Abstract
Empedobacter brevis (formerly designated Flavobacterium breve) is a gram-negative aerobe involved in nosocomial infections. The Ambler class B β-lactamase gene blaEBR-1 was cloned and expressed in Escherichia coli from E. brevis clinical strain ASS-1, which had reduced susceptibility to expanded-spectrum cephalosporins and carbapenems. Purified β-lactamase EBR-1 hydrolyzed penicillins, cephalosporins, and carbapenems efficiently but not aztreonam. Kinetic parameters of EBR-1 were similar to those of class B enzymes such as BlaB, IND-2, and GOB-1 identified from other Flavobacteriaceae species, except for meropenem, which was more hydrolyzed by β-lactamase GOB-1. EBR-1, with a pI of 8.0 and a relative molecular mass of ca. 25 kDa, was classified in functional subgroup 3a, which includes most of the class B β-lactamases. EBR-1, which belongs to molecular subclass B1 of metalloenzymes, shares 58, 57, and 42% amino acid identity with the most closely related β-lactamases, IND-1/IND-2 from Chryseobacterium indologenes, CGB-1 from Chryseobacterium gleum, and BlaB from Chryseobacterium meningosepticum, respectively.
Flavobacterium breve was included in the original description of the Flavobacterium genus in 1923 and was proposed as the type species of the Flavobacterium genus by Holmes and Owen in 1979 (13). Its classification was emended in 1994 (36), and F. breve was included in the novel species Empedobacter brevis belonging to the Flavobacteriaceae family (7). This species is the only member of the Empedobacter genus. E. brevis is a source of meningitis (10). Clinical strains of E. brevis have been also reported from blood infections and from other clinical sources (eye swabs, bronchial secretions, and urine) (14, 17). A few publications report the phenotype of resistance to β-lactams of E. brevis, E. brevis being of intermediate resistance to amoxicillin, ticarcillin, and narrow-spectrum cephalosporins and having a reduced susceptibility (but still being susceptible) to expanded-spectrum cephalosporins and imipenem (14, 17, 22). This species would be less resistant to β-lactams than other flavobacterial species (22).
Several carbapenem-hydrolyzing enzymes have been reported from bacterial species of the Flavobacteriaceae family, including BlaB and GOB from Chryseobacterium meningosepticum, IND from Chryseobacterium indologenes, CGB-1 from C. gleum, and a β-lactamase from Flavobacterium odoratum (1, 2, 4, 5, 32, 34). They are all Ambler class B enzymes.
We report here a novel chromosome-encoded Ambler class B β-lactamase from another Flavobacteriaceae species, E. brevis. We compared its activity to those of other metalloenzymes of the Flavobacteriaceae family.
MATERIALS AND METHODS
Bacterial strains.
E. brevis clinical isolate ASS-1, used in this study, was from a rectal swab of an 18-month-old boy hospitalized in the intensive care unit at the Bicêtre hospital (Le-Kremlin-Bicêtre, France) in 1999 for a chemical intoxication. E. brevis ASS-1 was identified by standard biochemical techniques and 16S rRNA sequencing (36). Escherichia coli DH10B, E. coli BL21(DE3), and nalidixic acid-resistant E. coli JM109 were used for cloning, subcloning, and conjugation assays, respectively. All strains were stored at −70°C in Trypticase soy (TS) broth (Becton Dickinson, Le Pont-de-Claix, France) supplemented with 15% glycerol until testing.
Antimicrobial agents and MIC determinations.
The antimicrobial agents used in this study were obtained in the form of standard laboratory powders and were used immediately after their solubilization. The agents and their sources have been described elsewhere (28). Antibiotic-containing disks (Bio-Rad, Marnes-la-Coquette, France) were used for routine antibiograms (www.sfm.asso.fr).
MICs were determined by an agar dilution technique on Mueller-Hinton plates with an inoculum of 104 CFU per spot as previously described (6). The plate contents were incubated at 35°C for 18 h at ambient atmosphere. MICs of β-lactams were determined alone or in combination with a fixed concentration of clavulanic acid (2 μg/ml). MIC results were interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (23).
