assess Number of Bacteria in Samples. We used three samples of aphid hemolymph, obtained similarly to that used in WGA, to estimate the number of bacterial chromosomes used as template for WGA. Using copy number of the single copy gene dnaK as an index of numbers of bacterial chromosomes in the samples, we estimate that 21,890-26,700 copies of H. defensa chromosomes were present in the WGA template.
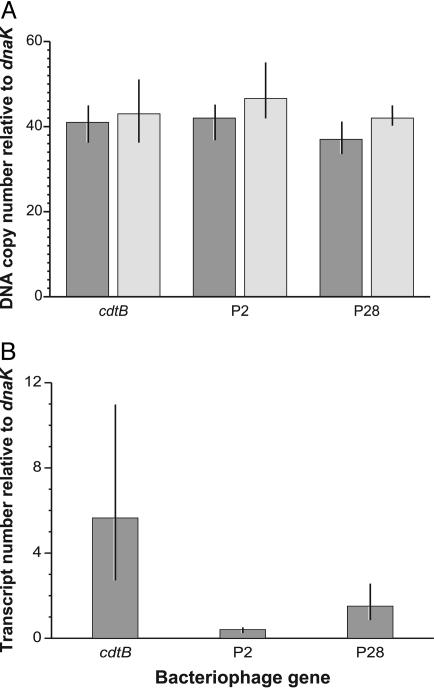
Quantitative PCR to Assess Phage Copy Number and Gene Expression. The APSE-2 phage genes P2, P28, and cdtB each showed similar copy numbers relative to the single copy bacterial chromosomal gene dnaK, indicating ≈40 phage copies per bacterial chromosome (Fig. 4A). This ratio was similar for different aphids, including individuals from the source aphid stock (8-2-B) and from the experimentally infected aphid stock (5AT). The high copy number of the phage was consistent with the abundance of phage sequences in the WGA clone library (Table 1) and with results from PFGE (below).
Fig. 4.
Relative abundance of phage-derived genes and transcripts to bacterial genes and transcripts. (A) Mean copy number of phage genes relative to the single copy bacterial chromosomal gene dnaK; range of values from three samples are indicated. Measurements made on two aphid clones infected with the same H. defensa strain. (B) Mean transcript abundance of phage genes relative to transcript abundance of the highly expressed bacterial gene dnaK; range of values from four samples are indicated.
RT-qPCR indicated that cdtB is expressed at high levels, with transcript numbers ≈5-fold those of dnaK, a highly expressed bacterial gene (Fig. 4B). Expression of P2 and P28, in contrast, was lower than that of dnaK. There was a 15-fold difference in transcript abundance between P2 and cdtB.
Possible Chimeric Sequences. Some clones were suspected of being chimeric, based on features such as presence of interrupted ORFs or conflicting assemblies for the APSE sequences. Of 10 such cases tested on genomic DNA by using PCR, none could be confirmed as corresponding to an actual region of genome. Approximately 15% of clones were deemed to be chimeric and eliminated from the final assemblies. Although GenomiPhi is useful for amplifying small quantities of DNA, our observations suggest that it is prone to producing chimeric molecules.
Amplification and Cloning of Variable Region of APSE-2 Genome. The APSE-2 assemblies included two versions of a region of particular interest based on the presence of cdtB and overall dissimilarity to the corresponding region in APSE-1. The targeted amplification of the APSE-2 sequences confirmed the two distinct haplotypes obtained from the four completely sequenced clones (three of one type and one of another). Both 6.5-kb sequences were identical to the assemblies from the WGA. We also amplified, cloned, and sequenced four clones from the aphid source stock of this H. defensa isolate (8-2-B). We again obtained identical sequences, indicating that both haplotypes were stably maintained for a period of at least 4 years (>100 aphid generations) in alternative aphid host lines.
