Summary
Methionine adenosyltransferase (MAT) I/III deficiency, caused by mutations in the MAT1A gene, is characterized by persistent hypermethioninemia without elevated homocysteine or tyrosine. Clinical manifestations are variable and poorly understood, although a number of individuals with homozygous null mutations in MAT1A have neurological problems, including brain demyelination. We analyzed MAT1A in seven hypermethioninemic individuals, to provide insight into the relationship between genotype and phenotype. We identified six novel mutations and demonstrated that mutations resulting in high plasma methionines may signal clinical difficulties. Two patients—a compound heterozygote for truncating and severely inactivating missense mutations and a homozygote for an aberrant splicing MAT1A mutation—have plasma methionine in the 1,226–1,870 μM range (normal 5–35 μM) and manifest abnormalities of the brain gray matter or signs of brain demyelination. Another compound heterozygote for truncating and inactivating missense mutations has 770–1,240 μM plasma methionine and mild cognitive impairment. Four individuals carrying either two inactivating missense mutations or the single-allelic R264H mutation have 105–467 μM plasma methionine and are clinically unaffected. Our data underscore the necessity of further studies to firmly establish the relationship between genotypes in MAT I/III deficiency and clinical phenotypes, to elucidate the molecular bases of variability in manifestations of MAT1A mutations.
Introduction
Methionine adenosyltransferase (MAT) (E.C.2.5.1.6) catalyzes an unusual two-step reaction that involves the transfer of the adenosyl moiety of ATP to methionine to form S-adenosylmethionine (AdoMet), with the concomitant formation of a tripolyphosphate that is then cleaved to PPi and Pi (Cantoni 1953; Mudd 1962). The importance of the product AdoMet cannot be overemphasized: it is the source of methyl groups for most biological methylations (Mudd et al. 1995a). All organisms have one or two genes that encode MAT isozymes (Kotb and Geller 1993; Mato et al. 1997). The two human genes, MAT1A and MAT2A, are located on chromosomes 10 and 2, respectively (De La Rosa et al. 1995; Chamberlin et al. 1996). The gene products have similar catalytic activities but vary in pattern of expression and kinetic parameters (Kotb and Geller 1993; Kotb et al. 1997; Mato et al. 1997). MAT isozymes can be separated by gel chromatography into three species—MAT I, MAT II, and MAT III (Kotb and Geller 1993). MAT I and MAT III are homotetramers and homodimers of the α1 subunit, encoded by the MAT1A gene (Ubagai et al. 1995; Kotb et al. 1997; Mato et al. 1997). MAT II is an oligomer containing two moieties of the highly homologous α2 catalytic subunit, encoded by the MAT2A gene and separately encoded regulatory subunit(s). MAT1A is expressed solely in adult liver (Alvarez et al. 1993), whereas MAT2A is expressed in virtually all tissues, including (at a low concentration) adult liver (Kotb and Kredich 1985; Mitsui et al. 1988; Horikawa and Tsukada 1992; De La Rosa et al. 1995). All three MAT isozymes therefore contribute to MAT activity in adult liver. For clarity, we refer to the gene products of MAT1A as “MAT I/III” (Kotb et al. 1997) rather than “hepatic MAT,” a term used formerly (Mudd et al. 1995a).
Our laboratory has demonstrated that mutations in MAT1A that abolish or greatly reduce MAT I/III activity cause a persistent form of hypermethioninemia (Ubagai et al. 1995). In MAT I/III deficiency, the hypermethioninemia is not accompanied by elevated plasma-free homocystine, as it is in cystathionine β-synthase deficiency; elevated tyrosine, as in tyrosinemia type I; or severe hepatocellular disease (Mudd et al. 1995b). Most MAT1A mutations are transmitted in an autosomal recessive manner, but autosomal dominant inheritance has also been observed (Blom et al. 1992; Mudd et al. 1995b; Chamberlin et al. 1997; Nagao and Oyanagi 1997). The dominant trait, in all cases described thus far, is due to heterozygosity for an R264H mutation (Chamberlin et al. 1997; Nagao and Oyanagi 1997), which seems to render dimers formed between mutant and wild-type subunits inactive (Chamberlin et al. 1997).
The clinical consequences of MAT I/III deficiency are poorly understood. In some individuals, this deficiency is apparently benign; in others, association of neurological problems with null mutations in the MAT1A gene has been observed (Surtees et al. 1991; Chamberlin et al. 1996). Thus, it is of interest to continue to define the molecular nature of MAT1A mutations in hypermethioninemic individuals and to investigate possible correlations between genotype and clinical severity. In the present study, we report molecular analyses of the MAT1A gene of seven previously uncharacterized hypermethioninemic individuals with neurodevelopmental status and neuroimaging ranging from normal to markedly abnormal.
Subjects and Methods
Subjects
We have analyzed the MAT1A genes of seven individuals with isolated persistent hypermethioninemia. Clinical and metabolic aspects of patients 1, 5, 7, and 14 have been reported elsewhere (Mudd et al. 1995b). The three individuals new to the study are patient 31, patient BII-1, and patient BI-1 (father of patient BII-1).