Cloning experiments and analysis of recombinant plasmids.
Genomic DNA from E. brevis ASS-1 was extracted as described earlier (15). Cloning experiments were performed with Sau3AI-restricted DNA fragments of E. brevis ASS-1 into the pBK-CMV plasmid (Stratagene, Amersham Pharmacia Biotech, Orsay, France) as previously described (25). Antibiotic-resistant colonies were selected onto TS agar plates containing amoxicillin (30 μg/ml) and kanamycin (30 μg/ml).
Recombinant plasmid DNAs were obtained from 100-ml TS broth cultures grown overnight in the presence of amoxicillin (30 μg/ml) at 35°C. Plasmid DNAs were recovered by using Qiagen columns (Qiagen, Courtaboeuf, France).
Conjugation assays, plasmid content, and hybridizations.
Direct transfer of resistance gene into nalidixic acid-resistant E. coli JM109 was attempted by liquid and solid conjugation assays at 35°C (1, 3, 6). Transconjugants were selected on TS agar plates containing nalidixic acid (100 μg/ml) and amoxicillin (30 μg/ml). Extraction of plasmid DNA from E. brevis ASS-1 was attempted as previously described (27). Southern hybridization experiments were performed as described earlier (33) by using a 0.8% electrophoresis gel containing either whole-cell or HindIII- and ClaI-restricted DNA of E. brevis ASS-1 transferred onto a nylon membrane that was hybridized with a PCR-obtained internal fragment of blaEBR-1 with primers EbA (5′-CACTTATTGCA TTGATAGGAAG-3′) and EbB (5′-TTTCGATA TGTCCAGTAGCC-3′). Visualization of hybridizations was made using the enhanced chemiluminescence nonradioactive labeling and detection kit as described by the manufacturer (Amersham Pharmacia Biotech).
DNA sequencing and protein analysis.
Both strands of the cloned DNA fragment of recombinant plasmid pEBR-1 were sequenced with an Applied Biosystems sequencer (ABI 377). The nucleotide and deduced protein sequences were analyzed with software available over the Internet (www.ncbi.nlm.nih.gov and www.cbs.dtu.dk/services/SignalP/) (24).
Subcloning of the blaEBR-1 gene in an expression vector.
The blaEBR-1 gene was amplified by PCR with primers EBR-1nde (5′-TTGA CGAGGACATATGAAG AAATTATTTTCAC-3′), which contained an NdeI restriction site (underlined), and EBR-1bam (5′-CTATTATATCGGATCCTTTT TTCAATTTTATTTC-3′), which contained a BamHI restriction site (underlined). Amplification was performed as described by the manufacturer (Perkin-Elmer) with an annealing temperature of 53°C for 1 min. After 38 amplification cycles, PCR product was purified, restricted with NdeI and BamHI endonucleases, and cloned into expression vector pET-9a (Merck EuroLab, Fontenay-sous-Bois, France) to obtain pET-EBR-1. After purification of plasmid pET-EBR-1, the NdeI-BamHI insert was sequenced to rule out any PCR-generated mismatches.
β-Lactamase purification.
Cultures of E. coli BL21(DE3) harboring recombinant plasmid pET-EBR-1 were grown at 37°C in 2 liters of TS broth containing amoxicillin (20 μg/ml) until an optical density at 600 nm of 1 was reached. The β-lactamase expression was induced for 5 h after addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside, as recommended by the manufacturer (Novagen, Fontenay-sous-Bois, France). Bacterial suspensions were pelleted, resuspended in 40 ml of 30 mM Tris-HCl (pH 9.0), disrupted by sonification (three times at 30 W for 1 min using a Vibra Cell 75022 Phospholyser from Bioblock, Illkirch, France), and centrifuged for 1 h at 48,000 × g at 4°C. The suspension was then ultracentrifuged at 100,000 × g for 1 h at 4°C, and the supernatant was dialyzed overnight against 30 mM Tris-HCl (pH 9.0).
