Abstract
Benzene is an important industrial chemical and environmental contaminant that causes leukemia. To obtain mechanistic insight into benzene's mechanism of action, we examined the impact of benzene on the human serum proteome in a study of exposed healthy shoe-factory workers and unexposed controls. Two sequential studies were performed, each using sera from 10 workers exposed to benzene (overall mean benzene air level >30 ppm) and 10 controls. Serum samples were subjected to anion-exchange fractionation and bound to three types of ProteinChip arrays (Ciphergen Biosystems, Fremont, CA) [hydrophobic (H50), metal affinity (IMAC3-Cu), and cation exchange (WCX2)]. Protein-expression patterns were detected by surface-enhanced laser desorption/ionization (SELDI)-TOF MS. Three proteins (4.1, 7.7, and 9.3 kDa) were consistently down-regulated in exposed compared with control subjects in both studies. All proteins were highly inversely correlated with individual estimates of benzene exposure (r > 0.75). The 7.7- and 9.3-kDa proteins were subsequently identified as platelet factor (PF)4 and connective tissue activating peptide (CTAP)-III. Initial proteomic results for PF4 and CTAP-III were subsequently confirmed in a single experiment using a ProteinChip-array-based immunoassay(Ciphergen Biosystems). The altered expression of the platelet-derived CXC-chemokines (40% and 63% for PF4 and CTAP-III, respectively) could not be explained by changes in absolute platelet counts. Thus, SELDI-TOF analysis of a limited number of exposed and unexposed subjects revealed that lowered expression of PF4 and CTAP-III proteins is a potential biomarker of benzene's early biologic effects and may play a role in the immunosuppressive effects of benzene.
Keywords: biomarker, leukemia, mass spectrometry, platelet, molecular epidemiology
Benzene is used in chemical manufacture and is a component of crude oil and gasoline. Benzene's widespread use and the fact that it is a product of incomplete combustion has led to contamination of the environment, especially in urban centers. Benzene is an established cause of acute myeloid leukemia and myelodysplastic syndromes and may play a role in lymphocytic leukemias and non-Hodgkin lymphoma in humans (1-6). Additionally, changes in blood and bone marrow consistent with hematotoxicity are recognized in humans and experimental animals (7-9). Benzene also produces immunosuppressive effects in exposed animals and humans and has been shown to increase susceptibility to tuberculosis and other infectious agents (10, 11). Despite extensive research, questions remain regarding the exact mechanisms by which benzene exerts its effects, and valid biomarkers of exposure and relevant early biologic effect are needed. To elucidate these mechanisms and better understand the risk benzene poses, we have examined the effects of benzene exposure on the serum proteome in a population of shoe-factory workers with well characterized occupational exposures to benzene and unexposed controls by using surface-enhanced laser desorption/ionization (SELDI)-TOF MS.
Proteome analysis provides information regarding the proteome's dynamic and rapid changes, which result from exogenous exposure and/or endogenous factors. These changes include DNA alterations, mRNA splicing, temporal and functional regulation of gene expression, and posttranslational modifications. The serum proteome would be expected to reflect what has been experienced or encountered by the blood during its constant perfusion through the body. SELDI-TOF MS has been demonstrated to be able to broadly explore the proteome and yield biomarkers that can be used individually or in combination to distinguish several forms of cancer from normal controls (12-15). However, for exogenous exposures, the application of SELDI-TOF MS as a discovery tool to identify differentially expressed proteins has been limited. We hypothesized that SELDI-TOF MS could identify changes in the serum proteome that could be used as markers of early biologic effect for benzene and provide further mechanistic insight into how benzene affects the body.
Materials and Methods
Study Population and Design. Subjects (n = 40) for proteomic analyses were selected from a molecular epidemiology study of workers exposed to benzene in Tianjin, China (7). Workers exposed to benzene were selected from a shoe factory, and unexposed controls were selected from a clothing manufacturing plant located in the same general geographical area. Institutional review boards at all participating institutions approved the study. Participation was voluntary, written informed consent was obtained, and the participation rate was ≈95%. Biological samples were collected in June, 2000.
Peripheral blood was collected from each subject and processed into serum within 6 h. White blood cell and platelet counts were measured in a Beckman Coulter T540 blood counter. Each subject was given a physical examination by a study physician. A questionnaire was administered, requesting detailed information on occupation, environmental exposures, medical history, current medications, and past and current tobacco and alcohol use.
Before phlebotomy, individual benzene and toluene exposure was monitored by the subject's wearing an organic vapor passive monitor badge as described in refs. 7 and 16. Personal full-shift air monitoring took place about every month over a 3-month period, resulting in ≈3-4 personal air measurements per person. Average individual benzene exposure was calculated for the whole observation period and separately for the last month before biological-sample collection. Benzene and toluene were not detected in air samples from the control factory (7).