Analysis of the MAT1A Gene by SSCP and DNA Sequencing
Genomic DNA was extracted from blood samples. All peripheral blood samples were obtained with the informed consent of the donors or their parents. Each exon of the MAT1A gene coding region was amplified as described elsewhere (Chamberlin et al. 1996), and the products were analyzed on mutation-detection–enhancement nondenaturing gels (AT Biochem) with or without 5% glycerol. Mutations were identified by sequencing at least five subclones of each. Direct sequencing was performed on MAT1A exon VIII of patient 14 and of her parents, by means of a Cyclist Exo-Pfu DNA sequencing kit (Stratagene).
Construction, Expression, and Enzymatic Activity of MAT1A Mutants
Mutant cDNAs were constructed as described elsewhere (Ubagai et al. 1995) and were subcloned into either pQE30 (QIAexpress system; QIAGEN), for expression in bacteria, or pSVL, for expression in COS-1 cells. Expression of wild-type or mutant MAT1A cDNAs in bacteria or COS-1 cells was achieved as described elsewhere (Chamberlin et al. 1996). MAT activity was determined from bacterial or COS-1 extracts containing approximately equal amounts of expressed MAT protein (as estimated by western-blot analysis), as described elsewhere (Chamberlin et al. 1996).
Minigene Construction
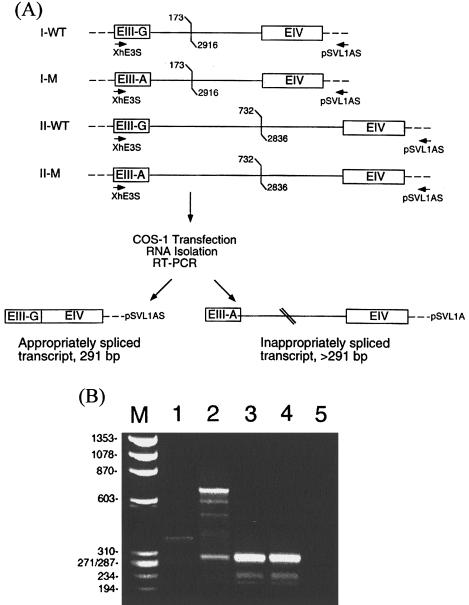
To examine a possible splice-donor mutation in exon III of patient 7, four MAT1A minigenes—containing 46 bp of exon III 3′-region, part of intron III, and 88 bp of exon IV 5′-region—were constructed as templates, by means of genomic DNAs from patient 7 and a normal subject. MAT1A intron III is 3,120 bp in length, containing MscI sites at nucleotides 171 and 2913 as well as PstI sites at nucleotides 728 and 2831. Minigene set I contained 378 bp of intron III (deletion of nucleotides 174–2915 of intron III), and minigene set II contained 1,017 bp of intron III (deletion of nucleotides 733–2835 of intron III) (see fig. 1A). Primers used to construct minigene set I are XhE3S (5′-GTGAGGGACACCATCAA-3′, nucleotides 247–263 of MAT1A cDNA) and XbE4AS (5′-TCATTTCTGTCCAGATG-3′, nucleotides 364–380 of MAT1A cDNA), containing either an XhoI or an XbaI site at the 5′ end, MscIAS (5′-TGGCCATCCTTATGCCAGAGT-3′, nucleotides 156–176 of MAT1A intron III), and MscIS (5′-TGGCCAAAGAGTTCTACATGA-3′, nucleotides 2913–2933 of MAT1A intron III), containing an MscI site at the 5′ end. Primers used to construct minigene set II are E3S, E4AS, PstIAS (5′-CTGCAGAATGTGATCGCATAC-3′, nucleotides 713–733 of MAT1A intron III), and PstIS (5′-CTGCAGGTCTCTGTAGGATGT-3′, nucleotides 2831–2851 of MAT1A intron III), containing a PstI site at the 5′ end. Each minigene was constructed by amplifying the exon III/intron III junction region and the intron III/exon IV region separately, by means of an Expand Hi Fidelity PCR system (Boehringer-Mannheim). The exon III/intron III fragments were digested with XhoI and MscI (set I) or XhoI and PstI (set II), and the intron III/exon IV fragments were digested with XbaI and MscI (set I) or XbaI and PstI (set II). The digested fragments were purified and used for three-way ligations in the presence of a pSVL vector digested with XhoI and XbaI. Clones with the correct inserts (512 and 1,151 bp for minigene set I and II, respectively) were sequenced at the exon/intron borders, to confirm their identity.
Figure 1.