This extract was loaded onto a preequilibrated Q-Sepharose column (Amersham Pharmacia Biotech). The enzyme recovered in the flowthrough was dialyzed overnight at 4°C against 50 mM phosphate buffer (pH 6.2). The enzyme fraction was then loaded onto a preequilibrated S-Sepharose column (Amersham Pharmacia Biotech). The enzyme was eluted by a 100-ml linear NaCl gradient (0 to 500 mM) in 50 mM phosphate buffer (pH 6.2). The β-lactamase was eluted at 310 mM NaCl. The fractions containing the highest β-lactamase activity were pooled, concentrated, and dialyzed overnight against 50 mM phosphate buffer (pH 7.0) containing 50 μM ZnCl2 and were stored at −80°C.
Biochemical analysis of β-lactamase.
Purified β-lactamase was used for kinetic measurements performed at 30°C in 50 mM sodium phosphate (pH 7.0) containing 50 μM ZnCl2. The rates of hydrolysis were determined with the spectrophotometer ULTROSPEC 2000 (Amersham Pharmacia Biotech). The wavelengths and absorption coefficients of β-lactams have been described previously (21).
Km and kcat values were determined by analyzing the β-lactam hydrolysis under initial rate conditions using the Eadie-Hoffstee linearization of the Michaelis-Menten equation (1). The Km values were expressed in micromolars, and kcat values were expressed in seconds−1. In the cases of low Km values, Ki values were determined with benzylpenicillin as the substrate.
Various concentrations of EDTA and clavulanic acid were preincubated with the enzyme for 3 min at 30°C before testing the rate of imipenem (100 μM) hydrolysis. The 50% inhibitory concentrations (IC50s) of these inhibitors were determined as the concentrations that inhibited hydrolysis activity by 50%. Results were expressed in micromolars.
Purified enzyme and nonpurified β-lactamase extracts of a 100-ml culture of E. brevis ASS-1 were subjected to isoelectric focusing analysis as previously described (6). The relative molecular mass of the purified β-lactamase and its purity were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis revealed by Coomassie blue staining or by overlaying the gel with 1 mM nitrocefin solution as previously described (6, 20). Specific activities of nonpurified enzymes from cultures of E. brevis ASS-1 and E. coli BL21(DE3) (pET-EBR-1) and purified enzyme were determined as previously reported with 100 μM imipenem as the substrate (1). One unit of enzyme activity was defined as the activity that hydrolyzed 1 μM imipenem/min; the total protein content was measured with bovine serum albumin as the standard (Bio-Rad DC protein assay kit).
Induction experiments.
E. brevis ASS-1 was grown at 37°C with or without cefoxitin (0.5 or 1 μg/ml) as the inducer, as reported previously (1). Briefly, overnight cultures were diluted 1:10 and were grown for 1 h and 30 min in a preincubated TS broth on a rotating shaker at 37°C. Then the cultures were grown for an additional 2 h in the presence of the inducer. Hydrolysis measurements were determined with 100 μM imipenem as the substrate.
Nucleotide sequence accession number.
The nucleotide sequence and deduced β-lactamase amino acid sequence reported in this work have been assigned to the GenBank and EMBL databases under accession no. AF416700.
RESULTS AND DISCUSSION
Cloning experiments and preliminary β-lactamase analysis.
Imipenem hydrolysis by a culture extract of E. brevis ASS-1 suggested the presence of a carbapenem-hydrolyzing-β-lactamase (specific activity, 0.022 U · mg of protein−1 with imipenem as the substrate). Cloning of Sau3AI-restricted DNA of E. brevis into the BamHI site of the pBK-CMV cloning vector gave 30 recombinant E. coli DH10B clones growing on ampicillin-containing plates and harboring plasmids with inserts that ranged from 1.7 to 9 kb. Among them, E. coli DH10B(pEBR-1) harbored a recombinant plasmid that possessed an 1.7-kb insert and was retained for further analysis.
Isoelectric focusing analysis revealed that E. coli DH10B(pEBR-1) produced a β-lactamase activity with a pI of 8.0 (data not shown). Only one β-lactamase activity with an identical pI was detected from culture extracts of E. brevis ASS-1. Induction studies with cefoxitin as a β-lactam inducer failed to detect β-lactamase induction with cultures of E. brevis ASS-1.