Discovery and Identification of Protein Biomarkers. To determine whether proteomic profiles differed between benzene-exposed individuals and unexposed controls, two sequential studies were performed, each using sera collected from 10 healthy workers exposed to benzene and 10 healthy, matched unexposed controls. The first set of samples was used as an initial screening to reveal proteins that were differentially expressed by exposure status (discovery set). The second set of samples was used to validate the previously observed proteins by trying to replicate the findings of the first screening (validation set). Demographic characteristics of the study population by exposure status and test set are presented in Table 4, which is published as supporting information on the PNAS web site.
Serum fractionation and preparation of ProteinChip arrays. Serum (20 μl) was mixed with 30 μl of U9 denaturing buffer [9 M urea/2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/50 mM Tris, pH 9.0] to reduce protein-protein interactions. U9-treated samples were applied to BioSepra Q Ceramic HyperD F resin (Pall) that had been preequilibrated in 50 mM Tris, pH 9.0. The flow-through was collected, and bound proteins were eluted in a step-wise gradient by using buffers with pH values of 7, 5, 4, and 3, followed by an organic wash (33% isopropanol/17% acetonitrile/0.1% trifluoroacetic acid). The fractionation process yielded a total of six fractions. For the purpose of this study, three of the six fractions were analyzed, including the pH 9 and pH 4 eluates and the organic fraction (fractions 1, 4, and 6). Duplicate 10-μl aliquots of each fraction were diluted with binding buffer and applied to H50, immobilized metal-affinity chromatography (IMAC)3-Cu, and WCX2 ProteinChip arrays (Ciphergen Biosystems). The binding buffers were 10% acetonitrile/0.1% trifluoroacetic acid for H50 arrays, 100 mM phosphate, pH 7.0/0.5 M NaCl for IMAC3-Cu arrays, and 100 mM sodium acetate, pH 4.0, for WCX2 arrays. Sinapinic acid was used as the energy-absorbing molecule. The fractionation and array preparation was fully automated and performed by using a Biomek 2000 robot with an integrated Micromix 5 shaker (Beckman Coulter). For quality control purposes, human reference serum samples and blind duplicates were included with the test serum samples. Hemolysis was noted as red coloration in some samples, and we quantitated the α- and β-chains of hemoglobin to determine the extent of this hemolysis.
Data acquisition and processing. Because of the time span between analysis of the discovery and validation samples, the data were collected on two different models of the ProteinChip reader (Ciphergen Biosystems). Arrays from the discovery and validation studies were analyzed on a PBSII and PBSIIC ProteinChip reader, respectively. Differences between the two models include the detector type and the presence of a mass deflector in the PBSIIC. Each array was read at two settings to optimize for low- and high-mass proteins. Data were collected up to a maximum m/z of 200,000. Processing included mass calibration, baseline subtraction, total ion current normalization, and peak detection. Samples were randomized within and across arrays to reduce analytical bias and run blind, so that exposure status was unknown.
Data analysis and statistics. The Biomarker Wizard feature of the proteinchip software (Ciphergen Biosystems) was used to create peak clusters and calculate P values for differences in peak intensity between exposed and unexposed subjects for each identified peak cluster by using the nonparametric test of mean (Mann-Whitney test). As a quality control, peak clusters with P values of <0.05 in the Biomarker Wizard were visually inspected and manually relabeled. After relabeling, the intensity values for the duplicates were averaged, and exact P values for differences in average peak intensity between exposed and unexposed subjects were calculated (Wilcoxon exact test). Peaks with a P value <0.01 in the discovery set were considered as potential markers.
To combine the results from the discovery and validation studies, relative average intensities (RAIs) were calculated for the identified peaks. RAIs were calculated by dividing the individual peak intensity by the average peak intensity among the controls by test, thereby artificially resetting the mean relative protein signal among the controls to 1 in both experiments.
Identification of protein biomarkers. Proteins were purified and subjected to tryptic digestion and the generated peptides sequenced by tandem MS (for details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Confirmation of Protein Biomarkers by Using ProteinChip Immunoassays. All samples from the initial experiments (n = 40) were reanalyzed for the identified proteins PF-4 and CTAP-III by using ProteinChip immunoassays (Ciphergen Biosystems). Samples were randomized and run blind, so that exposure status and previous proteomic results were unknown. Specific antibody arrays were prepared by covalently coupling the appropriate antibodies to preactivated ProteinChip arrays (Ciphergen Biosystems). Anti-PF4- and anti-NAP-2-affinity-purified rabbit polyclonal antibodies (Chemicon International, Temecula, CA) were covalently coupled to PS20 and RS100 arrays, respectively. After blocking with BSA and washing to remove uncoupled antibodies, serum samples were loaded on the antibody-coated arrays and incubated for 90 min. Binding buffers were optimized independently for each assay as follows: 0.1 M NaPO4, pH 7.2/0.5Murea/0.5%3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was used for the CTAP-III assay, and 0.1 M NaPO4, pH 7.2/0.5 M urea/0.25% CHAPS/75 mM NaCl was used for the PF-4 assay. Arrays were then washed twice with binding buffer, twice with 0.1 M NaPO4 (pH 7.2), and rinsed with water before drying. α-Cyano-4-hydroxycinnamic acid was used as the energy-absorbing molecule. Recombinant human PF-4 and NAP-2 (Chemicon) standards were analyzed along with the samples. The PF-4 standard curve was linear from 1 to 48 fmol PF-4 (0.19-9.3 μg/ml). In addition to capturing CTAP-III, the anti-NAP-2 antibody also captures NAP-2 (7,628 Da) and PBP (10,266 Da), which were also analyzed.