Distribution of APSE Genes in Different Isolates of H. defensa. To assess the extent of APSE and toxin-encoding genes in H. defensa isolates, we used PCR to screen for the presence of stx homologs and of three other phage genes in genomic DNA from insects known to possess H. defensa. One or more phage genes were present in all samples, extending previous evidence (2) that all H. defensa isolates contain an APSE-like phage. Given this broad distribution, we presume that negative reactions were due to sequence divergence at the primer sites or, possibly, genomic rearrangements or deletions, rather than to the absence of phage. The stx homolog, originally reported in APSE-1 from A. pisum in The Netherlands, was detected only in our single isolate from the aphid Aphis craccivora (Table 2). This H. defensa isolate and phage was maintained for at least 25 generations after experimental infection in an A. pisum laboratory colony.
Table 2. Presence of phage and toxin gene homologs in isolates of H. defensa from diverse insect hosts.
| APSE phage genes*
|
stx homolog
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
cdtB homolog
|
||||||||
| Species | Taxonomy | Collection location | P2 P2F&R | P4–P5 3.6F&4.2R | P4–P14 3.9F&8.1R | P7* P7F&R | F1&R1 | F1&R2 |
| A. pisum 5AT (A. pisum 8-2-B = donor) | Macrosiphini | New York | + | + | + | – | + | + |
| A. pisum 82B | Macrosiphini | New York | + | + | + | – | + | + |
| A. pisum A1A | Macrosiphini | Utah | + | + | – | – | – | – |
| A. pisum A2C | Macrosiphini | Utah | + | – | – | – | – | – |
| A. pisum A2F | Macrosiphini | Utah | + | – | – | – | – | – |
| A. pisum A2H | Macrosiphini | Utah | + | – | – | – | – | – |
| A. pisum 5ATac (Aphis craccivora = donor) | Macrosiphini (Aphidini) | Arizona | + | + | – | + | – | – |
| Macrosiphum euphorbiae (from Oenothera) | Macrosiphini | Arizona | + | – | – | – | – | – |
| Macrosiphum euphorbiae (from Penstemon) | Macrosiphini | Arizona | + | + | – | – | – | – |
| Uroleucon ambrosiae | Macrosiphini | Arizona | – | + | – | – | – | – |
| Uroleucon rudbeckiae | Macrosiphini | Arizona | + | + | + | – | – | – |
| Uroleucon astronomous | Macrosiphini | Minnesota | + | – | – | – | – | – |
| Geopemphigus sp. | Fordini | Texas | + | – | – | – | – | – |
| Bemisia tabaci | Aleyrodidae | Arizona | + | + | – | – | – | – |
| Cacopsylla pyri | Psyllidae | Switzerland | + | + | – | – | – | – |
All positives were confirmed by sequencing, but apparent negatives could result from sequence divergence.
Gene names follow the convention of van der Wilk et al. (7)
Estimates of H. defensa Genome Size. Pulsed-field gels resolved a set of DNA fragments that were visible in the aphids infected with H. defensa and absent from uninfected aphids. Fragments corresponding to the entire Buchnera genome (640 kb) were visible in both. DNAs from infected aphids produced a very dark-staining band at ≈40 kb, corresponding to the expected genome size of the APSE-1-like phage. This fragment constitutes about half of the DNA stained on the gel, consistent with other observations indicating very high copy number of the APSE-2 genome. Bands attributable to H. defensa yielded a combined length of ≈1.6 Mb from AscI digests (seven fragments ranging in size from 60 to 410 kb) and ≈1.7 Mb from NotI digests (eight fragments ranging from 50 to 555 kb).
Probing the PFGE fragments with P2 or with cdtB yielded similar results. With both probes, most of the hybridization signal occurred at the 40-kb band, corresponding to the intact APSE phage, with a fainter signal on identical H. defensa bands. This finding indicates that P2 and cdtB always occur together, sometimes on a temperate phage, but mostly on independent phage chromosomes. Our failure to detect lysogenic copies in the WGA assemblies is likely attributable to the low coverage of the H. defensa genome.
