Abstract
Homozygous zebrafish of the mutant relaxed (redts25) are paralyzed and die within days after hatching. A significant reduction of intramembrane charge movements and the lack of depolarization-induced but not caffeine-induced Ca2+ transients suggested a defect in the skeletal muscle dihydropyridine receptor (DHPR). Sequencing of DHPR cDNAs indicated that the α1S subunit is normal, whereas the β1a subunit harbors a single point mutation resulting in a premature stop. Quantitative RT-PCR revealed that the mutated gene is transcribed, but Western blot analysis and immunocytochemistry demonstrated the complete loss of the β1a protein in mutant muscle. Thus, the immotile zebrafish relaxed is a β1a-null mutant. Interestingly, immunocytochemistry showed correct triad targeting of the α1S subunit in the absence of β1a. Freeze-fracture analysis of the DHPR clusters in relaxed myotubes revealed an ≈2-fold reduction in cluster size with a normal density of DHPR particles within the clusters. Most importantly, DHPR particles in the junctional membranes of the immotile zebrafish mutant relaxed entirely lacked the normal arrangement in arrays of tetrads. Thus, our data indicate that the lack of the β1a subunit does not prevent triad targeting of the DHPR α1S subunit but precludes the skeletal muscle-specific arrangement of DHPR particles opposite the ryanodine receptor (RyR1). This defect properly explains the complete deficiency of skeletal muscle excitation-contraction coupling in β1-null model organisms.
Keywords: calcium channels, excitation-contraction coupling, tetrads, zebrafish
Excitation-contraction (EC) coupling is understood as the signal transduction process connecting membrane depolarization to the contraction of muscle cells. This process is initiated by the concerted action of two Ca2+ channels, the plasmalemmal voltage-gated dihydropyridine receptor (DHPR) and the sarcoplasmic reticulum (SR) ryanodine receptor (RyR). In junctions of the SR with the plasma membrane (peripheral couplings) or with the transverse tubules (triads), membrane depolarization is sensed by the DHPR, which then triggers RyR opening and Ca2+ release from the SR. In skeletal muscle cells, this signal-transduction is independent of Ca2+ influx through the DHPR (1) but depends on protein-protein interaction between the DHPR and the RyR1 (2, 3). This physical coupling requires the coordinated arrangement of DHPRs and RyR1s in the junctions. In skeletal muscle triads and peripheral couplings, groups of four DHPRs (tetrads) are arranged in orthogonal arrays matching the opposing RyR1 arrays (4). Formation of DHPR tetrads requires the presence of RyR1.
The skeletal muscle DHPR complex is composed of the voltage-sensing and pore-forming α1S subunit and the auxiliary subunits β1a, α2δ-1, and γ (5). Targeted deletions of the α2δ-1 and γ subunits do not critically interfere with EC coupling function (6, 7). In contrast, α1S and β1a subunit null-mutant mice display a lack of EC coupling and, thus, lethal muscle paralysis (8, 9). Although failure of EC coupling is obvious in the α1S-null (dysgenic) mouse muscle, which lacks the voltage-sensor, the molecular mechanism leading to the complete lack of EC coupling in the β1a-null mice is not fully understood (8). Myotubes from β1a-null mice show reduced L-type Ca2+ currents, charge movements, and 1,4-dihydropyridine binding (10). The staining intensity for α1S in immunocytochemistry was initially described to be below detectability (8), but later it was found to be significantly higher than that of dysgenic myotubes (11). Therefore, it remained unclear whether the loss of EC coupling in β1a-null myotubes was caused by decreased membrane expression of DHPRs, or by the lack of a specific contribution of the β subunit to the EC coupling mechanism (10, 12).
We addressed this question in the paralyzed zebrafish mutant relaxed (13), for which a defect in the EC coupling apparatus had been proposed (14). Using molecular biology, protein biochemistry, and immunocytochemistry techniques, we show that the zebrafish mutant relaxed lacks the DHPR β1a subunit. In contrast to previous studies (8, 11), we can use this novel model organism to demonstrate that the α1S is correctly targeted into skeletal muscle triad junctions in the absence of β1a. However, DHPRs lacked the skeletal muscle-specific arrangement in tetrads and displayed substantially reduced charge movement. Therefore, a disruption of functional DHPR-RyR interactions caused by the lack of the β1a subunit is responsible for the paralysis of skeletal muscle in β1-null muscle cells.
Materials and Methods
Zebrafish Strain. Zebrafish of the strain redts25 (relaxed) (13) were obtained from the Max Planck Institute in Tübingen, Germany. After cross breeding of heterozygous parents, homozygous relaxed larvae were identified by the inability to move in response to tactile stimuli. Larvae with a normal phenotype (heterozygous and wild type) were used for control experiments and will be collectively referred to as “normal.”
Isolation of Myocytes. Larvae were anesthetized with MS-222 (Sigma) and decapitated, and the tails were digested for 1 h in 200 units/ml collagenase in Hanks' solution (Sigma) with continuous trituration. Myotubes were plated on collagen-coated plastic dishes or glass cover slips and cultured in 60% L-15 medium (Sigma) with 3% FCS and 3% horse serum (Invitrogen) at 28°C for 12-72 h.
Biophysical Characterization. For field stimulation experiments, myotubes from 3- to 4-day-old larvae were incubated with 5 μM Fluo-4 AM plus 0.01% Pluronic F-127 (Molecular Probes) in 60% L-15 medium. Action potentials were elicited by applying extracellular electrical stimuli (0.5 Hz, 25 V/cm electrode distance, 1 ms). Ca2+ signals (Fluo-4) were recorded by a photometer system (PTI, South Brunswick, NJ) mounted on a Zeiss Axiovert epifluorescence microscope (15).