Results
Discovery of Protein Biomarkers. Sera from two sets of 10 exposed and 10 unexposed subjects were analyzed by SELDI-TOF MS. Demographic characteristics of the exposed and unexposed subjects within and between the two sets were very similar, as were the benzene exposure levels in the last month and last 3 months of the exposed subjects in both sets (t test; P values, 0.3291 and 0.3300, respectively; see Table 4).
Discovery set. In the discovery set, univariate analyses of group-dependent differences revealed 18 peaks, ranging from 4 to 74 kDa, that were differentially expressed and characterized by a P value of <0.01 (Table 1). Average intensity among exposed subjects was increased for 3 proteins and decreased for 15 proteins compared with unexposed controls. However, three of the samples from exposed subjects exhibited hemolysis, which globally reduced the intensity of non-hemoglobin-related peaks. Excluding the hemolyzed samples resulted in only 6 proteins being significantly differentially expressed at a significance level of 0.01.
Table 1. Exact P values of group differences between benzene-exposed and unexposed subjects for selected differentially expressed proteins in the discovery, validation, and combined data sets.
| Discovery set*
|
Validation set*
|
|||||
|---|---|---|---|---|---|---|
| m/z | Conditions fraction/surface | All samples (n = 20) | Excluding hemolyzed samples (n = 17) | All samples (n = 20) | Excluding hemolyzed samples (n = 17) | Combined set† (n = 34) |
| Up-regulated | ||||||
| 43,359 | F6-WCX2 | 0.0068 | 0.043 | 0.052 | 0.16 | 0.76 |
| 44,476 | F1-WCX2 | 0.0089 | 0.070 | 0.35 | 0.96 | 0.32 |
| 44,733 | F6-WCX2 | 0.0021 | 0.0097 | 0.63 | 0.54 | 0.12 |
| Down-regulated | ||||||
| 4,057‡ | F6-WCX2 | 1.3 E-4 | 0.0012 | 0.14 | 0.014 | 2.3 E-05 |
| 6,632 | F4-IMAC3 | 0.0021 | 0.019 | 0.17 | 0.744 | 0.32 |
| 6,435 | F4-IMAC3/H50§ | 4.9 E-4 | 0.0046 | 0.48 | 0.96 | 0.12 |
| 6,838 | F6-WCX2 | 0.0068 | 0.055 | 0.32 | 0.89 | 0.47 |
| 7,763 | F4/F6-IMAC3/WCX2¶ | 4.3 E-5 | 4.1 E-4 | 0.029 | 0.0031 | 1.8 E-06 |
| 8,206 | F6-WCX2 | 0.0011 | 0.0097 | 0.85 | 0.27 | 0.011 |
| 8,225 | F6-H50 | 0.0029 | 0.019 | 0.68 | 0.54 | 0.026 |
| 8,690 | F4-IMAC3 | 0.0068 | 0.043 | 0.25 | 0.96 | 0.092 |
| 8,765 | F6-WCX2 | 0.0052 | 0.033 | 0.74 | 0.42 | 0.053 |
| 9,287 | F6-IMAC3/WCX2∥ | 4.3 E-5 | 4.1 E-4 | 0.019 | 0.0031 | 3.8 E-06 |
| 1,2581 | F4-H50 | 0.0015 | 0.014 | 0.25 | 0.54 | 0.034 |
| 1,7310 | F4-IMAC3 | 0.0039 | 0.033 | 0.32 | 0.47 | 0.26 |
| 2,2229 | F4-WCX2 | 0.0052 | 0.043 | 0.74 | 0.47 | 0.11 |
| 2,6059 | F4-H50 | 0.0015 | 0.014 | 0.97 | 0.42 | 0.092 |
| 7,3128 | F6-WCX2 | 0.0089 | 0.070 | 0.85 | 0.23 | 0.020 |
P values were calculated based on the average intensities of the identified peak (Wilcoxon exact test)
P values were calculated based on the RAI (RAI = average intensity/mean of average peak intensity in control population) of the identified peak (Wilcoxon exact test)
Proteins in boldface type were significantly differentially expressed in the same direction between exposed and unexposed subjects in both the discovery and validation sets
Data presented based on F4-IMAC3; results of F4-H50 were essentially similar
Data presented based on F4-IMAC3; results of the F6-WCX2 were essentially similar
Data presented based on F6-WCX2; results of the F6-IMAC3 were essentially similar
Validation set. Three peaks with masses of 4.1, 7.7, and 9.3 kDa, identified as having the most significant P values in the first set, were also statistically significant in the second set, with P values ranging from 0.0031 to 0.014 after exclusion of three hemolyzed samples (Table 1). Grouping of the two sets, after normalization to the mean peak intensity among the unexposed controls, indicated extremely low P values (< 5 E-05) for group-dependent differences in RAI for these three proteins. In these analyses, as was done previously for the discovery and validation sets, six hemolyzed samples were excluded. Hemolysis in the samples was quantified by the average intensity of the α- and β-chain of hemoglobin from fraction 4 on the IMAC-Cu array. The hemolyzed samples that were excluded had, on average, a five-times-higher level of hemoglobin than the nonhemolyzed samples (RAI 1.1 vs. 5.8; P < 0.0001). In addition to simply excluding these samples, we also tested for differences in protein levels between exposed and unexposed subjects by linear regression, correcting for the RAI of the combined α- and β-chains of hemoglobin. These analyses resulted in essentially similar results, with the strongest differences in protein levels (P < 0.0001) for the 4.1-, 7.7-, and 9.3-kDa proteins. Additional multiple regression analyses correcting for possible confounding factors, such as age, sex, current smoking, recent infection, and alcohol use, did not alter the observed differences between exposed and unexposed subjects.