RT-PCR analysis of transcripts encoded by minigenes containing wild-type or mutant splice-donor site at exon III. A, Schematic showing four minigene constructs in pSVL. MAT1A sequence is represented by boxes (exons) and solid lines (introns). Oligonucleotide primer is represented by arrows; vector sequence is represented by dashes. The constructs contained 46 bp of exon III 3′-region with either a G (wild type [WT]) or an A (mutant [M]) as the last base of this exon, either 378 bp (set I) or 1,017 bp (set II) of intron III, and 88 bp of exon IV 5′-region, as described in the Methods section. The deleted intron III regions are denoted by forked lines. B, Agarose-gel electrophoretic analysis of RT-PCR products of transcripts expressed by minigene constructs. Lane 1, Products of minigene I-M. Lane 2, Products of minigene II-M. Lane 3, Products of minigene I-WT. Lane 4, Products of minigene II-WT. Lane 5, Products from COS-1 cells transfected with the vector alone. Correctly spliced products (291 bp) will include 46 bp of exon III, 88 bp of exon IV, 148 bp of vector sequence, and a 9-bp linker sequence at the 5′ end. Lane M, HaeIII digest of ΦX174 DNA. Sizes are listed to the left.
Assay for Correct Splicing of Minigenes
A minigene or a vector control was transfected into COS-1 cells, as described by Chamberlin et al. (1996), and total RNA was isolated by use of an RNEasy RNA isolation kit (QIAGEN), followed by DNase I digestion. First-strand synthesis was performed on ∼16 ng total RNA from each sample by means of a cDNA synthesis kit (Pharmacia) and 100 pmol of a primer derived from the pSVL sequence that includes the polyadenylation signal (5′-TTTATTGCAGCTTATAATGG-3′, nucleotides 1639–1659; see schematic, fig. 1A). PCR reactions were performed on the entire first-strand synthesis mix by addition of 40 pmol XhE3S, 33 μl H2O, and 1 μl Taq polymerase (Life Technologies) to each reaction. Samples were amplified for 30 cycles of 95°C for 1 min, 55°C for 30 s, and 72°C for 30 s, and products were analyzed by electrophoresis through a 2% agarose gel. Correctly spliced products should include 148 bp of vector sequence, 134 bp of MAT1A cDNA sequence, and 9 bp of linker sequence and should be 291 bp in length.
Results
Hypermethioninemic Individuals Carrying Autosomal Recessive MAT1A Mutations
Six of the seven individuals reported in this study were initially identified by routine newborn screening for hypermethioninemia, designed to detect homocystinuria due to cystathionine β-synthase deficiency. BI-1 was discovered during screening of the family of proband BII-1. All seven individuals were shown to have persistent isolated hypermethioninemia without evidence of secondary hypermethioninemia due to tyrosinemia type I, cystathionine β-synthase deficiency, or severe hepatocellular disease.
Patient 1 is one of the first infants described with confirmed deficiency of MAT activity in the liver. MAT activity in a crude liver extract, measured in the presence of 1 mM or 6 μM methionine, was 8%–39% of control values, respectively (Finkelstein et al. 1975; Gout et al. 1977). Magnetic resonance imaging (MRI) performed at age 20 years revealed anomalous signals in the gray matter and a heterogeneous hyposignal from the posterior part of the basal ganglia (A. Joannard, personal communication). At age 24 years, the patient continued to have an elevated methionine concentration of 1,030 μM (reference range 5–35 μM), as well as elevated methionine transamination metabolites (15 μM; reference range 0.20–0.54 μM) in plasma (Mudd et al. 1995b). Physical examination revealed that cognitive and neurological functions were normal, and the subject's overall health was good.
SSCP and sequence analyses showed that patient 1 is a compound heterozygote for two MAT1A mutations: 113G→A (fig. 2A), which converts a Ser at position 38 to an Asn (S38N; table 1), and 255delCA (fig. 2A), which introduces a premature termination codon at position 92 (92X; table 1). Both mutations completely abolished MAT activity (table 2). It has been reported that rat MAT2A cDNA contains an Asn at position 38 (Horikawa et al. 1990). However, sequencing of six independent exon II isolates of the rat MAT2A gene showed only a Ser at position 38 (data not shown), which suggests that the original sequence was either in error or derived from a rat strain that carries an S38N mutation. Our data explain the already documented deficit in MAT activity in the liver of patient 1, a deficit that was more severe when the assay was done at a higher methionine concentration, as would be expected when the relatively high Michaelis constant (Km), high maximal velocity (Vmax) MAT I/III activity is abnormally low, but the lower Km), lower Vmax MAT II activity is unaffected.
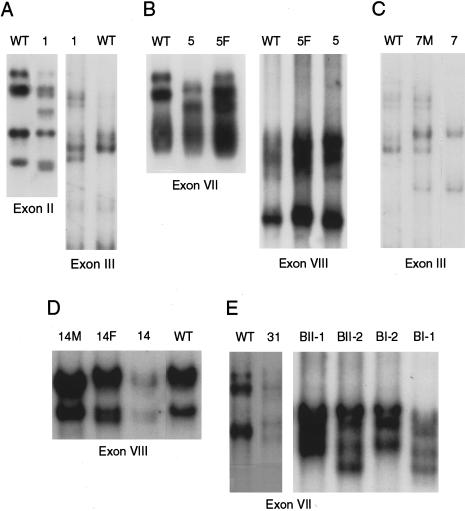
Figure 2.