Whole-cell patch clamp recordings of Ca2+ currents, intracellular Ca2+ transients, and intramembrane charge movements were performed as recently described for mouse myotubes (16, 17). The following modifications were applied to eliminate a robust Ca2+-activated Cl- current in zebrafish muscle cells¶: The bath solution contained 10 mM Ca(OH)2, 100 mM l-aspartate, and 10 mM Hepes (pH 7.4 with tetraethylammonium hydroxide). Contractions of myotubes were blocked by adding 100 μM N-benzyl-p-toluene sulfonamide (Sigma) (18). Patch pipettes had resistances of 3-5 MΩ when filled with 145 mM CsAspartate, 10 mM Hepes, 0.5 mM CsEGTA, and 3 mM MgATP (pH 7.4 with CsOH).
Sequence Analysis. α1S and β DHPR subunit cDNAs and genomic DNAs from wild type zebrafish and from homozygous relaxed larvae were PCR amplified by using the Pfu Turbo DNA polymerase (Stratagene) and sequenced (MWG Biotech). Total RNA was isolated by using the RNeasy Mini kit (Qiagen) and reverse transcribed by using the Ready-To-Go T-primed first-strand kit (Amersham Pharmacia). Genomic DNA was isolated according to the QIAamp DNA Mini kit protocol (Qiagen).
α1S cDNA. 12 overlapping fragments (≈650 bp) were obtained by RT-PCR. Primers were designed according to zebrafish whole genome shotgun (WGS)-trace exon sequences identified by a NCBI blast search with carp α1S cDNA (19) as template. The sequenced zebrafish α1S was deposited in the GenBank database (accession no. AY495698).
β1a cDNA. A zebrafish β1a-specific PCR primer pair was designed according to the outermost 5′ and 3′ exon sequences that could be identified by a NCBI zebrafish WGS-trace blast search using rabbit β1a (20) as template. The zebrafish β1a RT-PCR fragment (nucleotides 96-1493; GenBank accession no. AY952462) was derived from adult wild-type cDNA. For sequence analysis, a blunt-HindIII fragment (nucleotides 96-1353) was cloned into the SmaI/HindIII polylinker site of pBluescript SK+ (Stratagene). The derived sequence was used to identify exons 2-13 of β1a by a WGS-trace blast search. Exon 1 was identified using the Universal GenomeWalker kit (Clontech). Exon-flanking intron primer pairs were designed to sequence all 13 exons and exon-intron transitions from wild-type and mutant relaxed.
Quantitative TaqMan PCR. Normal and relaxed cDNA was generated as described above. The relative abundance of β1 subunit mRNA was assessed by TaqMan quantitative PCR (50 cycles) using the comparative CT method (Applied Biosystems) and the β-actin transcript as reference. Total RNA (7.6 ng) equivalents of cDNA and the specific TaqMan Gene Expression Assay were used for each 20-μl reaction in TaqMan Universal PCR Master Mix (Applied Biosystems). RNA samples without reverse transcriptase and samples without template were routine controls. Analysis was performed by using the Mx4000 Multiplex Quantitative PCR System (Stratagene). Custom TaqMan Gene Expression Assays were designed to span exon-exon boundaries: Exons 2/3 of zebrafish β1a mRNA and exons 3/4 of zebrafish β-actin mRNA (GenBank accession no. NM131031). Exon-intron structures of β-actin and Ca2+ channel β1a subunit genes were derived from NCBI blast searches of the zebrafish WGS trace database.
Immunostaining. Myotubes from 3- to 4-day-old larvae were cultured for 1 day on glass cover slips, fixed with either 100% methanol (10 min at -20°C) or 4% paraformaldehyde in 0.1 M phosphate buffer (20 min at room temperature) and immunostained as described (15). Antibodies were mAb 1A against α1S (Affinity Bioreagents, ref. 21) at 1:2,000-30,000 (see below), mAb 20A against α2δ-1 (22) at 1:1,000, sequence-directed pan-β antibody RCP6 (23) at 1:2,000, and mAB 34-C against RyR (Alexis Biochemicals, Lausen, Switzerland) at 1:2,000 Alexa-conjugated goat-anti-rabbit and goat-anti-mouse (Molecular Probes) were used as secondary antibodies. Omission of primary antibodies was routinely performed as controls. Experiments were repeated at least three times.
For quantification of α1S expression in normal and relaxed myotubes mAB 1A dilutions of 1:2,000, 1:10,000, 1:20,000, and 1:30,000 were used to confirm the linearity and reproducibility of the fluorescence signal. Cultures were immunostained simultaneously, and images were acquired with identical exposure times after background subtraction and shading correction. Average fluorescence intensity was recorded along a line across a row of α1S clusters (triadic junctions) in five measurements on each myotube, with 10 myotubes analyzed for each condition and antibody dilution in each of three different cultures. Statistical significance was determined by paired Student's t test (group: cell preparation/AB dilution).
