Summary
Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) is a monogenic autosomal disease with recessive inheritance. It is characterized by multiple autoimmune endocrinopathies, chronic mucocutaneous candidiasis, and ectodermal dystrophies. The defective gene responsible for this disease was recently isolated, and several different mutations in the novel gene, AIRE, have been identified, by us and by others, in patients with APECED. We have shown that the APECED protein is mainly localized, both in vitro and in vivo, to the cell nucleus, where it forms distinct speckles. This accords with the predicted structural features of the protein, which suggest involvement of AIRE in the regulation of gene transcription. Here, we report the results of mutational analyses of a series of 112 patients with APECED who were from various ethnic backgrounds. A total of 16 different mutations, covering 91% of disease alleles, were observed; of these, 8 were novel. The mutations are spread throughout the coding region of AIRE, yet four evident mutational hotspots were observed. In vitro expression of four different naturally occurring nonsense and missense mutations revealed a dramatically altered subcellular location of the protein in cultured cells. Interestingly, the wild-type APECED protein tethered to the Gal4 DNA-binding domain acted as a strong transcriptional activator of reporter genes in mammalian cells, whereas most of the analyzed mutant polypeptides had lost this capacity.
Introduction
Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED [MIM 240300]), also known as autoimmune polyglandular syndrome type I (APS I), is an autosomal recessive disorder. It is, to date, the only organ-specific human autoimmune disease described that affects multiple organs and that is known to be inherited in a Mendelian fashion. APECED is characterized by various circulating tissue-specific autoantibodies that cause the destruction of the target organs, mainly the various endocrine glands. The patients also manifest chronic mucocutaneous candidiasis and a wide spectrum of ectodermal dystrophies. The cardinal symptoms in APECED are hypoparathyroidism, primary adrenocortical failure, and chronic mucocutaneous candidiasis (Ahonen et al. 1990).
The defective gene in APECED disease was isolated and characterized by use of the positional cloning strategy, and it was named “AIRE” to denote autoimmune regulator (The Finnish-German APECED Consortium 1997; Nagamine et al. 1997). AIRE encodes a 545-amino-acid proline-rich protein with a molecular weight of 58 kD. The amino acid sequence of the APECED polypeptide includes a putative nuclear targeting signal between amino acids 113 and 133, and, recently, we and others have confirmed that, in transiently transfected mammalian cell lines, the APECED protein is mainly localized to the cell nucleus, where it forms distinct speckles (Björses et al. 1999; Heino et al. 1999a; Rinderle et al. 1999). Several predicted features of the protein suggest that it plays a role in the regulation of transcription. The protein harbors two zinc fingers of plant homeodomain (PHD) type—motifs that are found in many nuclear proteins, including chromatin-modulating proteins and transcription activators (Aasland et al. 1995). A putative DNA-binding domain, named “SAND” (Gibson et al. 1998), as well as four nuclear receptor–binding LXXLL motifs, an inverted LXXLL domain, and a variant form of the latter (FXXLL), hint that this protein functions as a transcription coactivator (Heery et al. 1997). Furthermore, a highly conserved N-terminal 100-amino-acid domain that was first described, in AIRE, by Gibson et al. (1998) and that has a significant homology to the homogeneously staining region (HSR) domain of Sp100 and Sp140 proteins is found in the amino acid sequence of the APECED protein. This domain has been shown to function as a dimerization domain in several Sp100-related proteins (Seeler et al. 1998; Sternsdorf et al. 1999). The domain has also been named the “ASS domain” (Mittaz et al. 1999). A mouse homologue of AIRE that has recently been cloned shows conservation of all the predicted functional domains (Blechschmidt et al. 1999; Mittaz et al. 1999; Wang et al. 1999). The AIRE gene seems to have a quite-limited pattern of expression, with detectable mRNA levels seen only in the thymus, lymph nodes, and fetal liver (Nagamine et al. 1997; Heino et al. 1999a). By means of immunohistochemical staining, the APECED protein was detected in the nuclei of cells from multiple immunologically relevant tissues, such as the thymus, spleen, lymph nodes, and peripheral leukocytes (Björses et al. 1999; Heino et al. 1999a).
