Summary
Isovaleric acidemia (IVA) is a recessive disorder caused by a deficiency of isovaleryl-CoA dehydrogenase (IVD). We have reported elsewhere nine point mutations in the IVD gene in fibroblasts of patients with IVA, which lead to abnormalities in IVD protein processing and activity. In this report, we describe eight IVD gene mutations identified in seven IVA patients that result in abnormal splicing of IVD RNA. Four mutations in the coding region lead to aberrantly spliced mRNA species in patient fibroblasts. Three of these are amino acid altering point mutations, whereas one is a single-base insertion that leads to a shift in the reading frame of the mRNA. Two of the coding mutations strengthen pre-existing cryptic splice acceptors adjacent to the natural splice junctions and apparently interfere with exon recognition, resulting in exon skipping. This mechanism for missplicing has not been reported elsewhere. Four other mutations alter either the conserved gt or ag dinucleotide splice sites in the IVD gene. Exon skipping and cryptic splicing were confirmed by transfection of these mutations into a Cos-7 cell line model splicing system. Several of the mutations were predicted by individual information analysis to inactivate or significantly weaken adjacent donor or acceptor sites. The high frequency of splicing mutations identified in these patients is unusual, as is the finding of missplicing associated with missense mutations in exons. These results may lead to a better understanding of the phenotypic complexity of IVA, as well as provide insight into those factors important in defining intron/exon boundaries in vivo.
Introduction
Isovaleric acidemia (IVA) (MIM 243500) is an organic acidemia caused by a deficiency of the enzyme isovaleryl-CoA dehydrogenase (IVD; E.C.1.3.99.10), which catalyzes the third step in the catabolism of the amino acid leucine. Deficiency of IVD results in the failure of isovaleryl-CoA to be oxidized to 3-methylcrotonyl-CoA (Sweetman and Williams 1995). It is usually characterized by severe, often fatal neonatal ketotic acidosis. Approximately 50% of patients with this disorder present in the newborn period with overwhelming acidosis. The remainder exhibit a less-severe clinical picture characterized by developmental delay or mental retardation, with or without episodes of intermittent acidosis. Infants who survive a severe neonatal crisis are subsequently clinically indistinguishable from patients with the more chronic form of the disease and continue to show a pattern of chronic intermittent acidotic episodes during intercurrent illnesses or other times of physiological stress. The mechanism for this clinical variability remains unclear.
IVD is a mitochondrial flavoenzyme and a member of the family of acyl-CoA dehydrogenases (ACDs), which includes short/branched chain acyl-CoA dehydrogenase (SBCAD), short chain acyl-CoA dehydrogenase (SCAD), medium chain acyl-CoA dehydrogenase (MCAD), long chain acyl-CoA dehydrogenase (LCAD), and very long chain acyl-CoA dehydrogenase (VLCAD) (Ikeda et al. 1986, 1987). These enzymes are encoded in the nuclear genome in precursor form. Precursor peptides are synthesized in the cytoplasm, transported into mitochondria, and processed to homotetramers (except VLCAD, which is processed to a homodimer), with each monomer containing a noncovalently but tightly bound flavin adenine dinucleotide molecule (Ikeda et al. 1986, 1987). All ACDs catalyze the α,β-dehydrogenation of their corresponding acyl-CoA thio-ester substrates, with each ACD having a distinct substrate specificity profile and all transfer electrons to the electron-transferring flavoprotein (ETF) (Crane and Beinert 1955; Ikeda et al. 1983, 1985a, 1985c).
[35S]methionine–protein-labeling studies of fibroblasts from IVA patients have revealed at least five classes of mutations within the IVD gene (Ikeda et al. 1985a). IVD precursor and mature proteins produced by type I mutants are indistinguishable in size from their normal counterparts. Types II, III, and IV mutants make IVD precursor proteins, which are 2–3 kDa smaller than normal. Subsequent processing in type III and IV mutants is normal but proceeds inefficiently in type II mutants. Type V mutants make no detectable IVD protein. We have elsewhere described nine point mutations in the IVD gene in fibroblasts from patients with IVA, which lead to Leu13Pro, Arg21Pro, Asp40Asn, Gly170Val, Ala282Val, Cys328Arg, Val342Ala, Arg363Cys, and Arg382Leu replacements in IVD protein (Vockley et al. 1991, 1992a, 1992b; Mohsen et al. 1998). The mutant enzymes exhibit decreased intramitochondrial stability and enzymatic activity. In other IVA cell lines, IVD mRNA is abnormal or absent, but a causative mutation could not be identified (Vockley et al. 1991, 1992a). We now report IVD gene mutations in IVA patients, which lead to abnormal processing of IVD RNA.