Western Blot. For total membrane protein preparation ≈300 tails of relaxed and normal larvae (4-6 days old), as well as skeletal muscle tissue from an adult zebrafish were ultrasonicated in 10 mM Tris·HCl with 10 mM EDTA and 5 μl/ml Proteinase Inhibitor Mixture for mammalian tissues (Sigma). After centrifugation at 400 × g, the supernatant was centrifuged at 50,000 × g to precipitate membrane fractions. Pellets were resuspended in the buffer described above. Rabbit muscle protein was isolated as described (24) and used as a control. Isolated protein (10 μg per lane) was loaded for β1a blots, and 15-50 μg was loaded for α1S blots and separated by SDS/PAGE (NuPage 4-12% Bis-Tris gel with NuPage Mops SDS running buffer, both Invitrogen) at 200 V for 60 min.
Proteins were blotted to an Immobilon-P polyvinylidene difluoride membrane (Millipore) using Tris-Glycine buffer with NuPAGE antioxidant (Invitrogen). For blotting of the β subunit protein (1.5 h at 200 mA), 10% methanol was added to the buffer; for the hydrophobic, high molecular weight α1S subunit (1 h at 100 V), 20% methanol and 0.1% SDS was added. After blocking (4% dry milk for 1 h), membranes were incubated overnight with primary antibodies RCP6 or mAB 1A, respectively, and detected with the HRP-system (ECL, Amersham Pharmacia).
Freeze Fracture Electron Microscopy. Myoblasts from 1-day-old larvae were cultured on Thermanox (Nalge Nunc) for 3 days until fusion, rinsed in PBS, and fixed with 6% glutaraldehyde in 0.1 M cacodylate buffer (both Sigma) at neutral pH at ≈23°C. Samples were stored in 2% glutaraldehyde at 4°C until processing for freeze-fracture. All other procedures and solutions have been recently described for fracturing mouse myotubes (25). For thin sectioning and freeze-fracture, 26- to 72-h-old larvae were fixed in 6% glutaraldehyde after removing the tail skin and processed as in ref. 4.
Statistics. Statistical significance from experimental approaches was determined by unpaired Student's t test, and data are reported as mean ± SEM, unless noted otherwise.
Results and Discussion
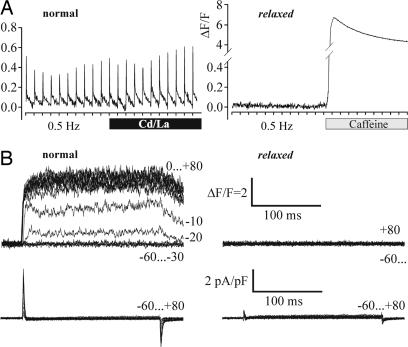
Phenotype and Genotype of the Zebrafish Mutant relaxed. The zebrafish mutant relaxed was obtained from a large-scale mutagenesis screen (26) and is characterized by complete skeletal muscle paralysis (13). Previous work indicated a defect in the EC coupling complex, most likely in the DHPR (14). To identify the molecular mechanism underlying the EC coupling defect, we first analyzed the response of myotubes from normal and relaxed zebrafish larvae to electrical field stimulation. As expected for skeletal muscle EC coupling, normal myotubes showed pronounced action-potential-induced intracellular Ca2+ transients, which were insensitive to L-type Ca2+ channel block by Cd2+ and La3+ (Fig. 1A). Relaxed myotubes did not respond to electrical stimulation, although the subsequent addition of 6 mM caffeine caused a strong Ca2+ transient, indicating that RyR-dependent SR Ca2+ release was fully functional. To exclude the possibility of a defect in muscle membrane excitability, whole-cell patch clamp analysis of L-type Ca2+ currents, combined with fluorometric recordings of depolarization-induced intracellular Ca2+ release, was performed (Fig. 1B). In normal myotubes, depolarizations to potentials between -20 and +80 mV induced robust Ca2+ transients with a mean ΔF/Fmax of 4.6 ± 0.6 (n = 9). However, depolarization-induced Ca2+ transients were never observed in relaxed myotubes (n = 6). Thus, the defect in EC coupling is located downstream of depolarization and upstream of SR Ca2+ release, pointing to a failure of voltage-sensing or the allosteric transmission of this signal to the RyR1.
Fig. 1.
Myotubes from zebrafish mutant relaxed lack EC coupling. (A) Recordings of action-potential-induced intracellular Ca2+ transients from dissociated myotubes of larval tail muscle loaded with the fluorescent Ca2+ indicator Fluo-4 AM. Tick marks on the x axes indicate electrical stimuli in 2-s intervals. Normal myotubes (Left) responded to 1-ms stimuli with Ca2+ transients that persisted after applying 0.5 mM Cd2+ and 0.1 mM La3+ (black bar). Myotubes isolated from zebrafish relaxed failed to display action-potential-induced Ca2+ transients (Right), even though Ca2+ release could be evoked with 6 mM caffeine (shaded bar), indicating intact loading of SR stores and functional RyR (n = 5 and 4, respectively). (B) Simultaneous recordings of depolarization-induced Ca2+ transients (Upper) and whole-cell Ca2+ currents (Lower) from normal (Left) and relaxed (Right) myotubes. Step depolarizations (200-ms pulses) to membrane potentials between -60 and +80 mV were applied in 10-mV increments from a holding potential of -80 mV following a prepulse protocol (17). Changes in the cytoplasmic free [Ca2+] were measured with Fluo-4 and are displayed as ΔF/F. Note that, in both normal and relaxed myotubes, ICa was never detected (n > 30).
To our surprise L-type Ca2+ currents were absent in both normal and relaxed myotubes (Fig. 1B Lower). Thus, the zebrafish DHPR is a non-Ca2+-conducting voltage sensor, and this organism provides in vivo evidence that skeletal muscle EC coupling is fully functional without Ca2+ influx through the DHPR.