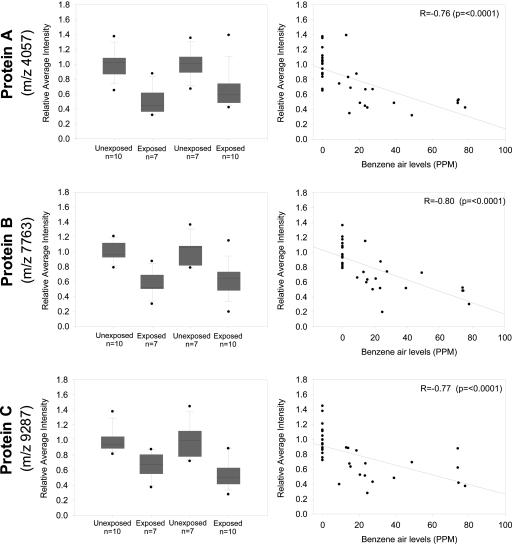
Representative SELDI-TOF MS spectra of exposed and unexposed subjects for the three differentially expressed proteins are shown in Fig. 4, which is published as supporting information on the PNAS web site. The average decrease in RAI for all proteins was similar between the two data sets and averaged ≈40% for all three proteins (Fig. 1). Furthermore, the proteins showed a strong negative correlation with individual benzene air levels in the last month, with correlation coefficients varying from -0.76 to -0.80.
Fig. 1.
RAI distributions for the three identified protein markers (m/z 4,057, protein A in Top; 7,763, protein B in Middle; and 9,287 protein C in Bottom) by test and exposure status (Boxplots, Left) and by benzene exposure (ppm) (Scatterplots, Right) for nonhemolyzed samples only (n = 34). (Left, Boxplots) The line within the box marks the median; the lower and upper boundary of the box indicates the 25th and 75th percentiles. Whiskers above and below the box indicate the 10th and 90th percentiles. (Right, Scatterplots) Scatter plots present RAI by benzene exposure (ppm) in the last month before blood collection. The solid line in the scatter plot represents the regression line between RAI and benzene exposure.
Identification of Protein Biomarkers. Two proteins (7.7 and 9.3 kDa) were chosen for purification and identification based on overall differential expression. The tryptic digest of the 7.7-kDa protein band yielded seven major unique peaks, all identified as fragments of PF4 (see Table 5, which is published as supporting information on the PNAS web site). The tryptic digestion of the 9.3-kDa protein band yielded two major unique peaks. Both peptides were identified as fragments of PBP. The tryptic fragments and the native mass of the 9.3-kDa protein correspond to a proteolytic fragment of PBP known as CTAP-III. Based on the amino acid sequences of PF4 and CTAP-III, the predicted masses of these proteins are 7,765 and 9,292 Da, respectively, which closely match the observed experimental masses of the identified proteins. Amino acid sequences of PF4 and CTAP-III and identified fragments are presented in Fig. 5, which is published as supporting information on the PNAS web site.