Material and Methods
Cell Lines and Mutation Detection
Fibroblast cell lines were obtained through contact with referring physicians who requested enzymatic testing for isovaleric acidemia. Approval for these studies was obtained through the Mayo Foundation Institutional Review Board. Cells were cultured in DMEM media (GibcoBRL Life Technologies), supplemented with 10% fetal calf serum. IVD activity in fibroblast extracts was measured by means of the electron transferring flavoprotein fluorescence reduction assay, as described elsewhere (Vockley et al. 1991; Mohsen et al. 1998). Point mutations in FB102, FB103, and FB834 have been described elsewhere (Vockley et al. 1991; Mohsen et al. 1998). Genomic DNA was isolated by means of the Puregene Kit, according to the manufacturer’s protocol (Gentra Systems). mRNA was isolated from confluent fibroblast cultures by means of the QuickPrep kit, according to the manufacturer’s instructions (Pharmacia Biotech). cDNA was synthesized by use of the SUPERSCRIPT Preamplification System, for first-strand cDNA synthesis (GibcoBRL Life Technologies). Mutations in the IVD gene were identified in genomic DNA and cDNA, made from mRNA from cell lines, as described elsewhere (Vockley et al. 1991; Mohsen et al. 1998). All exons and intron/exon boundaries were amplified from genomic DNA, and cDNA was amplified in overlapping fragments through the polymerase chain reaction (Vockley et al. 1991; Parimoo and Tanaka 1993; Mohsen et al. 1998). PCR-amplified products were subcloned into PCRScript (Stratagene), and at least six subclones were sequenced either manually, with the 7-deaza-dGTP Sequenase V2.0 kit (Amersham), or in automated fashion, by the Mayo Clinic Molecular Biology Core Facility. Alternatively, amplification products were sequenced directly by the Mayo Clinic Molecular Biology Core Facility after purification via a Qiaquick PCR purification kit (Qiagen), according to the manufacturer’s instructions.
Evaluation of Splicing Mutations by Use of an Exon-Trapping System
Cos-7 (1651-CRL) cells were obtained from American Type Culture Collection. The exon-trapping system, with the pSPL3 vector, LipofectACEΤΜ reagent, and the Superscript II kit, was obtained from GibcoBRL Life Technologies. All other reagents and chemicals, unless otherwise stated, were from Fisher, Sigma, or GibcoBRL Life Technologies. PCR products amplified from patient and control fibroblast genomic DNA were ligated into T-tailed-EcoRV-digested pSPL3 vector and transformed into DH5α cells. Amplified regions of IVD were FB103, FB118, FB136, and FB834, exon 1 to intron 3; FB136, intron 6 to intron 9. Primers to amplify exon 1 to intron 3 were 5′, nucleotides 103–123 of the coding sequence; 3′, 59–79 bp downstream of the splicing donor site for intron 3. Primers to amplify intron 6 to intron 9 were 5′, 93–113 nucleotides upstream from the splicing acceptor site for intron 6; 3′, 56–76 nucleotides downstream of the splicing donor site for intron 9. Transformants containing the desired inserts in the correct orientation were identified by restriction enzyme analysis and automated DNA sequencing. About 0.5–1 μg of the purified plasmid DNA was transfected, along with appropriate control DNAs, into Cos-7 cells by means of LipofectACEΤΜ reagent, according to the manufacturer’s protocol, and total RNA was isolated 48–72 h after transfection by means of RNA STAT 60 (Tel-Test). One microgram of total RNA (in 10-μl volume) was treated with DNAse I (Boehringer Mannheim) at 37°C for 15 min, followed by inactivation at 65°C for 15 min. Reverse transcription (RT) was done in a total volume of 20 μl by means of the treated RNA and the Superscript II kit. Five microliters of the RT product were amplified by use of AmpliTaq DNA polymerase (PE Biosystems), and the pSPL3-specific primers—SA2 and SD6 (GibcoBRL Life Technologies)—in a total volume of 25 μl in a GeneAmp PCR System 9600 (PE Biosystems), by means of the following thermal profile: initial denaturation for 5 min at 94°C; cycling for 1 min at 94°C, for 1 min at 60°C, and for 3 min at 72°C (35 cycles); and a final extension step for 10 min at 72°C. Five microliters of the RT-PCR product derived from the respective patient and control DNA were separated by electrophoresis on a 1% agarose gel for analysis. Bands from both normal and mutant samples were excised from the gel and purified by means of a Qiaquick gel extraction kit (Qiagen) and were sequenced by the Mayo Clinic Molecular Biology Core Facility either directly or after subcloning into pGEM-T vector (Promega).
Computer Analysis of Splicing Mutations
Genomic sequences spanning an interval of 50 nucleotides up- and downstream of each substitution were evaluated by information analysis of splice donor and acceptor sites (Rogan et al. 1998). By using the Scan program (from the Laboratory of Computational and Experimental Biology at the National Institutes of Health), we analyzed the individual information content of each natural and variant primary and secondary splice site by measuring their Ri value expressed in bits of information (Schneider 1997a). The programs MakeWalker and Lister were used to generate graphic displays of these findings (Rogan et al. 1998). The effects of nucleotide substitutions on splicing information were predicted from Ri values. The following criteria were validated in a previous study of 111 splice-site variants (Schneider 1997a, 1997b):
(1) Substitutions resulting in Ri values of <2.4 bits were predicted to completely inactivate the natural site.
(2) Substitutions with decreased Ri values of >2.4 bits were predicted to result in leaky splicing. To predict the residual amount of mRNA correctly spliced at such leaky sites, we calculated the minimum fold change in binding affinity as 2ΔRi (ΔRi is the difference between the Ri value of the natural site and that of the variant site); the result was expressed as the percentage of normally processed mRNA (Schneider 1997a; Rogan et al. 1998).
(3) Substitutions with secondary or cryptic splice sites near the primary site with Ri values greater than the natural splice site were predicted to result in cryptic splicing.
(4) Nucleotide changes that did not have significant impact on the ΔRi of the natural site were predicted not to affect splice site function (Schneider 1997a, 1997b). Significance of the change in Ri was assessed by means of a paired t test, with a cutoff at 5% significance (Rogan et al. 1998).