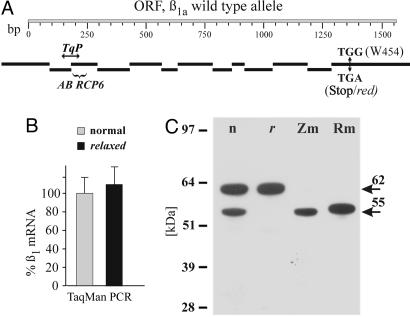
Because the functional evidence suggested a defective DHPR in relaxed muscle and because only two of the four DHPR subunits (α1S and β1a) cause paralysis in null-mutant mice, we searched for a mutation in these subunits. Sequencing of α1S cDNA revealed no sequence deviations between wild-type and relaxed zebrafish. However, cloning and sequencing of the β1a cDNA and, consequently, of all 13 β1a exons and exon-intron transitions from wild-type and relaxed zebrafish identified a point mutation (G1362A) in exon 13 (Fig. 2A). The ORF of the wild-type β1a subunit mRNA encodes for a 520-aa protein (Mr 57,445) with 76% sequence identity to rabbit β1a, but only 52, 43, and 42% to the rabbit β2, β4, and β3 subunits, respectively. The G-A point mutation in exon 13 of relaxed β1a DNA leads to the premature termination at the position of Trp-454. The observed fraction of 24.6% (n = 896) paralyzed offspring when mating heterozygous adults is consistent with the monogenetic heredity of the EC coupling defect (27).
Fig. 2.
The DHPR β1a subunit of zebrafish relaxed is transcribed as mRNA but not expressed as protein. (A) Schematic representation of the 13 β1a exons (black bars) aligned to the ORF (ORF, shaded bar) of its cDNA (GenBank accession no. AY952462). Numbers designate base pairs. TGG (W454) ↔ TGA (Stop/red) indicates the position of the point mutation in exon 13 leading to a premature stop in β1a of relaxed. TqP shows the position of the TaqMan probe. The parentheses indicate the epitope of pan-β antibody RCP6. (B) β1 mRNA transcription in relaxed normalized to normal zebrafish assessed by TaqMan quantitative PCR. Mean values of two individual experiments, from each of two separate cDNA preparations, are shown and are statistically indistinguishable (P = 0.87). Statistical significance was determined by using the ΔCT values in a paired student's t test (group, RNA preparation/TaqMan assay). (C) Western blot analysis (one representative experiment of nine is shown) with pan-β antibody RCP6 indicated that a 55-kDa β band (lower arrow) is expressed in normal larval tail (n) and adult zebrafish muscle (Zm) but is lacking in relaxed tail preparations (r). The 62-kDa protein band (upper arrow) in both larval tail preparations most probably represents another β isoform originating from other larval tissues (e.g., spinal cord). Pure skeletal muscle preparations from adult normal zebrafish (Zm) and from rabbit (Rm) do not contain this 62-kDa isoform.
The Zebrafish relaxed Is a β1a-Null Mutant. According to the recently published β subunit crystal structure (28, 29), the mutation would lead to a β1a protein that is truncated within the most C-terminal α-helix of the guanylate kinase domain. Because the consequences of this premature stop were not predictable, we analyzed the expression of the mutated β1a mRNA and protein. Using quantitative TaqMan PCR, we measured similar levels of β1 transcripts in normal and relaxed larvae (Fig. 2B), excluding a decreased stability of the message. Western blot analysis of preparations from normal zebrafish larvae (Fig. 2C) using the pan-β antibody RCP6 (23) directed against an epitope near the N terminus (see Fig. 2 A) revealed two specific bands at 55 and 62 kDa. Both bands could be completely blocked by antigen preincubation of the antibody (23). Most importantly, the 55-kDa band was absent in preparations from relaxed larvae (Fig. 2C). This band was identified as the skeletal muscle β1a homolog of zebrafish based on the following evidence: It was the exclusive protein labeled in pure muscle preparations from normal adult zebrafish, its size agrees with the published molecular mass of rabbit β1a (52-56 kDa) (23), as well as with the calculated Mr for zebrafish β1a (57 kDa), and it is clearly apart from the 62- to 88-kDa range of β1b-β4 (23). The higher molecular mass band found in the tail preparations from larvae most likely represents a homologue of another β isoform expressed in neuronal or smooth muscle tissue present in this preparation. Technical limitations did not allow isolation of pure muscle tissue from the ≈5-mm small larvae, which were used before relaxed larvae die at day 6-7.
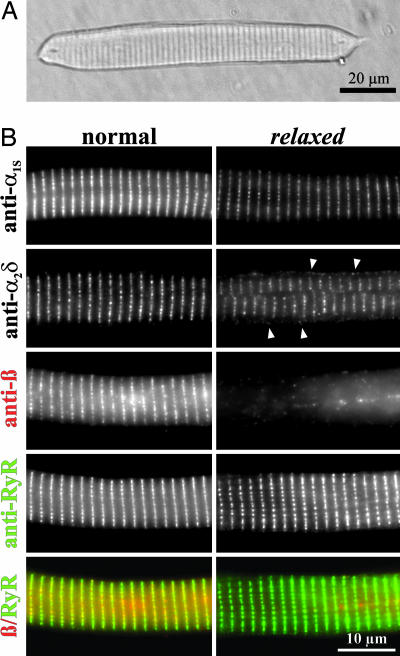
Next, expression of the DHPR β1a subunit was analyzed with immunolabeling of myotubes isolated from normal and relaxed zebrafish (Fig. 3). Double immunofluorescence labeling with antibodies against the β subunit and RyR1 showed that, in normal myotubes, the DHPR β1a subunit and the Ca2+ release channel are colocalized in transverse lines of clusters representing transverse tubule/SR junctions. Whereas the expression pattern of RyR1 is unaltered in myotubes of relaxed zebrafish, the specific β1a signal was absent.