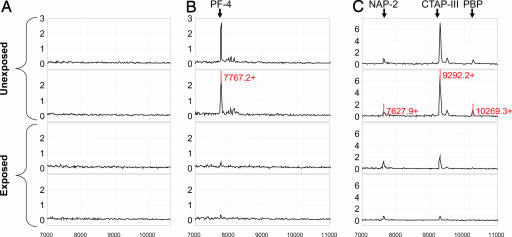
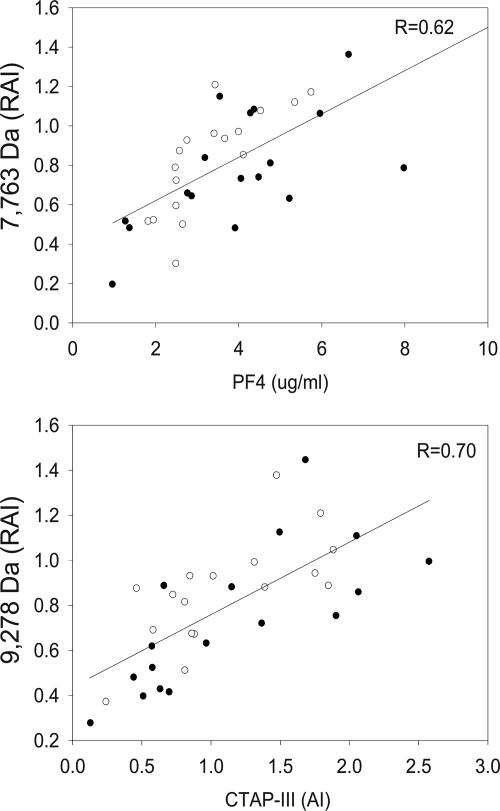
Confirmation of Protein Biomarkers. A ProteinChip-array-based immunoassay (Ciphergen Biosystems) was used to specifically capture PF4- and PBP-derived fragments from crude serum samples and confirm the significance of each marker. The anti-PF4 antibody specifically captured the previously identified 7.7-kDa protein (Fig. 2). A multiplexed antibody assay was developed to capture CTAP-III and other forms and products of PBP. The antibody against NAP-2 (the smallest derivative of PBP) specifically captured full-length PBP (10,266 Da), the previously identified 9.3-kDa protein biomarker (CTAP-III, 9,292 Da), and NAP-2 (7,628 Da). However, because of contamination of the anti-NAP2 with low concentrations of NAP-2 we could not accurately quantify NAP-2 levels, and, therefore, results are not shown. Note, as the ProteinChip-array-based immunoassay (Ciphergen Biosystems) was not influenced by hemolysis, the test samples results are shown for all 40 samples. PF4 levels, as quantified with the immunoassay, correlated well with the results of the first and second protein discovery experiment (r = 0.6, P < 0.0001) (Fig. 3) and were significantly (≈40%) lower among the exposed than the unexposed subjects (Table 2). The CTAP-III levels also correlated well with the initial experiment (r = 0.7, P < 0.0001) and were significantly decreased by 63% in samples from exposed individuals. PBP levels were decreased by 55%. As observed previously, all proteins showed a significant correlation with benzene exposure (r = -0.5; P = 0.0012, r = -0.8; P < 0.0001 and r = -0.5; P = 0.0011, respectively).
Fig. 2.
Spectra from ProteinChip array with immobilized antibodies against PF4 (B) and NAP-2 (C) for two exposed and two unexposed subjects and representative spectra of the negative control (nonspecific rabbit IgG; A). x axis depicts m/z. y axis depicts the average intensity of ion peaks.
Fig. 3.
Scatter plot and regression line between the RAI of the initial proteomic experiments and the PF4 and CTAP-III levels, as quantified by ProteinChip immunoassay (Ciphergen Biosystems). Open rounds are the results from the discovery set, and solid rounds are the results from the validation set of the initial experiment.
Table 2. Mean PF4, CTAP-III, and PBP levels in benzene-exposed (n = 20) and unexposed subjects (n = 20).
| Protein | Unexposed, mean (SD) | Exposed, mean (SD) | P* | Decrease, % |
|---|---|---|---|---|
| PF4, μg/ml | 5.19 (2.64) | 3.14 (1.49) | 0.0014 | 39.5 |
| CTAP-III (AI) | 1.58 (0.46) | 0.59 (0.28) | 3 E-9 | 62.7 |
| PBP (AI) | 0.11 (0.08) | 0.05 (0.03) | 0.0067 | 54.5 |
AI, average intensity.
Wilcoxon exact test
Both PF4 and CTAP-III are platelet-derived chemotactic cytokines (chemokines). Although they are also secreted by other cells, platelets remain the most abundant and the most rapidly available source for these CXC-chemokines (17, 18). It is well established that benzene lowers peripheral platelet counts (7-9), raising the possibility that our finding could be explained entirely or in part by a decrease in platelet counts among the exposed subjects. However, we determined that the absolute platelet count did not significantly impact on the difference in expression of PF4 and CTAP-III between benzene exposed and unexposed subjects by using linear regression on the natural logarithm of each protein adjusted and unadjusted for platelet count (Table 3). Additionally, total white blood cells and specific white blood cell types (e.g., monocytes, granulocytes, and lymphocytes) did not influence the relationship between exposure status and reduced expression of PF4 and CTAP-III.