APECED is enriched in three genetically isolated populations: the Finnish (estimated lifetime incidence 1/25,000), the Iranian Jewish (estimated lifetime incidence 1/9,000), and the Sardinian (estimated lifetime incidence 1/14,500) populations (Ahonen et al. 1990; Zlotogora and Shapiro 1992; Rosatelli et al. 1998). However, sporadic cases of APECED are observed worldwide. Twenty-one different mutations have previously been described in these patients. They include the common mutation in the Finnish population, R257X, which was observed in 85% of the Finnish APECED chromosomes (The Finnish-German APECED Consortium 1997; Nagamine et al. 1997; Björses et al. 1998), and the common Sardinian mutation, R139X, which is responsible for 92% of the disease alleles in that population (Rosatelli et al. 1998).
Here, we report the results of an analysis of mutations in the AIRE gene of 112 patients with APECED. In this series, we characterized 16 different mutations, 8 of which have not previously been reported. Four interesting mutations were further analyzed, by transient expression in tissue-culture cells, to examine the effects of the mutations on the subcellular targeting of APECED protein. We also performed preliminary functional studies, to examine the potential of the wild-type and mutant APECED proteins to activate transcription.
Patients and Methods
Patients
A series of 112 patients with APECED were systematically screened for mutations in the coding region of AIRE. Sixty-three of the patients were of Finnish origin; 13 were Iranian Jewish; 12, Italian; 5, British; 5, French; 5, German; 4, Dutch; 2, Swiss; 1, Swedish; 1, Hungarian; and 1, Canadian. No consanguinity was documented in any of the families. The diagnostic criterion for the patients was the unequivocal presence, documented by a physician, of at least two of the following symptoms: hypoparathyroidism (subnormal Ca and supranormal inorganic phosphate concentrations in serum, with renal failure excluded), primary adrenocortical failure (primary cortisol and/or aldosterone deficiency), and chronic mucocutaneous candidiasis (Ahonen et al. 1990). Genomic DNA was extracted from 10–20-ml blood samples, according to standard procedures (Vandenplas et al. 1984). All of the samples were drawn in accordance with the Helsinki Declaration, and the study was approved by the Helsinki University Hospital Ethics Committee.
Mutation Analysis
To detect the APECED mutations, all 14 exons of the AIRE gene (European Molecular Biology Laboratory Database and GenBank accession number AJ009610) were amplified, by means of PCR performed on patient DNA, with the use of primers located in the respective flanking introns. These primers and PCR conditions are described in table 1. PCR for SSCP was done according to the same protocols, with the exception that 0.8 μl α[32P]-dGTP per 50-μl reaction was added. The PCR-amplified exons were analyzed on nondenaturing polyacrylamide gels, both at room temperature with 10% glycerol and at 4°C without glycerol (Orita et al. 1989). Those exons that showed shifts on SSCP were amplified, with use of biotinylated primers, for solid-phase sequencing (Syvanen et al. 1989), and both DNA strands were sequenced with the use of PCR primers. The samples that showed no shifts on SSCP were amplified, by PCR, for ABI377 automated sequencing. Purification of the PCR product was done with the use of 0.25 μl (2.5 U) ExonucleaseI (Amersham Life Sciences) and 0.5 μl (0.5 U) Shrimp Alkaline Phosphatase (SAP) (Amersham Life Sciences), for 4.6 μl of the PCR reaction (Werle et al. 1994). Sequencing was done in both directions, according to the Big Dye Terminator Cycle Sequencing protocol (PE Biosystems), with minor modifications. Sequencing Analysis software, version 3.2 (PE Biosystems), was used to perform the initial basecalling of the sequencing traces. Sequence contigs were assembled with the use of Sequencher software (GeneCodes Corporation). To improve basecalling accuracy and heterozygote detection, a manual basecalling check was required, since dye-terminator chemistry produces uneven but predictable peak patterns that are dependent on local sequence context (Zakeri et al. 1998). All of the detected mutations were monitored against a control panel of 100 unrelated Finnish individuals and 32 CEPH parents, by means of minisequencing protocols (Syvanen et al. 1993), in which the primers listed in table 2 were used. Haplotype analyses for the markers JAI, D21S1912, PFKL, and D21S171 were done as described elsewhere (Björses et al. 1996).
Table 1.