Fig. 3.
Triad targeting properties of the DHPR subunits α1S, α2δ-1, and β1a in myotubes from zebrafish mutant relaxed differ from those in normal myotubes. (A) Typical appearance of an adherent zebrafish myotube isolated from larval tail muscle and visualized by Hoffman modulation contrast (36). No morphological differences could be observed between relaxed and normal myotubes. (B) Immunofluorescence labeling of normal (Left) and relaxed (Right) myotubes with mAB 1A against α1S (Top) showed correct triad clustering of the α1S subunits in relaxed, although at a reduced level. Staining of relaxed myotubes with mAB 20A against α2δ-1 (Upper Middle) showed that a fraction of the α2δ-1 subunit is normally clustered in triads whereas another fraction is mistargeted diffusely into the plasma membrane (arrowheads), presumably because of the reduction in α1S subunit levels (7, 33). In contrast to normal myotubes, immunostaining of relaxed myotubes with the pan-β antibody RCP6 (Middle) did not show any triadic staining, thus clearly demonstrating the β1a-null phenotype of the relaxed zebrafish. Double immunofluorescence labeling of the same myotubes with mAB 34-C against RyR (Lower Middle) showed identical triadic clustered staining in normal and relaxed. The merged image (Bottom) emphasizes the colocalization (yellow foci) of RyR1 (green) and β1a (red) in triadic clusters of normal myotubes.
Together, the specific lack of the lower β band in the Western blot analysis and the specific lack of the immunofluorescence signal in the samples from relaxed zebrafish clearly demonstrate the absence of the β1a protein in the mutant. Thus, the premature stop codon in the β1a transcript results either in deficient translation or in the immediate posttranslational degradation of the truncated protein. Truncation of up to 60 C-terminal residues had no adverse effect on expression of β chimeras in β1-null myotubes (30). In contrast, the data presented here show that the relaxed β1a protein, which lacks 67 C-terminal residues, is totally lost. The difference might be due to the fact that the mutant zebrafish β1a is truncated within the most C-terminal α-helical region of the guanylat kinase domain (28, 29), and thus affects a major structural element, which may be essential for proper expression. Alternatively, quality control mechanisms, which apply for endogenously expressed proteins, may have been bypassed in the heterologous expression experiments (30).
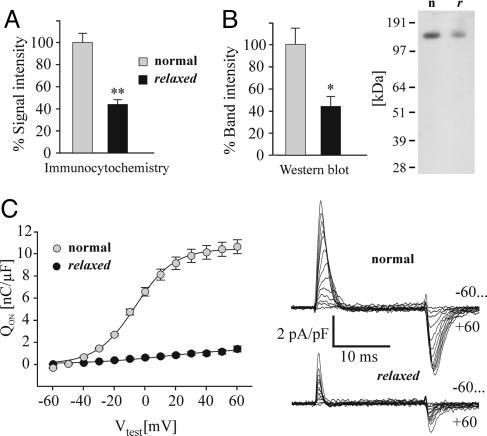
The Lack of β1a in Zebrafish relaxed Does Not Impede α1S Triad Targeting, but Results in a Failure of Skeletal Muscle-Specific Arrangement of DHPRs vis-à-vis the RyR1. The results presented so far demonstrated that the immotile zebrafish relaxed is a β1-null mutant. So, how does the lack of the β1a protein affect expression and function of the DHPR in skeletal muscle? Based on extensive coexpression experiments in heterologous systems (31, 32) and on results from the β1a-null mouse (8, 11), a lack of α1S triad expression would have been expected. However, immunofluorescence analysis of the fully differentiated zebrafish myotubes clearly demonstrated that β1a deficient cells are able to correctly target α1S into the triads (Fig. 3). This finding is in contrast to previous work describing α1S as only weakly expressed and located in a diffuse, nonclustered distribution. Thus, in native muscle, β1a is not essential for triad targeting of the DHPR. Nonetheless, quantification of the α1S immunofluorescence showed that in myotubes of the relaxed mutant the intensity of the signal was only about half (44 ± 4%) of that in normal myotubes (Fig. 4A). The reduction in α1S immunolabeling is consistent with a similar reduction of α1S protein measured in Western blots (45 ± 6%; Fig. 4B).
Fig. 4.