SSCP analyses of PCR-amplified MAT1A exons on MDE gels. Exons containing mutations, as confirmed by sequencing analyses, are shown here. A, Patient 1. B, Patient 5. C, Patient 7. D, Patient 14. E, Patients 31 and family B. F = father; M = mother.
Table 1.
MAT1A Mutations in Isolated Persistent Hypermethioninemic Patients
| Patient and Allele | Mutation | Location | Effect on Coding Sequence | Plasma Methioninea(μM) |
| 1: | ||||
| Allele 1 | 113G→A | Exon II | S38N | 770–1240 |
| Allele 2 | 255delCA | Exon III | 92X | |
| 5: | ||||
| Allele 1 | 791C→T | Exon VII | R264C | 1721–1870 |
| Allele 2 | 1006G→A | Exon VIII | G336R | |
| 14: | ||||
| Allele 1 | 966T→G | Exon VIII | I322M | 185–467 |
| Allele 2 | 1031A→C | Exon VIII | E344A | |
| 7: | ||||
| Allele 1 | 292G↓gt→A↓gt | Exon III/intron III | Splice donor | 1226–1664 |
| Allele 2 | 292G↓gt→A↓gt | Exon III/intron III | Splice donor | |
| 31: | ||||
| Allele 1 | 791G→A | Exon VII | R264Hb | 235–545 |
| Allele 2 | Normal | |||
| BII-1: | ||||
| Allele 1 | 791G→A | Exon VII | R264H | 233 |
| Allele 2 | Normal | |||
| BI-1: | ||||
| Allele 1 | 791G→A | Exon VII | R264H | 105 |
| Allele 2 | Normal |
Published in the study by Mudd et al. (1995b). Reference range for plasma methionine is 5–35 μM.
Mutation associated with dominantly inherited hypermethioninemia.
Table 2.
MAT Activity of Bacterial Extracts Transformed with Wild-Type or Mutant MAT1A cDNAs
| Construct | MAT Activitya(nmol/min/mg protein) |
| Mock | .38±.09 |
| Wild type | 37.52±2.77 |
| S38N | .00 |
| 92X | .00 |
| R264C | .50±.01 (.3%) |
| R264H | .43±.07 (.1%) |
| I322M | 17.39±1.25 (45.8%b) |
| G336R | 8.88±.94 (22.9%) |
| E344A | 4.87±.25 (12.1%) |
Data in parentheses are percent of wild-type activity.
As expressed in a bacterial system. In COS-1 cells, however, expression was 11% of wild-type activity (Ubagai et al. 1995). (We regard the result obtained with the bacterial system as more accurate, because of the markedly increased expression efficiency in bacteria; however, the differential activity may also be due to the inherent differences between the two expression systems.)
Patient 5, a female born in 1984, had plasma methionine concentrations in the range of 1,721–1,870 μM. Transient hypotonia has been observed in this patient (Mudd et al. 1995b), and she is considered to be mentally slow in comparison with other members of her family. At age 7.5 years she had, on the Wechsler Intelligence Scale for Children-Revised, a verbal score of 88 and a performance score of 84. At age 13 years, an MRI of the brain revealed a normal result, including normal myelination. Patient 5 was shown to be a compound heterozygote for two MAT1A mutations: 791C→T (fig. 2B), which converts an Arg at position 264 to a Cys (R264C; table 1), and 1006G→A (fig. 2B), which converts a Gly at position 336 to an Arg (G336R; table 1). Whereas the R264C mutation virtually abolished enzymatic activity, the G336R mutant retained 23% of wild-type activity (table 2).
Patient 14, a 6-year-old girl with moderately elevated plasma methionine concentrations of 185–467 μM, has no reported neurological problems (Mudd et al. 1995b). Molecular analysis of her MAT1A gene revealed that she is a compound heterozygote for two MAT1A mutations in exon VIII (fig. 2D). The 966T→G mutation, inherited from her father, converts an Ile at position 322 to a Met (I322M), and the 1031A→C mutation, inherited from her mother, converts a Glu at position 344 to an Ala (E344A) (table 1). Both mutations diminish, but do not completely abolish, MAT activity (table 2).