Discussion
Whole Genome Amplification of an Insect Endosymbiont. For most bacterial species, the inability to harvest pure cultures complicates analysis of their genomes. To overcome such difficulties, several methods have been developed in which DNA from minute quantities of cells is amplified enzymatically in an attempt at complete coverage of a genome. Among the most successful such procedures has been by multiple displacement amplification (MDA) (34), which has been shown to achieve minimal amplification biases of eukaryotic (35) and bacterial (36) genomes. To date, the most comprehensive application of MDA with bacteria surveyed genomes is of the cultivable insect-vectored plant pathogen, Xylella fastidiosa (36). To our knowledge, ours is the first description of a genome of a noncultivable bacterium, that is, a limited sample of H. defensa cells from the insect hemocoel, based on this method.
The H. defensa genome shows some features typical of chronic symbionts or intracellular pathogens. At about 1.7 Mb, its genome is small relative to those of free-living Enterobacteriaceae, which are typically 4-6 Mb, and it is compositionally biased (39% G+C). However, neither the size nor base composition is as extreme as those observed in long-term primary symbionts, such as B. aphidicola (22), Wigglesworthia glossinidia (37), Blochmannia (38), or Carsonella ruddii (39). All of these organisms have genome sizes <1 Mb and genomic base compositions of 18-28% G+C. Additionally, none of these highly reduced symbiont genomes possesses phage sequences, in contrast to H. defensa, which is always associated with a lambdoid phage. H. defensa also contains transposases typical of insertion sequence elements; these are generally absent from the genomes of the primary symbionts (40). Thus, H. defensa retains a larger and more plastic genome.
A Symbiont Using Toxins Known from Mammalian Pathogens. The most striking finding emerging from this study is the abundance and ubiquity of the APSE phage genomes among H. defensa isolates, coupled with the fact that these phage encode and express genes homologous to well known toxin-encoding genes in mammalian pathogens. Our observations suggest that this phage is an integral part of the life cycle and ecology of this bacterium.
We found two haplotypes of APSE-2, differing slightly in their nucleotide sequences and gene contents. Both contain intact copies of cdtB, which is highly expressed (41). In APSE-1, the corresponding genomic region instead contains a distinct gene set including a homolog of stx, encoding Shiga toxin (7); we detected a similar stx homolog in H. defensa originating from a different insect species. Both of these toxins are deployed by a variety of human pathogens, and their cellular activities have been well characterized: CdtB is a nuclease that interferes with DNA replication during the G2 phase of the eukaryotic cell cycle (42, 43), and Shiga toxin inhibits protein synthesis by disrupting 28S ribosomal RNA (44).
In addition to the presence of phage-encoded toxin genes, H. defensa further resembles mammalian enteric pathogens in the presence of two TTSS, homologous to those characterized in the SPI-1 and SPI-2 pathogenicity islands of S. enterica. TTSS have been shown to deliver effector proteins that mediate host cell invasion in S. enterica (45) and many other animal and plant pathogens (46, 47), and in some insect symbionts (15-17). Haghjoo and Galan (48) discerned a link between the TTSS and CdtB in S. enterica sv. Typhi. In other organisms that possess cdtB, products of the two flanking genes cdtA and cdtC mediate delivery of the CdtB toxin into the target cell (43). However, S. enterica sv. Typhi, which lacks cdtA and cdtC, is internalized into host cells via a TTSS, thereby providing a route for the delivery of CdtB into the host cytoplasm (48). Like S. enterica, H. defensa possesses cdtB and TTSS, but lacks cdtA or cdtC, suggesting the use of the same strategy for delivering the cdtB-encoded toxin.