Results
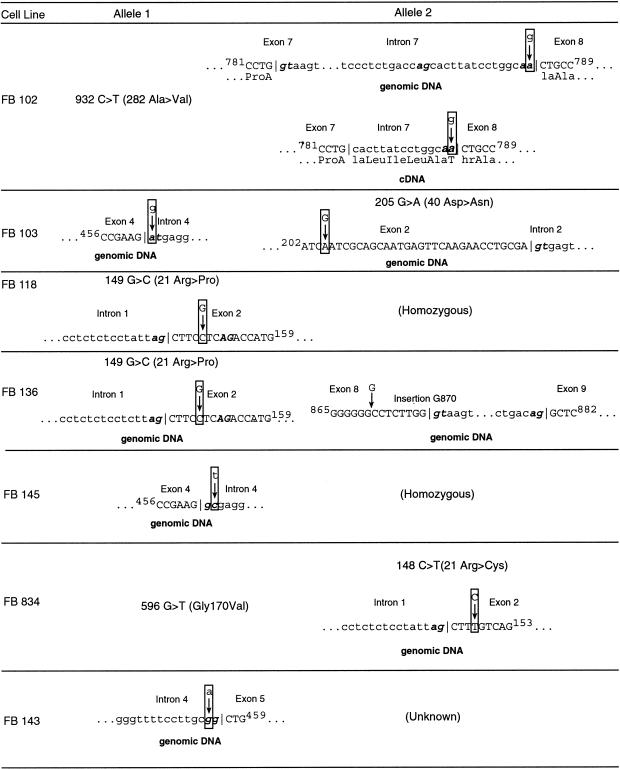
We have elsewhere identified nine point mutations in the IVD gene in fibroblasts from patients with enzymatically proved IVD deficiency (Vockley et al. 1991, 1992a, 1992b; Mohsen et al. 1998). In some cell lines, however, only one IVD coding mutation was found, whereas in others, multiple aberrant IVD cDNA species were present. The additional causative mutations in these cell lines were not identified. To examine these cell lines for mutations affecting IVD RNA processing, all exons and intron/exon boundaries of the IVD gene were amplified by PCR and sequenced from patient genomic DNA and cDNA. Wild-type IVD intron-exon boundary sequence information has been deposited in Genbank (accession numbers AF191214 to AF191218) and is also available on request. Figure 1 summarizes the sequence alterations identified in seven IVD-deficient cell lines. Five missense mutations were identified in cDNA and were confirmed to be present in genomic DNA (fig. 1–3: 148C→T, 149G→C, 205G→A, 596G→T, and 932C→T). In addition, a G insertion in exon 8 and four splice-junction mutations occurring at the conserved gt-or-ag dinucleotide were found. Of these cell lines, only FB102 shows any residual enzyme activity, and this can be attributed to the partial activity of the mutant 282 Ala→Val enzyme (Mohsen et al. 1998). During the past 10 years, we have analyzed >20 cell lines for IVD mutations. Only one (the 148G→C mutation) has ever been seen in more than one cell line. Thus, the other mutations have been identified only once in 40 alleles (J. Vockley, unpublished data, and Vockley et al. 1991, 1992a, 1992b; Mohsen et al. 1998).
Figure 1.
Summary of mutations in the IVD gene leading to abnormal splicing of IVD RNA. IVD sequences, including intron and exon boundaries, were amplified from genomic DNA isolated from cell lines from patients with IVA and shown to be deficient in IVD enzymatic activity. Mutations identified are listed. For FB102, the effect of the intron 7 mutation on cDNA sequence is also shown. FB118 and FB145 were homozygous for their respective mutations. Only one mutation could be identified in FB143, despite complete sequencing of all exons and extensive sequencing of the intron/exon boundaries. The remaining mutation in the IVD gene responsible for this patient’s IVA is unknown.
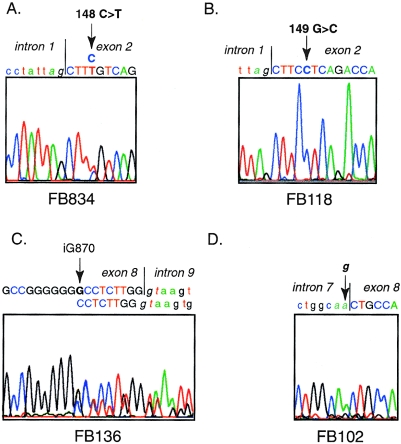
Figure 2.
Sequencing of DNA fragments amplified from IVD genomic DNA from IVA fibroblasts. A, Direct sequence of amplified genomic DNA from FB834, showing a 148C→T mutation. The patient is heterozygous at this position. B, 149G→C mutation in FB118 in products amplified from genomic DNA and sequenced directly. The patient is homozygous for this mutation, which was also identified in FB136. Control experiments confirmed that this was not related to contamination of PCR reactions with previously amplified template. C, Insertion of a G residue in genomic DNA from FB136 in an area that normally contains six Gs from cDNA positions 865–870. Direct sequence from amplified genomic DNA is shown. Because the patient is heterozygous for this mutation, a double sequencing pattern appears after the inserted nucleotide. This finding was confirmed in the reverse direction and by sequencing of subclones of the PCR amplified genomic DNA from this region (not shown). D, G→A point mutation of the intron 7 splice acceptor site in FB102 genomic DNA, identified by direct sequencing of amplified DNA. The patient is heterozygous at this location. The mutant A allele is easily visualized in the forward direction shown, whereas the wild-type G signal is more evident in the reverse direction (not shown).
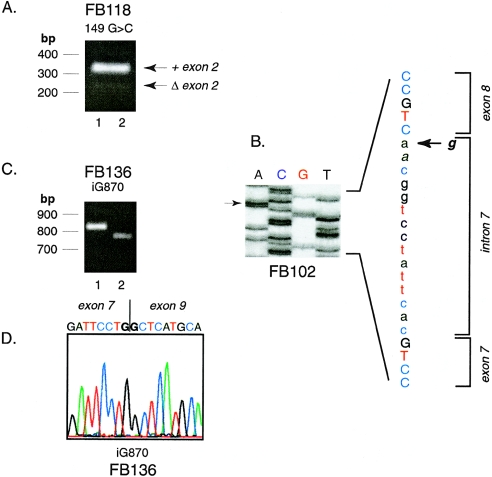
Figure 3.