DNA and protein sequence analysis.
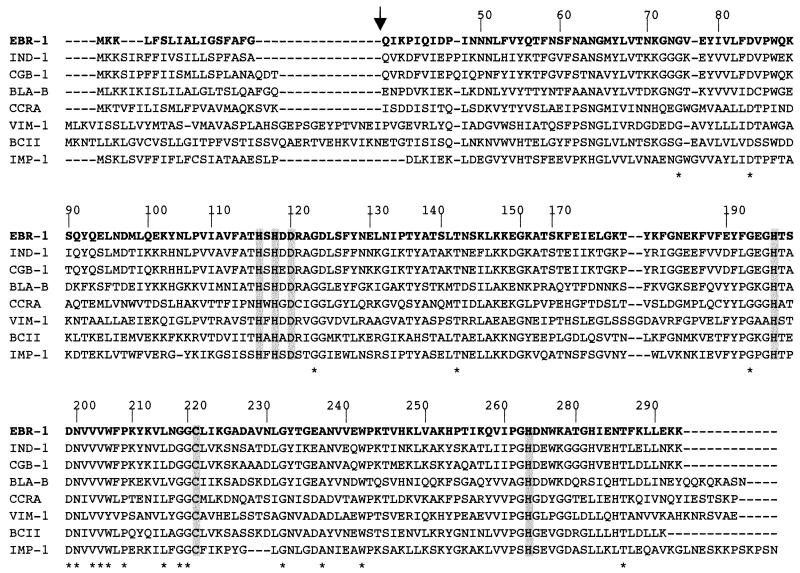
DNA sequence analysis of the 1,717-bp insert of pEBR-1 identified an open reading frame (ORF) of 708 bp encoding a 236-amino-acid preprotein. A putative cleavage site was found between the Ala-Phe-Gly and Gln-Ile-Lys motifs (Fig. 1). The overall GC content of this ORF was 31.2%, which lies close to the expected range of the G+C ratio of E. brevis genes (31 to 33%) (17).
FIG. 1.
Comparison of the amino acid sequence of EBR-1 with those of other β-lactamases of subclass B1 of metalloenzymes. The origin of the β-lactamases is as follows: IND-1 from C. indologenes 001 (2), CGB-1 from C. gleum (4), BlaB from C. meningosepticum CIP 6058 (32), CcrA from Bacteroides fragilis (31), VIM-1 from Pseudomonas aeruginosa VR-143/97 (18), BcII from Bacillus cereus 569/H (16), and IMP-1 from Serratia marcescens TN9106 (26). The BBL numbering scheme is indicated above the sequences (12). Broken lines indicate gaps introduced in the alignment. The vertical arrow indicates putative cleavage of the peptide leader site for EBR-1. Amino acid residues that may be involved in Zn2+ or/and water molecule binding are boxed. Identical amino acids in subclass B1 enzymes are indicated by an asterisk.
No plasmid was detected in E. brevis ASS-1, and direct conjugation assays failed to transfer any β-lactam resistance marker from E. brevis ASS-1 to nalidixic acid-resistant E. coli JM109. These negative results did not rule out a plasmid location of blaEBR-1. However, Southern experiments identified blaEBR-1 at the chromosomal position of migration using whole-cell DNA of E. brevis ASS-1 as a template (data not shown). Additionally, using HindIII and ClaI restriction enzymes that did not cut within the blaEBR-1 sequence, a 1,425-bp fragment of HindIII- and ClaI-restricted DNA of E. brevis ASS-1 hybridized with the internal probe for blaEBR-1 as expected, further underlining the origin of the cloned fragment.
The comparison of the EBR-1 amino acid sequence with those of other class B β-lactamases (1, 2, 4, 5, 8, 16, 18, 20, 26, 29, 31, 32, 37) revealed 58, 57, and 42% amino acid identity with the most closely related β-lactamases IND-1 and IND-2 from C. indologenes (2, 5), CGB-1 from C. gleum (4), and BlaB from C. meningosepticum (1, 32), respectively.