Primers and Conditions for PCR Amplification of Exons[Note]
|
Primer |
|||||
| Forward |
Reverse |
||||
| Exon | Position | Sequence | Position | Sequence | Conditionsa |
| 1 | 22473–22493 | 5′-CGTGGTCGCGGGGGTATAACA-3′ | 22811–22831 | 5′-TATCCCTGGCTCACAGGGCCT-3′ | 62°C; 1 |
| 2 | 23172–23190 | 5′-GGGACCCTCATGCCACCCC-3′ | 23375–23394 | 5′-GGCTGAGTCCACCAGGACCC-3′ | 65°C; 2 |
| 3 | 23564–23584 | 5′-CTACCCCTGGAGAAAACCCCT-3′ | 23904–23923 | 5′-GGTCTAGTACCCAGAGGAGA-3′ | 58°C; 2 |
| 4 | 24115–24134 | 5′-CACTGAGAGGGGAGGCCAGG-3′ | 24244–24263 | 5′-AAGGGAGGCGCTCCCACTGG-3′ | 64°C; 2 |
| 5 | 24928–24948 | 5′-TATGTGCTTGGGAACAGTCTT-3′ | 25150–25167 | 5′-CGTGGTCCTCCTTCCATC-3′ | 62°C; 0 |
| 6 | 26307–26326 | 5′-AGAAGTGCATCCAGGTTGGC-3′ | 26603–26622 | 5′-GGAAGAGGGGCGTCAGCAAT-3′ | 58°C; 3 |
| 7 | 26603–26622 | 5′-ATTGCTGACGCCCCTCTTCC-3′ | 26720–26739 | 5′-AGGGGCCTCTCCTGTGGTCT-3′ | 61°C; 1 |
| 8 | 27711–27730 | 5′-CATGGGCGTCTCTTGCCTGT-3′ | 27862–27880 | 5′-TGGTTGCGCTGGCAGAGGT-3′ | 62°C; 1 |
| 9 | 28913–28933 | 5′-CATGTCTCTGACTGGTGGACA-3′ | 29076–29094 | 5′-GTGGCCATGTGGACAGGAG-3′ | 62°C; 1 |
| 10 | 29609–29628 | 5′-CTGATGCTGACCCTTGGGTT-3′ | 29849–29868 | 5′-GTAGGTCCTGGGCTCCTTGA-3′ | 58°C; 1 |
| 11 | 30361–30380 | 5′-CTCGGGTTCGGGTTCAGCTA-3′ | 30553–30571 | 5′-CCCGCCGACCACGCTCACT-3′ | 59°C; 1 |
| 12 | 30970–30989 | 5′-CGGAGGTGGCACTCCTGCT-3′ | 31213–31232 | 5′-GAGGGCCCCTGCGGCACT-3′ | 60°C; 2 |
| 13 | 32980–33000 | 5′-CTGTGGGAGTTGGGCTGACCT-3′ | 33098–33117 | 5′-GGAGGAGCACCAGGAGGCCA-3′ | 63°C; 2 |
| 14 | 34263–34283 | 5′-AGGTTCTCACCGTCACTCTGT-3′ | 34461–34481 | 5′-ACTGACAAGAGGTGGCGCTGT-3′ | 64°C; 0 |
Note.—Intronic primers and conditions used for PCR and sequencing. The PCR reactions were done in a total volume of 25 μl containing 50 ng genomic DNA, 2.5 μl Dynazyme DNA polymerase 10 × buffer, 0.2 μM each nucleotide, 3 pmol each primer, and 0.25 U Thermus brochianus F501L DNA polymerase (Finnzymes). The PCR conditions were as follows: denaturation at 94°C for 4 min, then 30 cycles of denaturation at 94°C for 30 s, annealing at a specific temperature listed in the Conditions column, and extension at 72°C for 30 s, except for the final cycle, for which extension was for 4 min and 30 s.
The numbers “0”–“3” represent PCR reaction mixtures, with 0 denoting no dimethyl sulfoxide (DMSO)/glycerol; 1, 8.7% glycerol; 2, 5% DMSO; and 3, 7.5% DMSO.
Table 2.