Patient 7 is a 7-year-old boy of Hispanic and black origin. At the age of 2 wk his plasma methionine level was 1,664 μM. During the 1st mo of life his plasma total homocysteine levels ranged from 5 μM to a value as high as 47.2 μM (reference range 5.4–16.2 μM). Plasma B12 levels were not abnormally low, and several assays of plasma folate and methylmalonate had normal findings. At the age of 2 wk, pyridoxine treatment (500 mg/d) was begun for suspected cystathionine β-synthase deficiency. Two weeks later, the patient was hospitalized for apnea, and he required intubation and ventilator support for respiratory failure. Computed tomography and MRI studies of the brain had normal findings. On hospital day 19, B6 was discontinued, and respiratory function returned in 4 d. After discharge from the hospital, the patient was maintained on a methionine-restricted diet, with reduction of the plasma methionine concentration to ∼400 μM. Cystathionine β-synthase deficiency was ruled out by enzyme assays of cultured skin fibroblasts (J. Kraus, personal communication). Enzymatic activities assayed in the absence or presence of pyridoxal phosphate were 19.4 and 21.1 U/mg protein, respectively, in fibroblasts from patient 7 and were 8.5 and 10.4 U/mg protein, respectively, in fibroblasts from a normal individual. Assays of plasma-free homocysteine during the 1st year and at age 3.6 years revealed no abnormal elevation. Concern about growth retardation and dietary compliance led to discontinuance of methionine restriction at age 10 mo. Early gross motor milestones were delayed, but at age 4 years 2 mo the individual was developmentally appropriate. Although results of the neurological examination were normal, an MRI of the brain revealed diffusely abnormal white matter and patchy basal ganglia bilaterally, with an abnormally high signal in the inferior aspect and an abnormally high signal in the inferior midbrain. A repeat brain MRI 14 mo later revealed no change. At age 7 years, growth parameters were similar to those observed at age 4 years. Findings from the neurological examination were normal, but the patient is having learning difficulties. Results of a third brain MRI were no different from those of the prior examination.
Patient 7 was shown to be homozygous for a mutation at nucleotide 292, the last nucleotide in exon III of the MAT1A gene (fig. 2C): 292G↓gt→A↓gt (table 1). If normal splicing occurs, this mutation would result in a Gly→Ser conversion at position 98 (G98S). However, transient-expression assays showed that the G98S mutant retained nearly wild-type MAT activity (data not shown).
The 292G↓gt→A↓gt Mutation Causes Aberrant Splicing of the MAT1A Transcript
Because nucleotide 292 is the last base in exon III, a G→A change would alter the splice-donor site from GG↓gt→GA↓gt, which would lead to an inappropriately spliced MAT1A transcript. Because a liver sample from patient 7 was not available, we investigated the aberrant-splice hypothesis by constructing MAT1A minigenes containing the 3′ region of exon III, part of intron III, and the 5′ region of exon IV, cloned into the pSVL vector (fig. 1A). The exon III/intron III junction sequence in the minigenes contained either the normal splice-donor site (GG↓gt) or the mutated splice-donor site (GA↓gt) found in patient 7. Reverse transcription (RT)-PCR analysis of RNA from COS-1 cells transfected with minigenes containing normal splice-donor sequences gave the predicted splice product of 291 bp (fig. 1B, lanes 3 and 4), whereas minigenes containing the mutant splice site consistently yielded abnormally larger products (fig. 1B, lanes 1 and 2). COS-1 cells transfected with vector alone gave no discernible products (fig. 1B, lane 5). We conclude that the severe hypermethioninemia in patient 7 results from a MAT1A mutation that causes incorrect splicing of the MAT1A transcript.
R264C Behaves as an Autosomal Recessive Trait
In contrast with the autosomal dominant transmission of the hypermethioninemia associated with the R264H mutation (Chamberlin et al. 1997; Mudd et al. 1998), we found that the father of patient 5, a carrier of an R264C allele, had a normal plasma methionine level, 30 μM. Patient 5 is a compound heterozygote for two MAT1A mutations, R264C and G336R (table 1). Our data indicate that the R264C mutation is transmitted in an autosomal recessive mode. Further evidence supporting the autosomal recessive behavior of the R264C allele was obtained by examination of MAT activity after cotransfection of wild-type (R264) and mutant (R264C) MAT1A cDNAs into COS-1 cells. When R264 and R264C cDNAs were cotransfected at a ratio of 1:1, an MAT activity of 47%±4% (n=5 of two independent experiments) of wild type was observed. This is not significantly different from the 50% expected value, which suggests that formation of the enzymatically active wild-type MAT dimer was not adversely affected by the presence of the mutant R264C subunit.
Hypermethioninemic Individuals Carrying an Autosomal Dominant MAT1A Mutation
Patients 31 and BII-1, as well as patient BI-1, the father of BII-1, each carry the single-allelic–dominant R264H mutation (fig. 2E) (Chamberlin et al. 1997; Nagao and Oyanagi 1997). Patient 31, a girl born of unrelated German parents, was identified with plasma methionine elevated to 231–535 μM. In a crude liver extract, MAT activity was only questionably diminished (H. Przyrembel, personal communication; details reviewed in a study by Mudd et al. [1998]). Patient BII-1, a 1.6-year-old boy, with plasma methionine of 233 μM at age 2 mo, was shown to carry the single-allelic MAT1A R264H mutation with an exon VII haplotype of 1/4 (fig. 3). Members of his immediate family were screened for plasma methionine concentrations and for the presence of the R264H allele. Consistent with the dominant mode of inheritance, his father, BI-1, in good health at age 37 years, had moderately elevated plasma methionine levels (105 mM) and was a heterozygote for a normal allele and the R264H mutation, with exon VII haplotype 3/4 (fig. 3). The mother and a sister, with VII haplotypes 1/1 and 1/3, respectively, have wild-type MAT1A genes and normal plasma methionine levels (fig. 3).