The DHPR α1S subunit is significantly reduced in relaxed compared with normal zebrafish skeletal muscle. (A) Quantification of the α1S (antibody mAB 1A) immunofluorescence signal intensity in normal and relaxed myotubes (for details, see Materials and Methods) indicated a reduction of α1S expression in relaxed myotubes to 44 ± 4% (**, P < 0.001; n = 10; paired t test). (B) mAB 1A staining intensity quantification (TINA 2.09c, Raytest Isotopen-messgeräte) after Western blot analysis indicated that the α1S protein band intensity from relaxed tail muscle protein preparations was reduced to 45 ± 6% compared to normal (*, P < 0.02). A representative Western blot experiment out of five is shown at Right; n indicates normal, and r indicates relaxed zebrafish. (C) (Left) Average (±SE) intramembrane charge movements (Qon) obtained from normal (gray circle, n = 15) or relaxed (black circle, n = 12) myotubes. Qon was determined by integrating the “ON” outward current at each membrane potential and normalizing it to cell capacitance (nC/μF). Average (±SE) maximum intramembrane charge movement (Qmax) was 10.4 ± 0.6 (n = 15) for normal and 1.4 ± 0.2 (n = 12) nC/μF for relaxed myotubes (P < 0.001). (Right) Representative charge movement recordings from normal (Upper) and relaxed (Lower) myotubes in response to 20-ms depolarizations to membrane potentials between -60 and +60 mV applied in 10-mV increments.
Previously, we demonstrated that, in the absence of the α1S subunit, α2δ-1 is expressed but mistargeted into the plasma membrane (7, 33). Accordingly, in normal zebrafish myotubes, the α2δ-1 subunit is localized in the triads, whereas this subunit is also present in the plasma membrane in relaxed myotubes (Fig. 3B). The appearance of α2δ-1 in the plasma membrane of relaxed myotubes suggests that, because of reduced α1S expression in the absence of β1a, α2δ-1 is in excess of α1S, and some of it remains diffusely distributed in the plasma membrane. Thus, regulation of membrane expression of α1S and α2δ-1 is independent from each other, and only membrane expression levels of α1S, but not of α2δ-1, depend on β1a.
Electrophysiological recordings indicated a substantial reduction of the DHPR-specific maximum intramembrane charge movement (Qmax) (17) in relaxed myotubes (Fig. 4C). Qmax represents the maximum amount of α1S gating charges moving across the membrane upon depolarization and is considered to be an accurate measure of functionally expressed α1S. Qmax values in relaxed myotubes were reduced by ≈7.4-fold compared to normal.
Apparently, there is a discrepancy between the ≈2.2-fold reduction in α1S expression levels and the substantial reduction of Qmax. Because we cannot exclude the possibility that other ion channels, which are not blocked by our prepulse procedure (1 s to -30 mV; 70 ms to -50 mV; ref. 17), also contribute to the measured Qmax, the difference between DHPR-specific Q in normal and relaxed myotubes could be even more pronounced; this would indicate that without the β1a subunit many of the DHPRs in the membrane are deficient in voltage-sensing. Such a defect could be induced by the loss of an allosteric action of β1a on α1S or by the disintegration of a putative cooperative effect of α1S arranged in tetrads (see below). In contrast, if the possible contribution of Ca2+-sensitive ion channels to Qmax is less than in normal myotubes because of missing Ca2+ transients in relaxed muscle, the discrepancy between the reduction in α1S expression levels and DHPR-specific Q could be much smaller. In this case, most of the membrane expressed DHPRs in relaxed myotubes would still be functional voltage sensors.
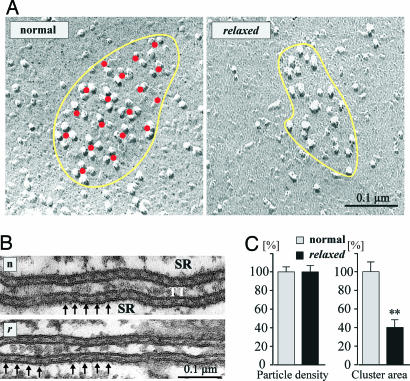
The skeletal muscle-specific characteristic of the DHPR is its direct coupling with the RyR, evident in Ca2+-independent EC coupling and the correlating structural organization of DHPRs in tetrad arrays in the junctional membranes of triads and peripheral couplings. Thus, an alternative explanation of the EC-coupling defect in relaxed zebrafish would be that the lack of β1a somehow disengages the coupling of α1S to the RyR1. Freeze-fracture analysis was performed on peripheral couplings because technical limitations do not allow obtaining images of the fractured transverse tubule membrane from the small larval myotubes. Similar results, as in isolated zebrafish myotubes, were obtained in 30-h-old whole embryo muscles (data not shown). The inner leaflet of the fractured plasmalemma of zebrafish myotubes revealed small domed membrane domains studded with groups of large intramembrane particles, which correspond to the DHPR clusters seen in immunofluorescence labeling (25, 34). In normal zebrafish myotubes, the DHPRs are arranged in tetrads that form regular orthogonal arrays (Fig. 5A). However, the corresponding domains of relaxed myotubes displayed a random distribution of particles, despite the normal organization of RyRs in arrays of “feet” as seen in sections of triads (Fig. 5B). The irregular distribution of DHPR particles seen in relaxed myotubes is therefore not due to a defect in the disposition of RyRs. This distribution is reminiscent of that detected in cardiac muscle and in α1S-null myotubes expressing skeletal/cardiac α1 subunit chimeras that are unable to support skeletal muscle EC coupling (25). The overall density of particles within these clusters was similar in normal and relaxed myotubes (Fig. 5C). However, in agreement with the reduced amount of α1S immunofluorescence and protein (Fig. 4 A and B), the area of the DHPR clusters, and consequently the number of DHPRs/cluster in relaxed myotubes, was reduced to 40 ± 7% (Fig. 5C). Thus, the lack of the β1a subunit in relaxed zebrafish partially reduces but does not eliminate membrane expression and triad targeting of the α1S subunit, but significantly affects the physical coupling of the DHPR to the RyR1.