The six conserved amino acids implicated in the Zn2+ or/and water molecule binding (His116, His118, Asp120, His196, Cys221, and His220, according to the BBL numbering of metallo-β-lactamases [12, 35, 37, 40]) were identified in the amino acid sequence of EBR-1 (Fig. 1). Additionally, 18 amino acid residues were conserved in metallo-β-lactamases of subclass B1 (30), to which EBR-1 belongs (Fig. 1).
Another ORF was detected in the same orientation and downstream of blaEBR-1. It encoded a 230-amino-acid protein that did not share significant identity with any amino acid sequence available in the GenBank database.
Susceptibility testing.
E. brevis ASS-1 was resistant or of intermediate susceptibility to amino- and carboxy-penicillins and to narrow-spectrum cephalosporins, susceptible to expanded-spectrum cephalosporins, and resistant to meropenem (Table 1). MICs of β-lactams were not lowered by the addition of clavulanic acid (Table 1). Once expressed from a recombinant E. coli DH10B clone, EBR-1 conferred a similar resistance phenotype to that for BlaB from C. meningosepticum expressed in the same E. coli DH10B background (1). IND-like β-lactamases conferred a higher degree of resistance to cefotaxime once expressed from E. coli DH10B (16 versus 0.5 μg/ml) (5). A slight decreased susceptibility to carbapenems was noticeable for E. coli DH10B(pEBR-1), highlighting a potential threat of undetectable dissemination of such metalloenzymes once expressed from Enterobacteriaceae. A similar low degree of resistance to carbapenems in Enterobacteriaceae has been reported for metalloenzymes of the IND and BlaB series of other flavobacterial species (1, 2, 5, 32).
TABLE 1.
MICs of β-lactams for E. brevis ASS-1, E. coli DH10B(pEBR-1), and reference strain E. coli DH10B
| β-Lactam(s) | MIC (μg/ml)
|
||
|---|---|---|---|
| E. brevis ASS-1 | E. coli DH10B (pEBR-1) | E. coli DH10B | |
| Amoxicillin | 64 | >512 | 2 |
| Amoxicillin-CLAa | 64 | >512 | 2 |
| Ticarcillin | 64 | >512 | 2 |
| Piperacillin | 2 | 8 | 1 |
| Cephalothin | 32 | 32 | 2 |
| Cefuroxime | 32 | 128 | 4 |
| Ceftazidime | 2 | 0.5 | 0.06 |
| Cefotaxime | 4 | 0.5 | 0.06 |
| Cefepime | 0.5 | 0.12 | 0.06 |
| Cefpirome | 0.25 | 0.12 | 0.06 |
| Ceftriaxone | 4 | 0.25 | 0.06 |
| Cefoxitin | 2 | 8 | 4 |
| Moxalactam | 2 | 1 | 0.12 |
| Aztreonam | 2 | 0.12 | 0.12 |
| Imipenem | 8 | 0.5 | 0.06 |
| Meropenem | 16 | 0.25 | 0.06 |
CLA, clavulanic acid at fixed concentration of 2 μg/ml.
Biochemical properties of EBR-1.
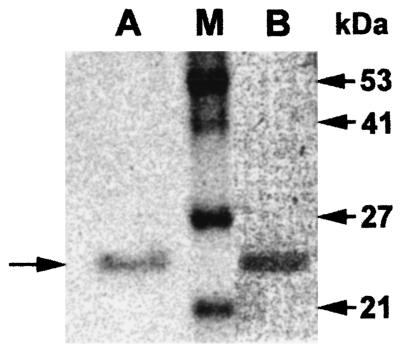
Specific β-lactamase activities of nonpurified extracts from cultures of E. brevis ASS-1 and E. coli BL21(DE3) (pET-EBR-1) were 0.022 and 0.4 U · mg of protein−1, respectively. This result corresponded to a β-lactamase overexpression in E. coli by 18-fold. EBR-1 purification from E. coli BL21(DE3) (pET-EBR-1) gave a specific activity of 132 μmol · min−1 · mg of protein−1 determined with 100 μM imipenem as substrate with a 330-fold purification factor. The total quantity of purified β-lactamase was 45 U, with a 12% purification yield. Its purity was estimated to be ca. 90% by SDS-PAGE analysis (Fig. 2).