The APSE-H. defensa-Aphid Consortium as a Complex Mutualistic System. Given the diverse life forms involved in this coinherited symbiosis, its long-term persistence relies on mutualistic interactions at several levels. Although lytic phage are usually considered to be detrimental to bacterial hosts, the persistence and ubiquity of APSE phages in H. defensa suggest that they are a required part of the ecology of this symbiosis. Indeed, these phages are selected to preserve rather than destroy their bacterial hosts as well as the aphid hosts, because these provide a reliable supply of future hosts through vertical transmission. Accordingly, APSE appears never to eliminate H. defensa from a host. Our H. defensa-infected aphid lines are >5 years old, spanning >150 aphid generations, and they retain both H. defensa and APSE.
In Shiga toxin-producing E. coli, the amplification of stx copy number, the regulation of stx transcription, and the delivery of Shiga toxin to host tissues are all achieved through phage-mediated processes, which culminate in the lysis of a subset of the bacterial cells (49-51). It has been proposed that the phage persist as mutualists of their bacterial hosts, as lysis of some bacteria improves the environment for the surviving bacterial cells, which retain lysogenic phage (50). The Shiga toxin-encoding lambdoid phages appear to undergo higher rates of spontaneous induction than do those lacking stx, which may represent a colony-level adaptation to increase the extent of the infection within the host (52). A similar progression likely occurs in H. defensa, whereby toxin production and delivery is regulated and achieved as part of the APSE life cycle.
A salient question is why H. defensa and its phage produce toxins that specifically disrupt eukaryotic cell processes. In enteric pathogens, such as S. enterica and enteropathogenic E. coli, the utility of phage-encoded toxins is clear: the toxic effects on host cells will enhance the extent and overall size of the infectious bacterial population, promoting replication of that portion of cells that carry the phage and continue to replicate. However, H. defensa, along with its toxin-producing phage, is vertically transmitted and expected to be harmless or beneficial to insect hosts.
At least one mutualistic effect of H. defensa has been documented: it confers substantial resistance to infection by parasitoid wasps, which are major natural enemies (6). In this regard, we propose two possible roles for the presence of phage-encoded CdtB in this system. First, it may allow H. defensa to establish a stable infection in aphids by causing distention of the cytoplasm of selected host cells, as described for mammalian host systems (43). However, cdtB is not present in all H. defensa isolates, yet they appear to show similar patterns of infection of host aphid cells and tissues (1, 2). Alternatively, APSE-encoded toxins may target and destroy the cells of attacking parasitoids or other eukaryotic enemies, thereby protecting their aphid hosts. The mosaic structure of APSE genomes, also observed in other lambdoid phage (20), reflects recombination among haplotypes and could promote hypervariability in toxin-encoding capacity of H. defensa isolates. Thus, the phages may enable H. defensa to mount a varied arsenal of toxins to defend aphid hosts against natural enemies, which vary among populations and over time (e.g., refs. 53 and 54). Different H. defensa isolates have been shown to confer very different degrees of resistance to a particular parasitoid enemy, possibly reflecting variation in phage-encoded genes (55). Under this view, the aphid-symbiont-phage system would comprise a coinherited symbiotic consortium.
The gene pool of bacteriophage is enormous and serves as a source of evolutionary novelty for bacterial hosts (56); through symbiosis, this novelty can extend to heritable properties of eukaryotic hosts. For example, the opportunistic symbiont Wolbachia also has high rates of infection by a bacteriophage (WO), and WO from Wolbachia in divergent insect hosts undergo genetic exchange and recombination (27, 57, 58), thus providing one of the main sources of variation among Wolbachia isolates (59). Similar mosaicism is evident in the genome of APSE-1 (8). Coinfection of host insects by different symbiont isolates, which occurs in both Wolbachia and in aphid secondary symbionts, may enable the exchange of genes through recombination.
Thus, bacterial symbionts and their phage present a mechanism for gene transfer among animal hosts, and these acquired genes can become stably inherited elements having major effects on ecological relationships. In pea aphids, acquired, heritable bacterial symbionts affect heat tolerance (60), parasitoid resistance (6), plant use (61), and possibly fungal pathogen resistance (62). As we further investigate the diversity of symbiotic systems, we can expect that many will consist of complex phenotypes emerging from the interactions of disparate life forms, including viruses, bacteria, and eukaryotes.