Identification of exon skipping in mRNA from IVA fibroblasts. A, Agarose gel of DNA fragments amplified from cDNA from FB118. The 5′ amplification primer was in the 5′ untranslated region of IVD (position −20 to −1) and the 3′ amplification primer was from position 265–286, spanning exons 1–3. Arrows mark the predicted size of a normal fragment (306 bp) and one with a deletion of exon 2 (90 bp). The migration positions of 100 bp molecular mass standards are shown. B, Manual DNA sequencing of a subcloned DNA fragment amplified from FB102 cDNA, including exons 7 and 8. The arrows show the position of the g→a mutation in the intron 7 splice acceptor site. Sequences corresponding to exon 7, intron 7, and exon 8 are marked. C and D, Analysis of subclones of amplified products from FB136 cDNA, spanning exons 3–11 (5′ primer, IVD coding nucleotides 318–347; 3′ primer, IVD coding nucleotides 1122–1138; anticipated size 820 bp). Two different-sized inserts are obtained after subcloning, as seen on agarose gel electrophoresis (C). The sequence of the insert from lane 1 is normal (not shown). The sequence of the insert from lane 2 shows a deletion of exon 8 (D).
Missplicing Caused by Mutations in the IVD Coding Region
Three of the IVD missense mutations and the insertion mutation in exon 8 are associated with exon-skipping. This rather unexpected finding led us to seek proof that these mutations were directly responsible for the splicing abnormalities. We therefore used an exon-trapping vector-based model RNA splicing system to verify the formation of abnormal mRNA species (Liu et al. 1997). Results are summarized in figure 4.
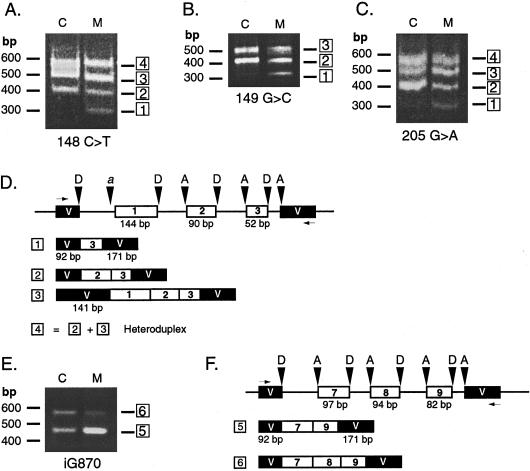
Figure 4.
Use of an exon-trapping system to examine the effects of IVD exon mutations on RNA splicing. IVD genomic DNA fragments, containing control or mutation sequences, spanning exon 1 to intron 3 (A–D) or intron 7 to intron 9 (E–F) were cloned into the exon-trapping vector pSPL3. Inserts for all clones were sequenced to confirm that no PCR errors were present. The vector was transiently transfected into COS-7 cells, and total RNA from transfected cells was used as template for RT-PCR, with vector-specific primers. Amplified products were separated by electrophoresis on agarose gels and visualized with ethidium bromide staining. The number of products listed below the vector maps correspond to the numbering of the bands on the gel photo. Lanes containing PCR products amplified from cDNA of cells transfected with control and mutant IVD sequences are marked as “C” and “M,” respectively. The mutation being analyzed is listed underneath, and the migration of molecular mass markers is indicated to the left of each gel photograph. A, 148C→T; B, 149G→C; C, 205G→A. D and F, Map of amplified products identified by sequencing subcloned products obtained with normal and mutant IVD exon-trapping experiments. The test IVD exon-trapping vector plus insert is depicted on the top line. IVD exons are shown as white boxes and exon sizes are as labeled. Vector exon sequences (V) are denoted by black boxes and intron sequences by a line. Donor and acceptor sites are indicated by D and A, respectively. The horizontal arrows indicate the position of PCR primers. D and F, Exon 2 splicing test vector. Cryptic acceptor site in vector is indicated by “a.” E, iG870 = insertion of G at position 870 in the IVD coding region. F, Exon 8 splicing test vector.
Previous analysis of FB834 mRNA suggested that skipping of exon 2 was occurring in this cell line, but the cause of this was not determined (Vockley et al. 1991, 1992a). We have now identified a C→T mutation at position 148, which results in a missense change (21 Arg→Cys, fig. 2A). This involves the same codon mutated in FB118 but at a different nucleotide position and appears to impair splicing to a greater extent than the 149 mutation, since no cDNA containing the point mutation in exon 2 could be amplified from cDNA. Exon-trapping analysis shows that the 148 mutation indeed causes skipping of exon 2 (fig. 4A and 4D).
cDNA amplification of DNA flanking exon 2 from FB118 yielded two products—a 306-bp–sized fragment and a less abundant species of 216 bp. Sequence analysis confirms that the larger fragment contains exon 2 with the coding mutation (149G→C), whereas the smaller is missing exon 2 (90 bp; fig. 3A). Apparently, the homozygous 149G→C mutation (21 Arg→Pro) in this patient leads to missplicing of exon 2. To evaluate this mutation, a fragment extending from exon 1 to intron 3, containing the mutation, was tested in an exon-trapping assay. Skipping of exon 2 was observed in the fragment containing 149G→C but not in the fragment with the normal sequence (fig. 4B and 4D).