Fig. 5.
Skeletal muscle of zebrafish β1a-null mutant relaxed is lacking DHPR tetrad formation. (A) Freeze-fracture replicas of zebrafish tail myotubes showing the cytoplasmic leaflet of the plasmalemma. In myotubes from normal zebrafish (Left), small, slightly domed membrane areas (indicated in yellow) are occupied by large integral membrane particles, representing DHPRs. DHPRs are grouped into tetrads (groups of up to four), and the tetrads in turn are regularly arranged in an orthogonal array, as indicated by the red dots marking their centers. The membrane between the tetrads is clear of additional particles. Relaxed zebrafish muscle cells also show clusters of large particles in domed membrane areas (Right). However, these particles are not arranged in tetrads or orthogonal arrays. (B) Sections along the transverse tubules and triads from normal (n) and relaxed (r) myotubes in 3-day-old larvae. RyR1 (feet, indicated by arrows) maintain their orderly disposition. (C) Relative particle densities and cluster area from normal and relaxed myotubes. Particle density was 1,813 ± 69/324 particles per μm2 (mean ± SE/SD; n, number of patches = 22) for normal and 1,814 ± 89/445 (n = 25) for relaxed myotubes. Average (± SE/SD) cluster area was 0.02 ± 0.002/0.009 μm2 (n = 22) for normal and 0.008 ± 0.0005/0.003 μm2 (n = 25) for relaxed myotubes. Whereas particle densities were indistinguishable, the reduction of cluster area to 40 ± 7% in relaxed is statistically significant (**, P < 0.001).
The observed reduction in the overall α1S membrane concentration might indicate an independent and nonessential role of β1a in DHPR membrane expression. However, it is also conceivable that DHPR complexes devoid of β1a are less stable, possibly because of a conformational modification or because they are not anchored to RyR1, and are therefore subject to a higher turnover rate than fully assembled channels. Another possibility is that the lack of Ca2+ signals during development somehow diminishes α1S expression, as observed in RyR1-null myotubes (35).
In summary, we distinguished three distinct effects of the β1a-null mutant in skeletal muscle: An ≈50% reduction of DHPR membrane expression, which by itself is not sufficient to explain the loss of EC coupling; severely reduced gating charge movement, which could indicate a possible role of the β1a subunit on the voltage-sensing function of the DHPR; and, most importantly, the complete loss of tetrad formation, which by itself can explain the failure of EC coupling in β1a-null muscle cells. The present findings demonstrate that β1a is absolutely required for tetrad formation and thus physical DHPR-RyR interaction. Whether it functions exclusively in physically coupling the α1S with the RyR1 and thus enabling EC coupling or whether β1a actively participates in the signal-transduction between the voltage-sensor and the Ca2+ release channel remains to be shown.
Acknowledgments
We thank Dr. H.-G. Frohnhöfer (Max Planck Institute, Tübingen, Germany) for the zebrafish mutant relaxed, Dr. J. Striessnig (University of Innsbruck, Innsbruck, Austria) for the pan-β antibody RCP6, and Dr. H. Glossmann for continuous support. This work was supported in part by the Austrian Science Fund and Austrian National Bank Grants P16098-B11 (to M.G.), P16532-B05 (to B.E.F.), and P17807-B05 (to G.J.O.); European Commission's IHP Grant HPRN-CT-2002-00331 (to B.E.F.); and National Institutes of Health Grant AR P01144650 (to C.F.-A.).
Author contributions: J.S., V.D.B., G.J.O., B.E.F., C.F.-A., and M.G. designed research; J.S., V.D.B., G.J.O., E.T.F., B.E.F., C.F.-A., and M.G. performed research; J.S., V.D.B., G.J.O., E.T.F., B.E.F., C.F.-A., and M.G. analyzed data; and J.S., B.E.F., C.F.-A., and M.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: EC, excitation-contraction; DHPR, dihydropyridine receptor; SR, sarcoplasmic reticulum; RyR, ryanodine receptor; WGS, whole genome shotgun.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY952462 and AY495698).
Footnotes
Schredelseker, J., Flucher, B. E., Kugler, G. & Grabner, M. (2004) Biophys. J. 86, 63a (abstr.).