Figure 3.
Pedigree of family B, showing dominant inheritance of hypermethioninemia. Blackened symbols represent individuals carrying the single-allelic R264H mutation; unblackened symbols represent normal subjects. Values in parentheses are plasma methionine concentrations (in μM [reference range 5–35 μM]). The MAT1A exon VII haplotype, described by Chamberlin et al. (1997), is indicated at the upper right corner of each symbol.
Discussion
The MAT1A gene is expressed primarily in the liver, the organ in which synthesis of the predominant portion of AdoMet occurs (Mudd and Poole 1975; Kotb et al. 1997). AdoMet serves as the methyl donor in the biosynthesis of ⩾100 metabolites. Thus, studies of genetic, metabolic, and clinical aspects of mutations in the MAT1A gene are of interest. Current evidence establishes that deleterious MAT1A mutations cause abnormal accumulation of methionine (reviewed in the studies by Mudd et al. [1995a, 1998]). In the present study, we report characterization of MAT1A mutations in seven hypermethioninemic individuals and uncover six novel mutations: 113G→A/S38N, 255delCA/92X, 791C→T/R264C, 1006G→A/G336R, 1031A→C/E344A, and 292G↓gt→A↓gt. To date, a total of 17 MAT1A mutations have been identified among 47 hypermethioninemic individuals (Ubagai et al. 1995; Chamberlin et al. 1996, 1997; Nagao and Oyanagi 1997; Hazelwood et al. 1998; present study).
The available evidence suggests that the metabolic and the clinical effects of MAT1A mutations are highly variable. The majority (32 of 47) of the genotyped individuals carry a dominant MAT1A mutation, R264H (Chamberlin et al. 1997; Nagao and Oyanagi 1997; Mudd et al. 1998). The metabolic result of this single-allelic mutation is relatively mild elevation of plasma methionine, which, clinically, appears to be entirely benign, as exemplified by the good health of individual BI-1 at age 37 years.
The remaining genotypes, all of which behave as autosomal recessive traits, are more heterogeneous with respect to the severity of the hypermethioninemia and the clinical states of the individuals carrying the mutations (Ubagai et al. 1995; Chamberlin et al. 1996; present study). Nine individuals (G1–G4, 5, 10, 11, 13, and 14) carry only missense mutations in their MAT1A gene (table 3). Most such individuals were clinically unaffected at last report, although some were very young at that time (Gaull et al. 1981; Mudd et al. 1995b, 1998). Transient expression assays demonstrate that most mutant MATs found in these patients retain significant enzymatic activity (Ubagai et al. 1995; Chamberlin et al. 1996). Within this group, patient 5 is perhaps exceptional in that, cognitively, she functions less well than her unaffected siblings. However, she had a normal MRI of the brain at age 13 years. She carries two mutations, R264C and G336R. R264C has virtually no enzymatic activity, whereas G336R retains significant MAT activity (23% of the level of the wild-type enzyme). Nevertheless, the persistently extremely high plasma methionine concentrations observed in this patient suggest that she may have less residual MAT I/III activity in vivo than do the other eight members of this group.
Table 3.
Mutations Identified in the MAT1A Gene that are Transmitted as Autosomal Recessive Traits
| Patient | Allele 1 | Allele 2 | Reference |
| G1 | 966T→G/I322M | 966T→G/I322M | Ubagai et al. (1995) |
| G2 | 914T→C/L305P | 966T→G/I322M | Ubagai et al. (1995) |
| G3 | 164C→A/A55D | 1070C→T/P357L | Ubagai et al. (1995) |
| G4 | 1068G→A/R356Q | 1132G→A/G378S | Chamberlin et al. (1996) |
| 5 | 791C→T/R264C | 1006G→A/G336R | Present study |
| 10 | 595C→T/R199C | 595C→T/R199C | Chamberlin et al. (1996) |
| 11 | 595C→T/R199C | 595C→T/R199C | Chamberlin et al. (1996) |
| 13 | 595C→T/R199C | 595C→T/R199C | Chamberlin et al. (1996) |
| 14 | 966T→G/I322M | 1031A→C/E344A | Present study |
| 9 | 539insTG/185X | 595C→T/R199C | Chamberlin et al. (1996) |
| 1a | 113G→A/S38N | 255delCA/92X | Present study |
| Ca | 827insG/351X | 827insG/351X | Chamberlin et al. (1996) |
| Mr C | 539insTG/185X | 539insTG/185X | Hazelwood et al. (1998) |
| 3 | 539insTG/185X | 539insTG/185X | Chamberlin et al. (1996) |
| 8a | 1043delTG/350X | 1043delTG/350X | Chamberlin et al. (1996) |
| 7a | 292G↓gt→A↓gt | 292G↓gt→A↓gt | Present study |
Manifested neurological problems and/or MRI abnormalities.