FIG. 2.
SDS-PAGE analysis of the purified β-lactamase EBR-1 from a culture of E. coli BL21(pET-EBR-1). Lane A, Coomassie blue staining. The horizontal arrow on the left side indicates the migration position of β-lactamase EBR-1. Lane M, size of relative molecular masses expressed in kilodaltons is indicated on the right. Lane B, zymogram of EBR-1.
The mature protein for EBR-1 had a relative molecular mass experimentally determined to be ca. 25 kDa (Fig. 2), which correlated with its calculated molecular mass (24.8 kDa).
Catalytic efficiency values (kcat/Km) of EBR-1 for penicillins were high, ranging from 760 to 2,500 s−1 · mM−1 and similar to those obtained for IND-2, BlaB, and GOB-1 (Table 2). Cephalothin, cefuroxime, and ceftriaxone were good substrates for EBR-1 due to high affinity for these substrates (low Km values). The hydrolysis spectrum of EBR-1 encompassed cefotaxime, ceftazidime, and cefpirome despite low catalytic efficiencies. As for all metallo-β-lactamases, the monobactam aztreonam was not hydrolyzed by EBR-1 (9). It was like BlaB and IND-2 β-lactamases from C. meningosepticum and C. indologenes (5, 32), respectively, but unlike the plasmid-mediated VIM- and IMP-type β-lactamases (19, 29). EBR-1 had high kcat values for imipenem with a high catalytic efficiency (kcat/Km; 250 s−1 · mM−1). These kinetic parameters showed that EBR-1 was a member of functional group 3a of the Bush classification for metallo-β-lactamases (9). This group includes most of the class B β-lactamases responsible for intrinsic or acquired resistance to carbapenems. As a zinc-dependent protein, EBR-1 activity was inhibited by EDTA (IC50 = 70 μM) but not by class A β-lactamase inhibitors, such as clavulanate (IC50 > 1 mM).
TABLE 2.
Kinetic parameters of EBR-1 from E. coli BL21(DE3) compared with those of IND-2 from C. indologenes CIP 101026 (5), BlaB from C. meningosepticum CIP 6058 (32), and GOB-1 from C. meningosepticum PINT (1)d
| β-Lactam | EBR-1 parameters
|
kcat/Km (s−1 · mM−1)
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (s−1 · mM−1) | IND-2 | BlaB | GOB-1 | |
| Benzylpenicillin | 115 | 47 | 2,500 | 4,600 | 8,800 | 1,900 |
| Amoxicillin | 640 | 690 | 900 | 700 | —a | 350 |
| Ticarcillin | 226 | 300 | 750 | — | — | 520 |
| Piperacillin | 410 | 490 | 850 | 2,000 | — | 1,700 |
| Cephalothin | 6 | 31 | 200 | 1,200 | — | 700 |
| Cephaloridine | 2 | 440 | 5 | — | 480 | — |
| Cefuroxime | 0.5 | 1b | 350 | — | — | 1,000 |
| Ceftriaxone | 3 | 7.5b | 400 | — | — | — |
| Ceftazidime | >2.5 | >1,000 | 2 | 5 | — | 800 |
| Cefotaxime | 1 | 50 | 20 | 900 | 220 | 850 |
| Cefepime | <0.01 | NDc | ND | <0.001 | — | 200 |
| Cefpirome | 0.5 | 370 | 2 | — | — | — |
| Cefoxitin | 0.2 | 140 | 2 | 50 | 250 | 250 |
| Moxalactam | 2 | 620 | 2 | — | — | 130 |
| Imipenem | 190 | 782 | 250 | 600 | 950 | 700 |
| Meropenem | >190 | >2,000 | 100 | 200 | — | 5,500 |
| Aztreonam | <0.1 | ND | ND | ND | ND | ND |
—, not available.