Table 3. Difference in PF4 and CTAP-III peak intensities between benzene-exposed and unexposed subjects by linear regression.
| Unadjusted* for platelet count
|
Adjusted† for platelet count
|
|||
|---|---|---|---|---|
| Protein | Regression coefficient | P | Regression coefficient | P |
| PF4 | –0.53 | 0.0008 | –0.54 | 0.0037 |
| CTAP-III | –1.09 | <0.0001 | –0.90 | <0.0001 |
Linear regression on the natural logarithim (ln) of each protein with exposure status (yes/no) as independent predictor. Analyses are unadjusted, because age, sex, current smoking, recent infections, and alcohol use did not weaken the effect of benzene exposure on any of the identified proteins. Relative difference between exposed and unexposed can be calculated as follows: Exposed/Unexposed = expβ = exp(–0.53) = 0.6
Linear regression on the ln of each protein adjusted for individual platelet count, which itself was not significant in the model
Discussion
This molecular epidemiological study uses SELDI-TOF MS for in vivo studies of the effects of a specific chemical exposure in humans. By using two sets of 10 exposed and 10 unexposed subjects, we were able to identify three differentially expressed proteins in the serum of benzene-exposed individuals. Two proteins were positively identified as PF4 and CTAP-III, both members of the CXC-chemokine family.
We used a two-step approach in which the first set was used to reveal differentially expressed proteins related to benzene exposure, and the second set was used to confirm the earlier findings. Although splitting the study population will inevitably result in the loss of power to detect more subtle differences in protein expression, it does permit evaluation of the reproducibility of the findings, especially relevant in this case, because the two studies were performed several months apart on slightly different platforms. Reproducibility, as assessed by blind quality control samples was good with coefficients of variance (CVs) ranging from 0.4% to 34% (mean = 13%) for the 18 peaks selected in the discovery phase (CVs for PF4, CTAP-III, and the 4.1-kDa protein were 14%, 10%, and 18%, respectively). Overall, these results indicate that acceptable reproducibility can be achieved with SELDI-TOF MS. However, the different absolute values of average peak intensities that arose from the two experiments using two different models of the ProteinChip reader (Ciphergen Biosystems) hindered the direct grouping of the results. We therefore arbitrarily set the mean peak among the controls to 1, achieved by dividing the individual peak intensities in both the exposed and unexposed subjects by the mean peak intensity of the controls in both the discovery and validation sets. We assumed that the average peak intensities in both sets were equal, because they came from the same control population and had been treated identically before analysis. After normalizing, no difference in RAI was observed for PF4, CTAPIII, and the unidentified 4.1-kDa protein between the exposed subjects of the discovery and validation study, indicating that this normalization worked for these peaks. Note that benzene exposure was essentially similar between the two sets, and the magnitude of the effect in the two sets should be comparable among the exposed subjects. Furthermore, the fact that the correlation between the RAI of the initial SELDI-TOF analyses, and the average intensities from the ProteinChip immunoassays (Ciphergen Biosystems) were not influenced by experiment (i.e., discovery and validation) indicates that this normalization was, indeed, adequate, at least for these proteins (see Fig. 3).
The peak intensities for the three identified proteins were markedly lower in the exposed group, with almost no overlap and extremely low P values. Accounting for multiple comparisons by the false discovery rate (19) resulted in P values <0.05 for all three proteins. Moreover, although we screened for approximately a thousand protein features in these samples, the observed P values (P < 5 E-05) also sustain a more conservative correction for multiple comparisons (Bonferroni; P < 0.05). In addition, given that the identified proteins showed a very clear association with individual benzene exposure levels, we are confident that the difference in protein expression for these proteins is, indeed, benzene-related, as was later confirmed by the essentially similar results based on the ProteinChip immunoassay (Ciphergen Biosystems).
Proteins were selected for identification based on the difference in expression and the amount of material required to purify the protein. Two proteins were successfully purified and were positively identified as PF4 and CTAP-III. Both proteins are platelet-derived chemokines that are secreted from activated platelets practically simultaneously (17), raising the possibility that the observed differences in protein levels may have been driven by depressed platelet counts among benzene-exposed subjects. Several observations suggested that this was not the case. Platelet counts among the benzene-exposed subjects were 28% lower than controls, compared with an ≈40-63% difference in platelet-derived cytokine levels. More importantly, linear regression revealed that the relationship between PF4 and CTAP-III was not markedly different when corrected for absolute platelet counts or any of the other measured white blood cell types. These results are an indication that the difference in serum levels of platelet-derived cytokines is not a simple reflection of the decrease in the absolute count of peripheral platelets or other blood cells, further corroborated by our recent observation that expression of PF4 RNA in peripheral blood mononuclear cells was consistently down-regulated in exposed individuals (mean decrease 58%) when compared with unexposed individuals (20).