Patients 9 and 1 each carry both missense and insertion or deletion mutations (table 3). The 539insTG mutation in patient 9 yields a severely truncated protein devoid of MAT activity, whereas his R199C retains 11% of wild-type activity (Chamberlin et al. 1996). At age 6 years, this patient attained a composite score on the Stanford-Binet Intelligence Scale that placed him in the lowest 1% of the population, but, on an MRI of his brain, white matter was termed unremarkable, and there was no evidence for gray-matter heterotopia (S. Richter, personal communication). In patient 1, the 255delCA mutation also yields a severely truncated protein without MAT activity, but, unlike the situation in patient 9, his S38N mutation abolishes MAT activity. An MRI study of the brain of this patient at age 20 years revealed anomalies of gray matter, the first demonstrated among patients with isolated hypermethioninemia.
Four patients (C, Mr C, 3, and 8) are homozygous for deletion/insertion mutations (table 3), yielding truncated MAT mutants devoid of enzymatic activity (Chamberlin et al. 1996; Hazelwood et al. 1998). Generally, these patients have the most extreme elevations of plasma methionine (Gahl et al. 1988; Surtees et al. 1991; Mudd et al. 1995b). Patient C is homozygous for a 827insG, predicted to encode a truncated protein of 350 amino acids; patient 8 is homozygous for a 1043delTG mutation, encoding a truncated protein of 349 amino acids (table 3); and both patients had neurological problems and demyelination documented by MRI scans at ages 11 and 9 years, respectively (Surtees et al. 1991; Mudd et al. 1995b). Patient 3 and Mr C are homozygous for a 539insTG mutation, yielding a truncated polypeptide of 184 amino acids (Chamberlin et al. 1996; Hazelwood et al. 1998). However, both are free of neurological problems and neither had suggestions of demyelination on MRI examinations at ages 43 years (Hazelwood et al. 1998) and 6.25 years (W. Wilson, personal communication), respectively. The possibility has been raised that deleterious interactions may take place between the abnormal, slightly truncated, MAT1A α1 subunits synthesized in patients 8 and C and the normal α2 subunits encoded by MAT2A, whereas similar interactions might be less likely with the more-severely truncated α1 subunits produced by patients 3 and Mr C (Hazelwood et al. 1998).
Patient 7 is the only affected individual demonstrated to be homozygous for a splicing mutation in the MAT1A gene. Although the concentration of MAT activity in the liver of this patient remains unknown, his consistently high plasma methionine concentrations, comparable to those found in patients with homozygous null mutations (Mudd et al. 1995b), suggests that his MAT I/III activity deficit may be virtually complete. In spite of normal neurological examinations at ages 4 years and 7 years, he was found, on MRIs of the brain, to have signs of demyelination and, at age 7 years, is said, by staff at his school, to be manifesting poor cognitive skills.
Assays of enzymatic activity of mutant MAT1A cDNA constructs in transient expression studies provide only provisional estimates of the contributions of such mutations to MAT I/III activity in vivo. Interactions among subunits, changes in the state of polymerization, and other variables may all affect the actual MAT activity in the liver. Nevertheless, the observations reviewed here seem to indicate that moderate impairments in MAT I/III activity are clinically benign. Among patients with severe impairments in MAT I/III activity, however, there seems to be a clustering of neurological problems, learning disabilities, and MRI abnormalities of white matter and, in one case, gray matter. Nevertheless, clinical problems are not universal in these individuals. Thus, the question of a possible causative relationship between severe deficiency of MAT I/III activity and clinical problems remains open. Further studies that assess the genetics and the clinical status of individuals with isolated persistent hypermethioninemia may help to answer this question.
Acknowledgments
The authors wish to thank A. Joannard, Richard Erbe, Hildegard Przyrembel, S. Richter, and William Wilson, for sending us specimens from, and/or information about, their patients. Victor Nikiforov and Loetta Bradley kindly provided amino acid analyses.