Supplementary Material
Acknowledgments
We thank Phat Tran for technical support in cloning and PCR screens, Kerry Oliver (University of Arizona, Tucson) for providing aphid clones, and Becky Nankivell for assistance in the preparation of the manuscript and figures. Scott O'Neill provided useful comments on an earlier draft of the manuscript. P.H.D. was a predoctoral fellow in the National Science Foundation-sponsored Integrative Graduate Education and Research Traineeship program in Genomics (to the University of Arizona). This research was supported by National Science Foundation Awards MCB0301829 (to H.O.) and DEB0313737 (Genome-Enabled Biocomplexity) (to N.A.M.) and Department of Energy Award DEFG0301ER63147 (to H.O.).
Author contributions: N.A.M., S.R.S., and H.O. designed research; N.A.M., P.H.D., S.R.S., and H.E.D. performed research; N.A.M., P.H.D., S.R.S., and H.O. analyzed data; and N.A.M., P.H.D., and H.O. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: TTSS, type III secretion system; WGA, whole genome amplification; qPCR, quantitative PCR; PFGE, pulsed-field gel electrophoresis.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ163092, DQ163903, and DQ092612-DQ092623).
See accompanying Profile on page 16916.
References
- 1.Moran, N. A., Russell, J. A., Fukatsu, T. & Koga, R. (2005) Appl. Environ. Microbiol. 71, 3302-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandström, J. P., Russell, J. A., White, J. P. & Moran, N. A. (2001) Mol. Ecol. 10, 217-228. [DOI] [PubMed] [Google Scholar]
- 3.Darby, A. C., Birkle, L. M., Turner, S. L. & Douglas, A. E. (2001) FEMS Microbiol. Ecol. 36, 43-50. [DOI] [PubMed] [Google Scholar]
- 4.Russell, J. A., LaTorre, A. L., Sabater-Muñoz, B., Moya, A. & Moran, N. A. (2003) Mol. Ecol. 12, 1061-1075. [DOI] [PubMed] [Google Scholar]
- 5.Haynes, S., Darby, A. C., Daniell, T. J., Webster, G., Van Veen, F. J., Godfray, H. C., Prosser, J. I. & Douglas, A. E. (2003) Appl. Environ. Microbiol. 69, 7216-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver, K., Russell, J., Moran, N. & Hunter, M. (2003) Proc. Natl. Acad. Sci. USA 100, 1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Wilk, F., Dullemans, A. M., Verbeek, M. & van den Heuvel, J. F. (1999) Virology 262, 104-113. [DOI] [PubMed] [Google Scholar]
- 8.Clark, A. J., Inwood, W., Cloutier, T. & Dhillon, T. S. (2001) J. Mol. Biol. 311, 657-679. [DOI] [PubMed] [Google Scholar]
- 9.Thao, M. L., Clark, M., Baumann, L., Brennan, E. B., Moran, N. A. & Baumann, P. (2000) Curr. Microbiol. 41, 300-304. [DOI] [PubMed] [Google Scholar]
- 10.Thao, M. L., Gullan, P. J. & Baumann, P. (2002) Appl. Environ. Microbiol. 68, 3190-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thao, M. L. & Baumann, P. (2004) Curr. Microbiol. 48, 140-144. [DOI] [PubMed] [Google Scholar]
- 12.Lefevre, C., Charles, H., Vallier, A., Delobel, B., Farrell, B. & Heddi, A. (2004) Mol. Biol. Evol. 21, 965-973. [DOI] [PubMed] [Google Scholar]
- 13.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T. G., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 14.Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Taourit, S., Bocs, S., Boursaux-Eude, C., Chandler, M., Dassa, E., Derose, R., et al. (2003) Nat. Biotechnol. 21, 1307-1313. [DOI] [PubMed] [Google Scholar]