FB136 harbors coding mutations in both IVD alleles, one of which has a 149G→C mutation (as in FB118). Exon 2 was deleted in one of seven subclones of PCR-amplified cDNA derived from this cell line. As shown for FB118, this mutation leads to exon 2 skipping. The other mutation is a single G insertion in exon 8 within a polyguanine run nine nucleotides upstream from the donor site (fig. 1 and 2C). Of nine amplified cDNA subclones analyzed from this patient, one contained the inserted guanine residue in exon 8, another was missing exon 8, and six had the normal sequence (fig. 3C and 3D). Mutations were not found in adjacent splice junctions, indicating that the exon skipping is related to this insertion. Genomic fragments consisting of introns 6–9 were tested with the exon-trapping system (fig. 4E and 4F); bands 6 and 5 correspond to cDNA fragments with and without exon 8, respectively. Fragments missing exon 8 were also identified when wild-type sequence was present, but insertion of a G at position 870 leads to a much larger proportion of skipped exon 8 product than a control (58% vs 10%, by densitometry). Skipping of exon 8 has never been identified in control cDNAs made from human fibroblasts or liver (data not shown), and thus, the low level of exon 8 skipping seen with wild-type sequence may be an artifact of the exon-trapping system.
Several subclones of amplified cDNA from FB103 were missing exon 2, presumably related to the 205G→A (40 Asp→Asn) mutation in this exon reported elsewhere (Mohsen et al. 1998). This observation was confirmed with our exon-trapping assay (fig. 4B). When a normal genomic IVD fragment extending from the middle of exon 1 to intron 3 is present in the vector, either two or three products are obtained after amplification of cDNA from transfected cells (fig. 4B, bands 2 and 3). Band 2 consists of exons 2 and 3 spliced correctly to the vector donor and acceptor sites (fig. 4D). Band 3 contains exons 1–3, joined appropriately but with a cryptic acceptor site in the vector being used. Both bands 2 and 3 are anticipated products of the splicing reaction; because no acceptor site precedes exon 1, the vector donor site selects either the nearest acceptor site, in exon 2, or a cryptic acceptor site in the vector. Heteroduplexes of these two products are sometimes also detected (band 4). When a similar fragment containing the 205G→A mutation is used, an additional product that contains only exon 3 is detected—that is, exons 1 and 2 have been skipped (band 1). Because skipping of exon 1 is seen with the wild-type IVD fragment, our results suggest that the 205G→A mutation is responsible for the skipping of exon 2.
Mutations in the IVD Gene Splice Junctions
Mutations at the conserved gt-or-ag dinucleotides of splice junctions were identified in four IVD-deficient cell lines. These mutations were not tested in an exon-trapping system. IVD mRNA from FB102 contains an insertion of 15 nucleotides from the 3′ end of intron 7 (fig. 1). This missplicing is the result of a g→a mutation in the intron 7 acceptor site, altering the ag acceptor site to aa (fig. 2D). Consequently, a cryptic splice acceptor site in intron 7, located 15 nucleotides upstream of exon 8, is used, accounting for the 15-nt insertion in the cDNA. This insertion maintains the correct reading frame. FB145 harbors a homozygous intron 4 splicing donor site t→c mutation that alters the conserved gt to gc. IVD sequences could not be amplified from cDNA derived from FB145, which suggests that the IVD mRNA is unstable in this cell line. A g→c mutation at the donor site in intron 4 of the IVD gene in FB103 converts the conserved gt dinucleotide to at. cDNA missing exon 4 was identified in a minority of clones made from amplified cDNA. This patient also had a coding mutation in the other allele, as described elsewhere (Mohsen et al. 1998). Finally, an intron 4 acceptor site mutation (a→g) was identified in FB143 that converted the conserved ag to gg. As with FB145, IVD sequences could not be amplified from the cDNA of FB143, which suggests that the IVD mRNA is also unstable in this cell line.
Information Analysis of Splice Junction Mutations
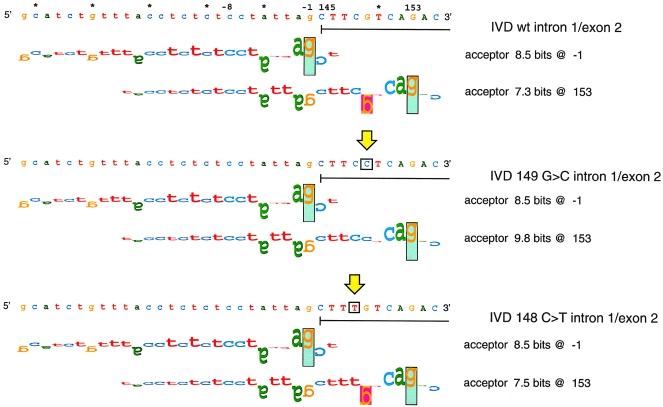
Because missplicing related to mutations in IVD exons was unexpected, we conducted information analysis on the predicted effect of these and the splice junction mutations, on the strength of the splicing signal, by using the Scan program (from the Laboratory of Computational and Experimental Biology at the National Institutes of Health) (Rogan et al. 1998). An example of these analyses is shown in figure 5, and results for all the mutations are summarized in table 1. The 149G→C mutation in FB118 and FB136 strengthens a cryptic acceptor site (7.3→9.8 bits) at position 153, nine nucleotides downstream from the natural splice site (8.5 bits). The strengthened cryptic acceptor overlaps the natural site and apparently interferes with recognition and assembly of the spliceosome at this site. The mechanism of exon skipping for the C→T mutation at position 148 (FB834) appears to be similar; however, the cryptic site at position 153 is only marginally strengthened (7.3→7.5 bits). The insertion of a guanine residue within a polyguanine run in exon 8 of the IVD gene in FB136 does not alter the Ri value of the adjacent donor site (9.2 bits) and does not create a cryptic site, although it leads to the accumulation of both normal and abnormally sized IVD mRNA species in this cell line. Of the eight subclones of amplified cDNA from this region analyzed, one contained the single nucleotide insertion, one was deleted of exon 8, and six were of normal sequence (fig. 3C and 3D). The abnormal IVD mRNA identified in FB102 results from a g→a mutation, one nucleotide upstream of exon 8, which inactivates the natural acceptor site for intron 7 splicing (5.4→−2.2 bits) and uses a pre-existing 5.6-bit cryptic site upstream of exon 8. The intron 4 splice-junction mutations in FB103 and FB145 weaken the intron 4 donor site (9.2→−1.9 and 7.9→−0.3 bits, respectively).