References
- Alvarez L, Corrales F, Martin-Duce A, Mato JM (1993) Characterization of a full-length cDNA encoding human liver S-adenosylmethionine synthetase: tissue-specific gene expression and mRNA levels in hepatopathies. Biochem J 293:481–486 [DOI] [PMC free article] [PubMed]
- Blom HJ, Davidson AJ, Finkelstein JD, Luder AS, Bernardini I, Martin JJ, Tangerman A, et al (1992) Persistent hypermethioninaemia with dominant inheritance. J Inherit Metab Dis 15:188–197 [DOI] [PubMed]
- Cantoni GL (1953) S-adenosylmethionine: a new intermediate formed enzymatically from L-methionine and adenosinetriphosphate. J Biol Chem 204:403–416 [PubMed] [Google Scholar]
- Chamberlin ME, Ubagai T, Mudd SH, Levy HL, Chou JY (1997) Dominant inheritance of isolated hypermethioninemia is associated with a mutation in the human methionine adenosyltransferase 1A gene. Am J Hum Genet 60:540–546 [PMC free article] [PubMed]
- Chamberlin ME, Ubagai T, Mudd SH, Wilson WG, Leonard JV, Chou JY (1996) Demyelination of the brain is associated with methionine adenosyltransferase I/III deficiency. J Clin Invest 98:1021–1027 [DOI] [PMC free article] [PubMed]
- De La Rosa J, Ostrowski J, Hryniewicz MM, Kredich NM, Kotb M, LeGros HL Jr, Valentine M, et al (1995) Chromosomal localization and catalytic properties of the recombinant α subunit of human lymphocyte methionine adenosyltransferase. J Biol Chem 270:21860–21868 [DOI] [PubMed]
- Finkelstein JD, Kyle WE, Martin JJ (1975) Abnormal methionine adenosyltransferase in hypermethioninemia. Biochem Biophys Res Commun 66:1491–1497 [DOI] [PubMed]
- Gahl WA, Bernardini I, Finkelstein JD, Tangerman A, Martin JJ, Blom HK, Mullen KD, et al (1988) Transsulfuration in an adult with hepatic methionine adenosyltransferase deficiency. J Clin Invest 81:390–397 [DOI] [PMC free article] [PubMed]
- Gaull GE, Tallan HH, Lonsdale D, Przyrembel H, Schaffner F, Von Bassewitz DB (1981) Hypermethioninemia associated with methionine adenosyltransferase deficiency: clinical, morphological and biochemical observations on four patients. J Pediatr 98:734–741 [DOI] [PubMed]
- Gout J-P, Serre J-C, Dieterlen M, Antener I, Frappat P, Bost M, Beaudoing A (1977) Une nouvelle cause d'hypermethioninemie de l'enfant: le deficit en S-adenosyl-methionine-synthetase. Arch Fr Pediatr 34:416–423 [PubMed]
- Hazelwood S, Bernardini I, Tangerman A, Guo J, Shotelersuk V, Mudd SH, Gahl WA (1998) Lack of brain demyelination in a patient homozygous for a mutation encoding a severely truncated methionine adenosyltransferase I/III. Am J Med Genet 75:395–400 [DOI] [PubMed]
- Horikawa S, Sasuga J, Shimizu K, Ozasa H, Tsukada K (1990) Molecular cloning and nucleotide sequence of cDNA encoding the rat kidney S-adenosylmethionine synthetase. J Biol Chem 265:13683–13686 [PubMed]
- Horikawa S, Tsukada K (1992) Molecular cloning and developmental expression of a human kidney S-adenosylmethionine synthetase. FEBS Lett 312:37–41 [DOI] [PubMed]
- Kotb M, Geller AM (1993) Methionine adenosyltransferase: structure and function. Pharmacol Ther 59:125–143 [DOI] [PubMed]
- Kotb M, Kredich NM (1985) S-Adenosylmethionine synthetase from human lymphocytes: purification and characterization. J Biol Chem 7:3923–3930 [PubMed] [Google Scholar]
- Kotb M, Mudd SH, Mato JM, Geller AM, Kredich NM, Chou JY, Cantoni GL (1997) Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet 13:51–52 [DOI] [PubMed]
- Mato JM, Alvarez L, Ortiz P, Pajares MA (1997) S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther 73:265–280 [DOI] [PubMed]
- Mitsui K, Teraoka H, Tsukada K (1988) Complete purification and immunochemical analysis of S-adenosylmethionine synthetase from bovine brain. J Biol Chem 263:11211–11216 [PubMed]
- Mudd SH (1962) Activation of methionine for transmethylation. V. The mechanism of action of the methionine-activating enzyme. J Biol Chem 237:PC1372–PC1375 [PubMed] [Google Scholar]
- Mudd SH, Chamberlin ME, Chou JY (1998) Isolated persistent hypermethioninemia: genetic, metabolic, and clinical aspects. In: Mato JM, Caballero A (eds) Methionine metabolism: molecular mechanisms and clinical implications. Consejo Superior Investigaciones Cientificas, Madrid, pp 3–13 [Google Scholar]
- Mudd SH, Levy HL, Skovby F (1995a) Disorders of transsulfuration. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 1279–1327 [Google Scholar]
- Mudd SH, Levy HL, Tangerman A, Boujet C, Buist N, Davidson-Mundt A, Hudgins L, et al (1995b) Isolated persistent hypermethioninemia. Am J Hum Genet 57:882–892 [PMC free article] [PubMed]
- Mudd SH, Poole JR (1975) Labile methyl balances for normal humans on various dietary regimens. Metabolism 24:721–735 [DOI] [PubMed]
- Nagao M, Oyanagi K (1997) Genetic analysis of isolated persistent hypermethioninemia with dominant inheritance. Acta Paediatr Jpn 39:601–606 [DOI] [PubMed]
- Surtees R, Leonard J, Austin S (1991) Association of demyelination with deficiency of cerebrospinal-fluid S-adenosylmethionine in inborn errors of methyl-transfer pathway. Lancet 338:1550–1554 [DOI] [PubMed]
- Ubagai T, Lei K-J, Huang S, Mudd SH, Levy HL, Chou JY (1995) Molecular mechanisms of an inborn error of methionine pathway: methionine adenosyltransferase deficiency. J Clin Invest 96:1943–1947 [DOI] [PMC free article] [PubMed]