References
- 1.Armstrong, C. M., Bezanilla, F. M. & Horowicz, P. (1972) Biochim. Biophys. Acta 267, 605-608. [DOI] [PubMed] [Google Scholar]
- 2.Schneider, M. F. & Chandler, W. K. (1973) Nature 242, 244-246. [DOI] [PubMed] [Google Scholar]
- 3.Rios, E. & Brum, G. (1987) Nature 325, 717-720. [DOI] [PubMed] [Google Scholar]
- 4.Block, B. A., Imagawa, T., Campbell, K. P. & Franzini-Armstrong, C. (1988) J. Cell Biol. 107, 2587-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arikkath, J. & Campbell, K. P. (2003) Curr. Opin. Neurobiol. 13, 298-307. [DOI] [PubMed] [Google Scholar]
- 6.Freise, D., Held, B., Wissenbach, U., Pfeifer, A., Trost, C., Himmerkus, N., Schweig, U., Freichel, M., Biel, M., Hofmann, F., et al. (2000) J. Biol. Chem. 275, 14476-14481. [DOI] [PubMed] [Google Scholar]
- 7.Obermair, G. J., Kugler, G., Baumgartner, S., Tuluc, P., Grabner, M. & Flucher, B. E. (2005) J. Biol. Chem. 280, 2229-2237. [DOI] [PubMed] [Google Scholar]
- 8.Gregg, R. G., Messing, A., Strube, C., Beurg, M., Moss, R., Behan, M., Sukhareva, M., Haynes, S., Powell, J. A., Coronado, R., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanabe, T., Beam, K. G., Powell, J. A. & Numa, S. (1988) Nature 336, 134-139. [DOI] [PubMed] [Google Scholar]
- 10.Strube, C., Beurg, M., Powers, P. A., Gregg, R. G. & Coronado, R. (1996) Biophys. J. 71, 2531-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strube, C., Beurg, M., Sukhareva, M., Ahern, C. A., Powell, J. A., Powers, P. A., Gregg, R. G. & Coronado, R. (1998) Biophys. J. 75, 207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beurg, M., Sukhareva, M., Ahern, C. A., Conklin, M. W., Perez-Reyes, E., Powers, P. A., Gregg, R. G. & Coronado, R. (1999) Biophys. J. 76, 1744-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granato, M., van Eeden, F. J., Schach, U., Trowe, T., Brand, M., Furutani-Seiki, M., Haffter, P., Hammerschmidt, M., Heisenberg, C. P., Jiang, Y. J., et al. (1996) Development (Cambridge, U.K.) 123, 399-413. [DOI] [PubMed] [Google Scholar]
- 14.Ono, F., Higashijima, S., Shcherbatko, A., Fetcho, J. R. & Brehm, P. (2001) J. Neurosci. 21, 5439-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flucher, B. E., Andrews, S. B. & Daniels, M. P. (1994) Mol. Biol. Cell 5, 1105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss, R. G., O'Connell, K. M., Flucher, B. E., Allen, P. D., Grabner, M. & Dirksen, R. T. (2004) Am. J. Physiol. 287, C1094-C1102. [DOI] [PubMed] [Google Scholar]
- 17.Adams, B. A., Tanabe, T., Mikami, A., Numa, S. & Beam, K. G. (1990) Nature 346, 569-572. [DOI] [PubMed] [Google Scholar]
- 18.Cheung, A., Dantzig, J. A., Hollingworth, S., Baylor, S. M., Goldman, Y. E., Mitchison, T. J. & Straight, A. F. (2002) Nat. Cell. Biol. 4, 83-88. [DOI] [PubMed] [Google Scholar]
- 19.Grabner, M., Friedrich, K., Knaus, H. G., Striessnig, J., Scheffauer, F., Staudinger, R., Koch, W. J., Schwartz, A. & Glossmann, H. (1991) Proc. Natl. Acad. Sci. USA 88, 727-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruth, P., Rohrkasten, A., Biel, M., Bosse, E., Regulla, S., Meyer, H. E., Flockerzi, V. & Hofmann, F. (1989) Science 245, 1115-1118. [DOI] [PubMed] [Google Scholar]
- 21.Morton, M. E. & Froehner, S. C. (1987) J. Biol. Chem. 262, 11904-11907. [PubMed] [Google Scholar]
- 22.Morton, M. E. & Froehner, S. C. (1989) Neuron 2, 1499-1506. [DOI] [PubMed] [Google Scholar]
- 23.Pichler, M., Cassidy, T. N., Reimer, D., Haase, H., Kraus, R., Ostler, D. & Striessnig, J. (1997) J. Biol. Chem. 272, 13877-13882. [DOI] [PubMed] [Google Scholar]
- 24.Glossmann, H. & Ferry, D. R. (1985) Methods Enzymol. 109, 513-550. [DOI] [PubMed] [Google Scholar]
- 25.Takekura, H., Paolini, C., Franzini-Armstrong, C., Kugler, G., Grabner, M. & Flucher, B. E. (2004) Mol. Biol. Cell 15, 5408-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haffter, P., Granato, M., Brand, M., Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., van Eeden, F. J., Jiang, Y. J., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 1-36. [DOI] [PubMed] [Google Scholar]
- 27.Mendel, J. G. (1866) Verh. Nat. Vereins Brünn 4, 3-47. [Google Scholar]
- 28.Van Petegem, F., Clark, K. A., Chatelain, F. C. & Minor, D. L., Jr. (2004) Nature 429, 671-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opatowsky, Y., Chen, C. C., Campbell, K. P. & Hirsch, J. A. (2004) Neuron 42, 387-399. [DOI] [PubMed] [Google Scholar]
- 30.Sheridan, D. C., Cheng, W., Ahern, C. A., Mortenson, L., Alsammarae, D., Vallejo, P. & Coronado, R. (2003) Biophys. J. 84, 220-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolphin, A. C. (2003) J. Bioenerg. Biomembr. 35, 599-620. [DOI] [PubMed] [Google Scholar]
- 32.Bichet, D., Cornet, V., Geib, S., Carlier, E., Volsen, S., Hoshi, T., Mori, Y. & De Waard, M. (2000) Neuron 25, 177-190. [DOI] [PubMed] [Google Scholar]
- 33.Flucher, B. E., Phillips, J. L. & Powell, J. A. (1991) J. Cell Biol. 115, 1345-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Protasi, F., Franzini-Armstrong, C. & Allen, P. D. (1998) J. Cell Biol. 140, 831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avila, G., O'Connell, K. M., Groom, L. A. & Dirksen, R. T. (2001) J. Biol. Chem. 276, 17732-17738. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman, R. & Gross, L. (1975) Nature 254, 586-588. [DOI] [PubMed] [Google Scholar]