Mutations and Primers Used For Minisequencing[Note]
| Exon | Mutation | Primer | Position |
| 2 | G358T | 5′-GCCATCCTGGACTTCTGGAGG-3′ | 23282–23302 |
| 2 | A374G | 5′-GAGGGTGCTGTTCAAGGACT-3′ | 23299–23318 |
| 4 | 13 bp ins 628–629 | 5′-CCAAGAAGCCGGAGAGCAGCG-3′ | 24182–24202 |
| 4 | C637T | 5′-CGGAGAGCAGCGCAGAGCAG-3′ | 24191–24210 |
| 5 | C727T | 5′-CCGGGGACGTCCCGGGAGCC-3′ | 25034–25053 |
| 6 | C889T | 5′-GTGGCCCGAAGCCTCTGGTT-3′ | 26394–26413 |
| 8 | T1051del | 5′-ACGGCGGGGAGCTCATCTGC-3′ | 27767–27786 |
| 8 | G1052A | 5′-CGGCGGGGAGCTCATCTGCT-3′ | 27768–27787 |
| 8 | 4-bp ins 1086–1087 | 5′-CTTCCACCTGGCCTGCCTGT-3′ | 27807–27826 |
| 8 | 1085–1097del | 5′-GGGCCTTCCACCTGGCCTGC-3′ | 27803–27822 |
| 8 | C1094A | 5′-ACCTGGCCTGCCTGTCCCCTC-3′ | 27812–27832 |
| 10 | A1284ins | 5′-TGGGGAACCCCTAGCCGGCAT-3′ | 29680–29700 |
| 10 | C1309del | 5′-CGACTCTTGTCTACAAGCAC-3′ | 29706–29725 |
| 10 | C1313del | 5′-TCTTGTCTACAAGCACCTGC-3′ | 29710–29729 |
| 10 | C ins 1364–1365 | 5′-CTCCTCGGGCCCTGCACCCCC-3′ | 29767–29787 |
| 14 | A1758T | 5′-GCGGCCCCCTTCCCCTCTG-3′ | 34342–34361 |
Antibodies
The raising and purification of a specific antibody against the synthetic peptide of 17 amino acids representing amino acids 151–167 of the APECED polypeptide have been described elsewhere (Björses et al. 1999). To produce an antiserum against the N-terminal half of the APECED polypeptide (amino acids 1–209), a DNA construct coding for a GST-fusion polypeptide was prepared in pGEX-2T plasmid (Amersham Life Sciences; modified and donated by Dr. T. Sareneva, National Public Health Institute, Helsinki). The expressed fusion protein was purified from the bacterial lysate, by use of a preparative 8% SDS-PAGE (Bio-Rad). The fractions containing the fusion protein were detected by means of western blotting and were pooled, concentrated, and used as an antigen for the immunization of rabbits. Two rabbits were immunized by means of subcutaneous injection with the fusion protein in Freund’s complete adjuvant. The primary injection of 150 μg fusion protein was followed by three additional boosts of 100 μg after 10 d and 6 and 10 wk, respectively. The blood was collected 1 wk after the last immunization, and the immunoglobulin G fraction of the antibodies was isolated by Protein A Sepharose Column (Amersham Life Sciences). The antibody titers and specificity were determined by means of immunofluorescence and western blotting. For double stainings, a commercially available Anti-FLAG M2 monoclonal mouse antibody binding to FLAG fusion proteins (Eastman Kodak) was used.
Expression Constructs
The AIRE cDNA was previously isolated from a human adult thymus cDNA library (The Finnish-German APECED Consortium 1997). This cDNA was cloned into EcoRI sites of the mammalian expression vectors SV-poly (Stacey and Schnieke 1990) and pcDNA3.1(+) (Invitrogen). For double immunofluorescence labeling, AIRE cDNA was tagged, at the N-terminal, with the 8-amino-acid FLAG epitope recognized by the M2 monoclonal antibody (Eastman Kodak). In vitro mutagenesis, performed with the Chameleon Double-Stranded, Site-Directed Mutagenesis Kit (Stratagene), was used to produce this construct, with the full-length AIRE cDNA in SV-poly used as a template. Mutant clones were identified by PCR amplification and were analyzed by means of solid-phase sequencing (Syvanen et al. 1989). Four different AIRE constructs were produced that mimicked naturally occurring APECED-causing mutations. These included the Finnish and Iranian Jewish major mutations C889T and A374G, a mutation in the first PHD finger that substituted tyrosine for one of the conserved cysteines (Cys311), and a single C-nucleotide deletion at nucleotide (nt) 1313. These mutations were introduced into the wild-type AIRE by means of in vitro mutagenesis with QuikChange Site-Directed Mutagenesis Kit (Stratagene), and both the full-length AIRE in SV-poly and the N-terminal FLAG-tagged construct were used as templates. The mutagenesis primers are listed in table 3.
Table 3.