- 15.Dale, C., Young, S. A., Haydon, D. T. & Welburn, S. C. (2001) Proc. Natl. Acad. Sci. USA 98, 1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dale, C., Jones, T. & Pontes, M. (2005) Mol. Biol. Evol. 22, 758-766. [DOI] [PubMed] [Google Scholar]
- 17.Dale, C., Plague, G., Wang, B., Ochman, H. & Moran, N. A. (2002) Proc. Natl. Acad. Sci. USA 99, 12397-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brugirard-Ricaud, K., Givaudan, A., Parkhill, J., Boemare, N., Kunst, F., Zumbihl, R. & Duchaud, E. (2004) J. Bacteriol. 186, 4376-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman, E. A. & Ochman, H. (1993) EMBO J. 12, 3779-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen, B. K., Wasteson, Y., Granum, P. E. & Brynestad, S. (2001) Microbiology 147, 1929-1936. [DOI] [PubMed] [Google Scholar]
- 21.Wagner, P. L. & Waldor, M. K. (2002) Infect. Immun. 70, 3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. (2000) Nature 407, 81-86. [DOI] [PubMed] [Google Scholar]
- 23.Tamas, I., Klasson, L., Näslund, K., Eriksson, A. S., Wernegreen, J. J., Sandström, J. P., Moran, N. A. & Andersson, S. G. E. (2002) Science 296, 2376-2379. [DOI] [PubMed] [Google Scholar]
- 24.van Ham, R. C., Kamerbeek, J., Palacios, C., Rausell, C., Abascal, F., Bastolla, U., Fernández, J.M., Jiménez, L., Postigo, M., Silva, F. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcox, J. L., Dunbar, H. E., Wolfinger, R. D. & Moran, N. A. (2003) Mol. Microbiol. 48, 1491-1500. [DOI] [PubMed] [Google Scholar]
- 26.Zientz, E., Dandekar, T. & Gross, R. (2004) Microbiol. Mol. Biol. Rev. 68, 745-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., McGraw, E. A., Martin, W., Esser, C., Ahmadinejad, N., et al. (2004) PLoS Biol. 2, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchida, T., Koga, R., Meng, X. Y., Matsumoto, T. & Fukatsu, T. (2005) Microb. Ecol. 49, 126-133. [DOI] [PubMed] [Google Scholar]
- 29.Sutton, G., White, O., Adams, M. & Kerlavage, A. (1995) Genome Sci. Technol. 1, 9-19. [Google Scholar]
- 30.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 31.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran, N. A., Dunbar, H. E. & Wilcox, J. L. (2005) J. Bacteriol. 187, 4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran, N. A., Dale, C., Dunbar, H., Smith, W. & Ochman, H. (2003) Environ. Microbiol. 5, 116-126. [DOI] [PubMed] [Google Scholar]
- 34.Dean, F. B., Nelson, J. R., Giesler, T. L. & Lasken, R. S. (2001) Genome Res. 11, 1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dean, F. B., Hosono, S., Fang, L., Wu, X., Faruqi, A. F., Bray-Ward, P., Sun, Z., Zong, Q., Du, Y., Du, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Detter, J. C., Jett, J. M., Lucas, S. M., Dalin, E., Arellano, A. R., Wang, M., Nelson, J. R., Chapman, J., Lou, Y., Rokhsar, D., et al. (2002) Genomics 80, 691-698. [DOI] [PubMed] [Google Scholar]
- 37.Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M. & Aksoy, S. (2002) Nat. Genet. 32, 402-407. [DOI] [PubMed] [Google Scholar]