In cases of low Km values, Ki values were determined with benzylpenicillin as the substrate.
ND, not determinable.
Data are means of three independent measurements. Standard deviations were within 15%.
Conclusion.
In addition to GOB, BlaB, IND, and CGB of other flavobacterial species, we report a novel metalloenzyme from the same flavobacterial reservoir, which constitutes the most important source of known class B β-lactamase genes. Identification of EBR-1 adds to the diversity of class B enzymes of this reservoir. Class B enzymes are the only β-lactamases produced by C. indologenes and E. brevis. Most of the natural producers of class B enzymes also produce class A β-lactamases, such as C. meningosepticum (6), C. gleum (3), Stenotrophomonas maltophilia (38), or class D β-lactamases, such as Legionella gormanii (11) and Aeromonas spp. (39). The reason why several environmental gram-negative aerobes produce multiple β-lactamases with complementary or partially overlapping substrate profiles remains to be determined.
EBR-1 is distantly related to the plasmid-mediated class B β-lactamases increasingly reported worldwide in clinically significant gram-negative rods (19). Finally, identification of the wide-spectrum hydrolysis profile of EBR-1 underlines that therapies that include β-lactam antibiotics for treating clinical infections due to Flavobacteriaceae should be precautionary.
Acknowledgments
This work was financed by a grant from the Ministères de l'Education Nationale et de la Recherche (grant UPRES-EA), Université Paris XI, Paris, France.
REFERENCES
- 1.Bellais, S., D. Aubert, T. Naas, and P. Nordmann. 2000. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing β-lactamases in Chryseobacterium meningosepticum. Antimicrob Agents Chemother. 44:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellais, S., S. Léotard, L. Poirel, T. Naas, and P. Nordmann. 1999. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol. Lett. 171:127-132. [DOI] [PubMed] [Google Scholar]
- 3.Bellais, S., T. Naas, and P. Nordmann. 2002. Molecular and biochemical characterization of Ambler class A extended-spectrum β-lactamase CGA-1 from Chryseobacterium gleum. Antimicrob. Agents Chemother. 46:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellais, S., T. Naas, and P. Nordmann. Genetic and biochemical characterization of CGB-1, a novel Ambler class B carbapenem-hydrolyzing β-lactamase from Chryseobacterium gleum. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 5.Bellais, S., L. Poirel, S. Léotard, T. Naas, and P. Nordmann. 2000. Genetic diversity of carbapenem-hydrolyzing metallo-β-lactamases from Chryseobacterium (Flavobacterium) indologenes. Antimicrob. Agents Chemother. 44:3028-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellais, S., L. Poirel, T. Naas, D. Girlich, and P. Nordmann. 2000. Genetic-biochemical analysis and distribution of the Ambler class A β-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob. Agents Chemother. 44:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernardet, J. F., P. Segers, M. Vancanneyt, F. Berthe, K. Kersters, and P. Vandamme. 1996. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the familly Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 46:128-148. [Google Scholar]
- 8.Boschi, L., P. S. Mercuri, M. L. Riccio, G. Amicosante, M. Galleni, J. M. Frère, and G. M. Rossolini. 2000. The Legionella (Fluoribacter) gormanii metallo-β-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 β-lactamases. Antimicrob. Agents Chemother. 44:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush, K. 1998. Metallo-β-lactamases: a class apart. Clin. Infect. Dis. 27(Suppl. 1):S48-S53. [DOI] [PubMed] [Google Scholar]
- 10.Duong, M., K. Mourier, N. Peyrard, V. Magnin, G. Couillaud, and P. Chavanet. 1997. Méningite à Chryseobacterium (Flavobacterium). Med. Mal. Infect. 27:802-803. [Google Scholar]
- 11.Franceschini, N., L. Boschi, S. Pollini, R. Herman, M. Perilli, M. Galleni, J.-M. Frère, G. Amicosante, and G. M. Rossolini. 2001. Characterization of OXA-29 from Legionella (Fluoribacter) gormanii: molecular class D β-lactamase with unusual properties. Antimicrob. Agents Chemother. 45:3509-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J.-M. Frère. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes, B., and R. J. Owen. 1979. Proposal that Flavobacterium breve be substituted as the type species of the genus in place of Flavobacterium aquatile and emended description of the genus Flavobacterium: status of the named species of Flavobacterium. Int. J. Syst. Bacteriol. 29:416-426. [Google Scholar]