Figure 5.
Walker diagrams of IVD exon 2 splice acceptor sites. Splice sites are shown by walkers (Schneider 1997b), in which the height of each letter is the contribution of that base to the total conservation of the site (in bits). The upper bound of the vertical rectangles is at +2 bits, and their lower bound is at −3 bits. Letters that are upside down represent negative contributions. The top panel depicts the natural splice site one nucleotide upstream of exon 2 and a cryptic acceptor site at position 153. Neither cryptic site is used. The vertical arrows indicate the locations of mutations in the sequence. The middle panel corresponds to the G→C change at position 149, and the lower panel to C→T at position 148. The mutations alter the Ri value of the cryptic acceptor at position 153.
Table 1.
Information Analysis of IVD Gene Mutations
|
Primary Splice Site |
Secondary Splice Site |
|||||
| Patient | Splice Junction Affected | NT Positiona | Ri, Natural → Ri, Mutant (bits) | NT Position | Ri, Natural → Ri, Mutant (bits) | Predicted Residual Splicingb(% normal) |
| FB102 | Intron 7, acceptor | −1, G→A | 5.4 → −2.2 | −15 | 5.6 → 5.6 | |
| FB103 | Intron 4, donor | +1, G→A | 9.2 → −1.9 | (0) | ||
| FB118, FB136 | Intron 1, acceptor | 149G→C | 8.5 → 8.5 | 153 | 7.3 → 9.8 | |
| FB136 | Intron 8, donor | −8 insertion G | 9.2 → 9.2 | + (100) | ||
| FB145 | Intron 4, donor | +2, T→C | 11.0 → 3.5 | – (.05) | ||
| FB834 | Intron 1, acceptor | 148C→T | 8.5 → 8.5 | 153 | 7.3 → 7.5 | |
| FB143 | Intron 4, acceptor | −2, A→G | 7.9 → −.3 | (0) | ||
Mutation position for coding mutations is listed in terms of the cDNA position. Noncoding mutations are referenced to the nearest exon/intron junction (a minus sign [−] indicates distance from a natural acceptor site, and a plus sign [+] indicates distance from a natural donor site; see fig. 1).
Nucleotide substitutions may result either in a complete loss of primary site function (−) or in leaky splicing (+). Ri-based predictions of mutated primary splice-site binding are shown with the predicted amount of correctly spliced mRNA, given as a percentage of binding of the common allele (in parentheses).
Polymorphic Tandem Repeat in Intron 1 of the IVD Gene
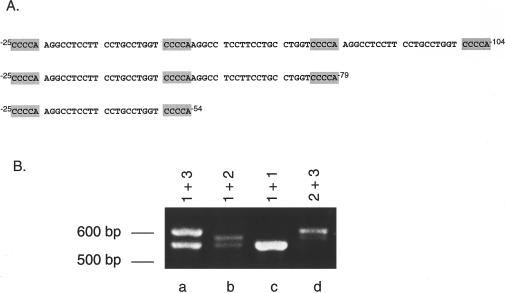
While sequencing intron 1 from FB834, we noticed a sequence variant 25 nucleotides downstream of exon 1, which consisted of three tandem copies of a 20-bp sequence flanked by 5-bp direct repeats in half the subclones from this region (fig. 6A). The remaining clones had one copy of the repeat. The patient was heterozygous at this locus, having inherited a single repeat from his homozygous father and three repeats from his heterozygous mother (whose alleles had two and three repeats; data not shown).
Figure 6.
Polymorphic repeats in intron 1 of the IVD gene. A, Sequence of three, two, and one repeat alleles in intron 1, beginning 25 bp from the junction of exon 1 and intron 1. B, Amplification of intron 1 repeats from fibroblast cDNA. Fragments corresponding to each number of repeats are shown.
To determine the frequency of the intron 1 repeats, the flanking region of genomic DNA was amplified from 70 control fibroblasts (table 2). A representative PCR analysis from four samples is shown in figure 6B. The variable repeat sequence is polymorphic with a heterozygosity value of 0.26. Additional sequence analysis of subclones from FB834 reveals that the 148C→T mutation is linked to the IVD allele with three repeats in intron 1 in the father and patient (not shown). To examine the effect of the intron 1 polymorphism on splicing, exon-trapping vectors, consisting of normal exons 1–3 but with a variable number of intron 1 repeats, were analyzed. Regardless of whether one, two, or three repeats were present, IVD mRNA produced from all three vectors is correctly spliced in COS cells (data not shown).
Table 2.