PF4, also known as CXCL4, is an ELR(-) CXC-chemokine present in platelet α-granules that is released during platelet aggregation and inhibits heparin-mediated reactions. PF4 is also released from activated T lymphocytes and mast cells (21). It has been shown to influence numerous other biological properties, including inhibiting endothelial cell proliferation, migration, and angiogenesis (22-24), inhibiting T cell function by down-modulating cell proliferation and cytokine release (25), and supporting the survival of normal hematopoietic precursors and protecting them from the toxicity of chemotherapeutic agents (26). CTAP-III is an N-terminal cleavage product of PBP, which is synthesized in megakaryocytes. The different products resulting from proteolysis of PBP, also called β-thromboglobulins, include a group of homologous and immunologically cross-reactive platelet α-granule-derived proteins, CTAP-III, β-thromboglobulin (β-TG), and NAP-2. All except NAP-2 are secreted by platelets. Whereas NAP-2 may initiate neutrophil infiltration at sites of inflammation, PBP and, especially, CTAP-III have been shown to counteract the stimulatory potential of NAP-2 and other CXC-chemokines (27). These precursors might, therefore, act as potent agents to down-modulate neutrophil activation, thus having antiinflammatory properties. In addition, Krijgsveld et al. (28) have demonstrated that C-terminally truncated variants of CTAP-III/NAP-2 act as microbicidal proteins. CTAP-III has also been reported to support stem-cell-derived hematopoiesis and, like PF4, to protect early cells from the toxic effects of various chemotherapeutic agents (26). Thus, down-regulation of CXC-chemokines may play a role in benzene hematotoxicity and diseases related to benzene exposure, such as leukemia and myelodysplastic syndromes.
In conclusion, we have used SELDI-TOF MS-based proteomics to discover potential biomarkers of exposure and early effect for benzene in a molecular epidemiology study. Our studies revealed lower expression of platelet-derived CXC-chemokines in benzene-exposed subjects that may contribute to the immuno- and hematotoxic effects of benzene and serve as a potential biomarker of early effect. We were able to obtain these findings in a relatively small number of suitably matched exposed and control individuals and confirm the results from the independent discovery and validation phases by reanalyzing all samples by using a ProteinChip immunoassay (Ciphergen Biosystems). This study, therefore, provides a model for biomarker discovery in chemically exposed human populations, although, with lower-exposed populations or less toxic chemicals, it may be necessary to study larger populations.
Supplementary Material
Acknowledgments
We thank Dr. Lee Moore for helpful review of the manuscript; Enrique A. Dalmasso, Regis Perichon, and Stacy Moore for skillful technical assistance; and Dr. Joseph F. Fraumeni, Jr., for long-term support of collaborative research studies in China, which provided the foundation for this project. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and by NIH grants R01ES06721 and P30ES01896 (to M.T.S.) from the National Institute of Environmental Health Sciences.
Author contributions: R.V., Q.L., L.Z., G.L., S.Y., N.R., and M.T.S. designed research; R.V., Q.L., L.Z., L.G., D.M., R.L.W., M.M., V.N.P., G.L., R.M., S.Y., N.R., and M.T.S. performed research; D.M., R.L.W., M.M., and V.N.P. contributed new reagents/analytic tools; R.V., D.M., N.C., N.R., and M.T.S. analyzed data; and R.V., D.M., N.R., and M.T.S. wrote the paper.
Conflict of interest statement: M.T.S. has received consulting and expert testimony fees from law firms representing both plaintiffs and defendants in cases involving exposure to benzene. G.L. has received funds from the American Petroleum Institute for consulting on benzene-related health research.
Abbreviations: CTAP, connective-tissue-activating peptide; NAP, neutrophil-activating peptide; PF, platelet factor; PBP, platelet basic protein; RAI, relative average intensity; SELDI, surface-enhanced laser desorption/ionization.
References
- 1.Aksoy, M. (1985) Am. J. Ind. Med. 7, 395-402. [DOI] [PubMed] [Google Scholar]
- 2.Austin, H., Delzell, E. & Cole, P. (1988) Am. J. Epidemiol. 127, 419-439. [DOI] [PubMed] [Google Scholar]