- 14.Holmes, B., J. J. S. Snell, and S. P. Lapage. 1978. Revised description, from clinical strains, of Flavobacterium breve (Lustig) Bergey et al. 1923 and proposal of the neotype strain. Int. J. Syst. Bacteriol. 28:201-208. [Google Scholar]
- 15.Honoré, N., M.-H. Nicolas, and S. T. Cole. 1986. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 5:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain, M., A. Carlino, M. J. Madonna, and J. O. Lampen. 1985. Cloning and sequencing of the metallothioprotein β-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J. Bacteriol. 164:223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jooste, P. J., and C. J. Hugo. 1999. The taxonomy, ecology and cultivation of bacterial genera belonging to the family Flavobacteriaceae. Int. J. Food Microbiol. 53:81-94. [DOI] [PubMed] [Google Scholar]
- 18.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 20.Massidda, O., G. M. Rossolini, and G. Satta. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among metallo-β-lactamases. J. Bacteriol. 173:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matagne, A., A. M. Misselyn-Baudoin, B. Joris, B. Erpicum, B. Granier, and J.-M. Frère. 1990. The diversity of the catalytic properties of class A β-lactamases. Biochem. J. 265:131-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulin, V., J. Freney, W. Hansen, and A. Philippon. 1992. Comportement phénotypique des Flavobacterium vis-à-vis de 39 antibiotiques. Med. Mal. Infect. 22:902-908. [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Nielsen, H., J. Engelbrecht, S. Brunak, and G. Von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 25.Nordmann, P., E. Ronco, T. Naas, C. Duport, Y. Michel-Briand, and R. Labia. 1993. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel, L., T. Naas, M. Guibert, B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen, B. A., and K. Bush. 1997. Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen, B. A., Y. Gluzman, and F. P. Tally. 1990. Cloning and sequencing of the class B β-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob. Agents Chemother. 34:1590-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossolini, G. M., N. Franceschini, M. L. Riccio, P. S. Mercuri, M. Perilli, M. Galleni, J.-M. Frère, and G. Amicosante. 1998. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem. J. 332:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sato, K., T. Fujii, R. Okamoto, M. Inoue, and S. Mitsuhashi. 1985. Biochemical properties of β-lactamase produced by Flavobacterium odoratum. Antimicrob. Agents Chemother. 27:612-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullah, J. H., T. R. Walsh, I. A. Taylor, D. C. Emery, C. S. Verma, S. J. Gamblin, and J. Spencer. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7A resolution. J. Mol. Biol. 284:125-136. [DOI] [PubMed] [Google Scholar]
- 36.Vandamme, P., J.-F. Bernardet, P. Segers, K. Kersters, and B. Holmes. 1994. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int. J. Syst. Bacteriol. 44:827-831. [Google Scholar]
- 37.Walsh, T. R., L. Hall, S. J. Assinder, W. W. Nichols, S. J. Cartwright, A. P. MacGowan, and P. M. Bennett. 1994. Sequence analysis of the L-1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218:199-201. [DOI] [PubMed] [Google Scholar]
- 38.Walsh, T. R., A. P. MacGowan, and P. M. Bennett. 1997. Sequence analysis and enzyme kinetics of the L2 serine β-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1460-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh, T. R., R. A. Stunt, J. A. Nabi, A. P. MacGowan, and P. M. Bennett. 1997. Distribution and expression of β-lactamase genes among Aeromonas spp. J. Antimicrob. Chemother. 40:171-178. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Z., W. Fast, A. M. Valentine, and S. J. Benkovic. 1999. Metallo-β-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3:614-622. [DOI] [PubMed] [Google Scholar]