Frequency of Intron 1 Variable Repeats in the IVD Gene
| No. of Intron 1 Repeats | No. of Alleles (Frequency) |
| 1 | 17 (.12) |
| 2 | 119 (.85) |
| 3 | 4 (.03) |
Discussion
Mutations leading to missplicing and exon skipping account for 15%–20% of mutations leading to human disease (Krawczak et al. 1992). Thus, the high frequency of splicing mutations in IVD in patients with IVA is unusual (to date, 9 of 20 mutant alleles characterized; this report and Mohsen et al. 1992). In addition, exon skipping due to single-base mutations in exons has only rarely been reported as a cause of human disease (Ligtenberg et al. 1991; Berg et al. 1992; Steingrimsdottir et al. 1992; Li et al. 1995; Liu et al. 1997; O'Neill et al. 1998). A 2055C→T mutation in exon 8 of the ATP7A gene encoding a Ser637Leu substitution leads to skipping of exon 8 in splicing (Ronce et al. 1997). Several single nucleotide substitutions in exons 3 and 8 of the HPRT gene in patients with Lesch-Nyhan syndrome have been identified, which lead to deletion of these exons in splicing (Steingrimsdottir et al. 1992; O'Neill et al. 1998), whereas a similar effect on exon 2 has been described with a single-base polymorphism of that exon in the MUC1 gene (Ligtenberg et al. 1991). Single-base changes in exons leading to exon skipping have been described in patients with Laron dwarfism, Crouzon syndrome, and Marfan syndrome (Berg et al. 1992; Li et al. 1995; Liu et al. 1997).
Exon substitutions may affect splicing by altering the adjacent splice site, creating or activating secondary splice sites, or concurrently mutating a splicing enhancer element. Computer analysis reveals the presence of two potential acceptor sites in the vicinity of the exon 2/intron 1 boundary—the natural one and a cryptic one about seven nucleotides downstream. Both sites are of approximately equal strength and overlap considerably. The 148C→T and 149G→C mutations do not significantly alter the information content of the natural site or activate secondary sites. Rather, the mutations appear to prevent either site from being recognized appropriately, leading to loss of definition of the exon and skipping of this exon during splicing. Although we cannot exclude the possibility that these changes may have a corollary effect on a splicing enhancer sequence, it seems more likely that these mutations represent another previously unrecognized class of splicing mutation.
The cause for missplicing related to the remaining exon 2 mutation (205G→A) is not clear. No unusual secondary structures are obvious in this region of the IVD gene. The polymorphic repeat in intron 1 is interesting but unlikely to be significant in relation to splicing, given our population data. In the case of the HPRT gene, mutations in exon 8 have been postulated to affect secondary structure of the primary RNA transcript, perhaps masking a splicing enhancer, and thus leading to missplicing of exon 8 sequences (Steingrimsdottir et al. 1992; O'Neill et al. 1998). Overall, our findings are consistent with the hypothesis that exon sequences can play a crucial role in determining the integrity of the splicing mechanism and guiding the processing of IVD RNA. Moreover, the magnitude of the disruption of exon 2 splicing appears to be related to the position of the mutation within the exon, because the mutation at position 148 leads predominantly to a product lacking exon 2, whereas the 205G→A and 149G→C mutations result in accumulation of both normal and abnormally spliced products.
Insertion of a G residue at position 870 in exon 8 (FB136) (fig. 2C) leads to skipping of this exon. Because most of the IVD subclones identified in these experiments contain a normal sequence in this location, it is conceivable that the misspliced RNA is less stable than the wild type. The Scan program (from the Laboratory of Computational and Experimental Biology at the National Institutes of Health) does not predict a change in the information content of the exon 8 donor site by this mutation, so an alternative mechanism for missplicing must be hypothesized. It has recently been shown that RNA must form loops for different splicing factors to interact with one another at splicing-enhancer sites (Graveley et al. 1998). It is possible that the insertion of another guanine residue into the poly-G tract already present may lead to an alteration of secondary structure that interferes with the splicing machinery, resulting in the production of unstable message. Indeed, even a silent nucleotide substitution has been shown to induce exon skipping (Liu et al. 1997). Alternatively, frameshift mutations and mutations creating a new stop codon have elsewhere been suggested to lead to instability of the nuclear RNA, perhaps related to a nuclear scanning function or direct coupling of RNA processing and translation (Dietz et al. 1993; Dietz and Kendzior 1994).
In summary, we have identified an unusually high frequency of splicing errors in IVD mRNA in patients with IVA, including missplicing related to coding mutations. The causative nature of these latter mutations has been substantiated in a cellular-based model splicing system. In general, stable mRNAs appear to be produced by exon skipping if the resulting message maintains an open frame (as shown for the exon 2 mutations), whereas exon-skipping events that result in a frameshift (such as the splicing mutations affecting exon 4, or the G insertion in exon 8) either do not occur or produce unstable RNA species. Additional analysis of splicing in this system should prove helpful in further elucidating the normal mechanism of splicing of IVD mRNA.