- 38.Gil, R., Silva, F. J., Zientz, E., Delmotte, F., González-Candelas, F., Latorre, A., Rausell, C., Kamerbeek, J., Gadau, J., Holldöbler, B., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark, M. A., Baumann, L., Thao, M. L., Moran, N. A. & Baumann, P. (2001) J. Bacteriol. 183, 1852-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moran, N. A. & Plague, G. R. (2004) Curr. Opin. Genet. Dev. 14, 627-633. [DOI] [PubMed] [Google Scholar]
- 41.Lara-Tejero, M. & Galán, J. E. (2000) Science 290, 354-357. [DOI] [PubMed] [Google Scholar]
- 42.Pickett, C. L. & Whitehouse, C. A. (1999) Trends Microbiol. 7, 292-297. [DOI] [PubMed] [Google Scholar]
- 43.Lara-Tejero, M. & Galán, J. E. (2002) Trends Microbiol. 10, 147-152. [DOI] [PubMed] [Google Scholar]
- 44.Acheson, D. W. & Keusch, G. (1996) Am. Soc. Microbiol. News 62, 302. [Google Scholar]
- 45.Galán, J. E. (2001) Annu. Rev. Cell Dev. Biol. 17, 53-86. [DOI] [PubMed] [Google Scholar]
- 46.Hueck, C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galán, J. E. & Collmer, A. (1999) Science 284, 1322-1328. [DOI] [PubMed] [Google Scholar]
- 48.Haghjoo, E. & Galán, J. E. (2004) Proc. Natl. Acad. Sci. USA 101, 4614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, P. L., Neely, M. N., Zhang, X., Acheson, D. W. K., Waldor, M. K. & Friedman, D. I. (2001) J. Bacteriol. 183, 2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, P. L., Livny, J., Neely, M. N., Acheson, D. W. K., Friedman, D. I. & Waldor, M. K. (2002) Mol. Microbiol. 44, 957-970. [DOI] [PubMed] [Google Scholar]
- 51.Tyler, J. S., Mills, M. J. & Friedman, D. I. (2004) J. Bacteriol. 186, 7670-7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livny, J. & Friedman, D. I. (2004) Mol. Microbiol. 51, 1691-1704. [DOI] [PubMed] [Google Scholar]
- 53.Cardinale, B. J., Harvey, C. T., Gross, K. & Ives, A. R. (2003) Ecol. Lett. 6, 857-865. [Google Scholar]
- 54.Ferrari, J., Muller, C. B., Kraaijeveld, A. R. & Godfray, H. J. C. (2001) Evolution (Lawrence, Kans.) 55, 1805-1814. [DOI] [PubMed] [Google Scholar]
- 55.Oliver, K. A., Moran, N. A. & Hunter, M. S. (2005) Proc. Natl. Acad. Sci. USA 102, 12795-12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casjens, S. (2003) Mol. Microbiol. 49, 277-300. [DOI] [PubMed] [Google Scholar]
- 57.Masui, S., Kamoda, S., Sasaki, T. & Ishikawa, H. (2000) J. Mol. Evol. 51, 491-497. [DOI] [PubMed] [Google Scholar]
- 58.Bordenstein, S. R. & Wernegreen, J. J. (2004) Mol. Biol. Evol. 21, 1981-1991. [DOI] [PubMed] [Google Scholar]
- 59.Simkins, S. P., Walker, T., Lund, A. R., Steven, A. R., Makepeace, B. L., Godfray, H. C. J. & Parkhill, J. (2005) Nature 436, 257-260. [DOI] [PubMed] [Google Scholar]
- 60.Montllor, C. B., Maxmen, A. & Purcell, A. H. (2002) Ecol. Entomol. 27, 189-195. [Google Scholar]
- 61.Tsuchida, T., Koga, R. & Fukatsu, T. (2004) Science 303, 1989. [DOI] [PubMed] [Google Scholar]
- 62.Ferrari, J., Darby, A. C., Daniell, T. J. & Godfray, H. J. C. (2004) Ecol. Entomol. 29, 60-65. [Google Scholar]
- 63.Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G. & Thompson, J. D. (2003) Nucleic Acids Res. 31, 3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 65.Swofford, D. L. (2002) paup*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA), version 4.0 Beta.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.