- 3.Hayes, R. B., Yin, S. N., Dosemeci, M., Li, G. L., Wacholder, S., Chow, W. H., Rothman, N., Wang, Y. Z., Dai, T. R., Chao, X. J., et al. (1996) Environ. Health Perspect. 104, Suppl. 6, 1349-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes, R. B., Yin, S. N., Dosemeci, M., Li, G. L., Wacholder, S., Travis, L. B., Li, C. Y., Rothman, N., Hoover, R. N. & Linet, M. S. (1997) J. Natl. Cancer Inst. 89, 1065-1071. [DOI] [PubMed] [Google Scholar]
- 5.Savitz, D. A. & Andrews, K. W. (1997) Am. J. Ind. Med. 31, 287-295. [DOI] [PubMed] [Google Scholar]
- 6.Yin, S. N., Hayes, R. B., Linet, M. S., Li, G. L., Dosemeci, M., Travis, L. B., Zhang, Z. N., Li, D. G., Chow, W. H., Wacholder, S., et al. (1996) Environ. Health Perspect. 104, Suppl. 6, 1339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan, Q., Zhang, L., Li, G., Vermeulen, R., Weinberg, R. S., Dosemeci, M., Rappaport, S. M., Shen, M., Alter, B. P., Wu, Y., et al. (2004) Science 306, 1774-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu, Q., Shore, R., Li, G., Jin, X., Chen, L. C., Cohen, B., Melikian, A. A., Eastmond, D., Rappaport, S. M., Yin, S., et al. (2002) Am. J. Ind. Med. 42, 275-285. [DOI] [PubMed] [Google Scholar]
- 9.Rothman, N., Li, G. L., Dosemeci, M., Bechtold, W. E., Marti, G. E., Wang, Y. Z., Linet, M., Xi, L. Q., Lu, W., Smith, M. T., et al. (1996) Am. J. Ind. Med. 29, 236-246. [DOI] [PubMed] [Google Scholar]
- 10.Rosenthal, G. J. & Snyder, C. A. (1985) Toxicol. Appl. Pharmacol. 80, 502-510. [DOI] [PubMed] [Google Scholar]
- 11.White, W. C. & Gammon, A. M. (1914) Trans. Assoc. Amer. Phys. 29, 332-337. [Google Scholar]
- 12.Tolson, J., Bogumil, R., Brunst, E., Beck, H., Elsner, R., Humeny, A., Kratzin, H., Deeg, M., Kuczyk, M., Mueller, G. A., et al. (2004) Lab. Invest. 84, 845-856. [DOI] [PubMed] [Google Scholar]
- 13.Wadsworth, J. T., Somers, K. D., Stack, B. C., Jr., Cazares, L., Malik, G., Adam, B. L., Wright, G. L., Jr., & Semmes, O. J. (2004) Arch. Otolaryngol. Head Neck Surg. 130, 98-104. [DOI] [PubMed] [Google Scholar]
- 14.Lehrer, S., Roboz, J., Ding, H., Zhao, S., Diamond, E. J., Holland, J. F., Stone, N. N., Droller, M. J. & Stock, R. G. (2003) BJU Int. 92, 223-225. [DOI] [PubMed] [Google Scholar]
- 15.Zhukov, T. A., Johanson, R. A., Cantor, A. B., Clark, R. A. & Tockman, M. S. (2003) Lung Cancer 40, 267-279. [DOI] [PubMed] [Google Scholar]
- 16.Vermeulen, R., Li, G., Lan, Q., Dosemeci, M., Rappaport, S. M., Bohong, X., Smith, M. T., Zhang, L., Hayes, R. B., Linet, M., et al. (2004) Ann. Occup. Hyg. 48, 105-116. [DOI] [PubMed] [Google Scholar]
- 17.Brandt, E., Ludwig, A., Petersen, F. & Flad, H. D. (2000) Immunol. Rev. 177, 204-216. [DOI] [PubMed] [Google Scholar]
- 18.Brandt, E., Petersen, F., Ludwig, A., Ehlert, J. E., Bock, L. & Flad, H. D. (2000) J. Leukocyte Biol. 67, 471-478. [DOI] [PubMed] [Google Scholar]
- 19.Hochberg, Y. & Benjamini, Y. (1990) Stat. Med. 9, 811-818. [DOI] [PubMed] [Google Scholar]
- 20.Forrest, M. S., Lan, Q., Hubbard, A. E., Zhang, L., Vermeulen, R., Zhao, X., Li, G., Wu, Y. Y., Shen, M., Yin, S., et al. (2005) Environ. Health Perspect. 113, 801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehlen, F. & Clemetson, K. J. (2001) Transfus. Med. 11, 403-417. [DOI] [PubMed] [Google Scholar]
- 22.Strieter, R. M., Belperio, J. A., Phillips, R. J. & Keane, M. P. (2004) Novartis Found. Symp. 256, 173-184. [PubMed] [Google Scholar]
- 23.Szekanecz, Z., Kim, J. & Koch, A. E. (2003) Semin. Immunol. 15, 15-21. [DOI] [PubMed] [Google Scholar]
- 24.Szekanecz, Z. & Koch, A. E. (2001) Curr. Opin. Rheumatol. 13, 202-208. [DOI] [PubMed] [Google Scholar]
- 25.Fleischer, J., Grage-Griebenow, E., Kasper, B., Heine, H., Ernst, M., Brandt, E., Flad, H. D. & Petersen, F. (2002) J. Immunol. 169, 770-777. [DOI] [PubMed] [Google Scholar]
- 26.Han, Z. C., Lu, M., Li, J., Defard, M., Boval, B., Schlegel, N. & Caen, J. P. (1997) Blood 89, 2328-2335. [PubMed] [Google Scholar]
- 27.Ehlert, J. E., Ludwig, A., Grimm, T. A., Lindner, B., Flad, H. D. & Brandt, E. (2000) Blood 96, 2965-2972. [PubMed] [Google Scholar]
- 28.Krijgsveld, J., Zaat, S. A., Meeldijk, J., van Veelen, P. A., Fang, G., Poolman, B., Brandt, E., Ehlert, J. E., Kuijpers, A. J., Engbers, G. H., et al. (2000) J. Biol. Chem. 275, 20374-20381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.