Acknowledgments
This work was supported in part by grant RO1-DK45482 (J.V.), from the U.S. Public Health Service, and by a research grant from the Merck Genome Research Foundation (P.K.R.). Special thanks to the many physicians who referred clinical samples for analysis.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (accession numbers AF191214 to AF191218)
- Laboratory of Computational and Experimental Biology at the National Institutes of Health, http://www.lecb.ncifcrf.gov/~toms/ (for the Scan program)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for IVA [MIM 243500])
References
- Berg MA, Guevara-Aguirre J, Rosenbloom AL, Rosenfeld RG, Francke U (1992) Mutation creating a new splice site in the growth hormone receptor genes of 37 Ecuadorean patients with Laron syndrome. Hum Mutat 1:24–32 [DOI] [PubMed]
- Crane F, Beinert H (1955) On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A: part II. The electron-transferring flavoproteins. J Biol Chem 218:717–731 [PubMed] [Google Scholar]
- Dietz HC, Kendzior RJ Jr. (1994) Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet 8:183–188 [DOI] [PubMed]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ Jr., Pyeritz RE, Cutting GR (1993) The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259:680–683 [DOI] [PubMed]
- Graveley BR, Hertel KJ, Maniatis T (1998) A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J 17:6747–6756 [DOI] [PMC free article] [PubMed]
- Ikeda Y, Dabrowski C, Tanaka K (1983) Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. J Biol Chem 258:1066–1076 [PubMed]
- Ikeda Y, Hale DE, Keese SM, Coates PM, Tanaka K (1986) Biosynthesis of variant medium chain acyl-CoA dehydrogenase in cultured fibroblasts from patients with medium chain acyl-CoA dehydrogenase deficiency. Pediatr Res 20:843–847 [DOI] [PubMed]
- Ikeda Y, Hine DG, Okamura-Ikeda K, Tanaka K (1985a) Mechanism of action of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases: direct evidence for carbanion formation as an intermediate step by using enzyme-catalyzed C-2 proton/deuteron exchange in the absence of C-3 exchange. J Biol Chem 260:1326–1337 [PubMed]
- Ikeda Y, Keese S, Fenton WA, Tanaka K (1987) Biosynthesis of four rat liver mitochondrial acyl-CoA dehydrogenases. Import into mitochondria and processing of their precursors in a cell-free system and in cultured cells. Arch Biochem Biophys 252:662–674 [DOI] [PubMed]
- Ikeda Y, Keese S, Tanaka K (1985b) Molecular heterogeneity of variant isovaleryl acyl-CoA dehydrogenase from cultured isovaleric acidemia fibroblasts. Proc Natl Acad Sci USA 82:7081–7085 [DOI] [PMC free article] [PubMed]
- Ikeda Y, Okamura-Ikeda K, Tanaka K (1985c) Spectroscopic analysis of the interaction of rat liver short chain, medium chain and long chain acyl-CoA dehydrogenases with acyl-CoA substrates. Biochemistry 24:7192–7199 [DOI] [PubMed]
- Krawczak M, Reiss J, Cooper DN, Milland J, Christiansen D, Thorley BR, McKenzie IF, et al (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90:41–54 [DOI] [PubMed]
- Li X, Park WJ, Pyeritz RE, Jabs EW (1995) Effect on splicing of a silent FGFR2 mutation in Crouzon syndrome. Nat Genet 9:232–233 [DOI] [PubMed]
- Ligtenberg MJL, Gennissen AMC, Vos HL, Hikens J (1991) A single polymorphism in an exon dictates allele dependent differential splicing of episialin mRNA. Nucleic Acids Res 19:297–301. [DOI] [PMC free article] [PubMed]
- Liu W, Qian C, Franke U (1997) Silent mutation induces exon skipping of fibrillin-1 gene in Marfan syndrome. Nat Genet 16:328–329 [DOI] [PubMed]
- Mohsen A-W, Anderson B, Volchenboum S, Battaile K, Tiffany K, Roberts D, Kim J-J, et al (1998) Characterization of molecular defects in isovaleryl-CoA dehydrogenase in patients with isovaleric acidemia. Biochemistry 37:10325–10335 [DOI] [PubMed]
- O'Neill JP, Rogan PK, Cariello N, Nicklas JA (1998) Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat Res 411:179–214 [DOI] [PubMed]
- Parimoo B, Tanaka K (1993) Structural organization of the human Isovaleryl-CoA dehydrogenase gene. Genomics 15:582–590 [DOI] [PubMed]
- Rogan P, Fauz B, Schneider T (1998) Information analysis of human splice site mutations. Hum Mutat 12:153–171 [DOI] [PubMed]
- Ronce N, Moizard M-P, Robb L, Toutain A, Villard L, Moraine C (1997) A C2055T transition in exon 8 of the ATP7A gene is associated with exon skipping in an occipital horn syndrome family. Am J Hum Genet 61:233–238 [DOI] [PMC free article] [PubMed]
- Schneider T (1997a) Information content of individual genetic sequences. J Theor Biol 189:427–441 [DOI] [PubMed]
- Schneider T (1997b) Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res 25:4408–4415 [DOI] [PMC free article] [PubMed]
- Steingrimsdottir H, Rowley G, Dorado G, Cole J, Lehmann AR (1992) Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res 20:1201–1208 [DOI] [PMC free article] [PubMed]
- Sweetman L, Williams JD (1995) Branched chain organic acidurias. In: Scriver C, Beaudet AL, Sly W, Valle D (eds) The Metabolic and Molecular Basis of Inherited Disease. McGraw-Hill, New York, pp 1387–1422 [Google Scholar]
- Vockley J, Nagao M, Parimoo B, Tanaka K (1992a) The variant human isovaleryl-CoA dehydrogenase gene responsible for type-II isovaleric acidemia determines an RNA splicing error, leading to the deletion of the entire 2nd coding exon and the production of a truncated precursor protein that interacts poorly with mitochondrial import receptors. J Biol Chem 267:2494–2501 [PubMed]
- Vockley J, Parimoo B, Tanaka K (1991) Molecular characterization of four different classes of mutations in the isovaleryl-CoA dehydrogenase gene responsible for isovaleric acidemia. Am J Hum Genet 49:147–157 [PMC free article] [PubMed]
- ——— (1992b) Identification of the molecular defects responsible for the various genotypes of isovaleric acidemia. Prog Clin Biol Res 375:533–540 [PubMed]