Primers for Mutagenesis Constructs[Note]
|
Primer |
||
| Mutation | Mutagenesis | Selection (for SV-poly vector) |
| 5′ FLAG | 5′-CGCGCCAACACCATGGACTATAAGGACGACGACGACAAGGCGACGGACGCGGCGC3′ | 5′-CCGCATGCGAATTCGAGCTCTCC-3′ |
| C889T | 5′-CCGAAGCCTCTGGTTTGAGCCAAGGGAGCCC-3′ |
5′-CCGCATGCGAATTCGAGCTCTCC-3′ |
| Mutagenesis Forward |
Mutagenesis Reverse |
|
| A374G | 5′-TGCTGTTCAAGGACTGCAACCTGGAGCGCT-3′ | 5′-TAGCGCTCCAGGTTGCAGTCCTTGAACAGCA-3′ |
| G1052A | 5′-GACGGCGGGGAGCTCATCTGCTATGACGGCTGCCCTC-3′ | 5′-GAGGGCAGCCGTCATAGCAGATGAGCTCCCCGCGCT-3′ |
| C1313del | 5′-CACGACTCTTGTCTACAAGCACTGCCGCCTTC-3′ | 5′-GAAGGCGGAGCCGGCAGTGCTTGTAGACAAGAGTCGTG-3′ |
Note.—The construct with the 5′ FLAG tag and the plasmids carrying the common Finnish mutation C889T were prepared by use of the Chameleon Double-Stranded Site-Directed Mutagenesis Kit (Stratagene); the selection primer is located in the polylinker region of the SV-vector. The remaining mutation plasmids were prepared by use of the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
Cell Culture, Transfections, and Immunological Detection
African green-monkey kidney cell line COS-1 (ATCC CRL 1650), the human cervical carcinoma cell line HeLa (ATCC CCL 2), and the mouse fibroblast cell line NIH3T3 (ATCC CRL 1658) were cultured and were transfected, according to standard protocols in which the FuGene transfection system (Boehringer-Mannheim) was applied. Western blotting of COS-1 cells transfected with different APECED constructs, as well as immunofluorescent stainings and confocal microscopy, were done as described elsewhere (Björses et al. 1999).
Metabolic Labeling and Immunoprecipitation
COS-1 cells were seeded on 3-cm cell-culture dishes (4 × 105 cells/well) and were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. At 48 h after transfection with the different AIRE cDNA constructs, COS-1 cells were starved, for 1 h, in cysteine-free Dulbecco's minimal essential medium (DMEM) and were then labeled with 200 μCi [35S]-Cys for 1 h. The cell-culture medium was then collected, and the cells were lysed by freezing and melting three times in the presence of protease inhibitor cocktail (Boehringer Mannheim). To avoid nonspecific precipitants, we first incubated the samples with the preimmune serum for 1 h and then with 1:1 (v/v) solution of Protein A Sepharose (Amersham Life Sciences) for 1 h. Immunoprecipitation was then done, by adding to the supernatant, at 1:2,000 dilution, the antibody against the N-terminal half of the APECED protein and by incubating overnight at 4°C. An equal volume of Protein A Sepharose solution was again added, and the samples were incubated for 1 h at 4°C; this was followed by four washes. After immunoprecipitation, the polypeptides were electrophoresed on an 11% SDS–polyacrylamide gel and were visualized by autoradiography.
Analysis of Transcriptional Activation
Both the full-length AIRE cDNA and the APECED-mutation constructs were subcloned into the EcoRI site of the pM2 vector (from I. Sadowski, University of British Columbia, Vancouver, Canada) located both downstream of and in frame with the region encoding the Gal4 DNA-binding domain (DBD) (Silver et al. 1984). The reading frame was verified by DNA sequencing. Two reporter constructs were used, both of which contained the luciferase gene under the control of either the thymidine kinase (tk) promoter with four upstream Gal4-binding sites (pUAS4tk-LUC) (from T. Pearlmann, Ludwig Cancer Research Institute, Stockholm, Sweden) or a minimal promoter with five Gal4 responding elements in front of the adenovirus E1b TATA sequence (pG5LUC) (Promega). COS-1 cells were plated on 12-well plates (6.6 × 104 cells/well), and, after 24 h, they were cotransfected with 40 ng pCMVβ (Clontech), 200 ng either pUAS4tk-LUC or pG5LUC, and 220 ng of Gal4 DBD construct, by use of the FuGene transfection system (Boehringer-Mannheim). At 18 h after transfection, the medium was changed to DMEM containing 2% (v/v) fetal bovine serum. Luciferase and β-galactosidase activities were assayed as described elsewhere (Moilanen et al. 1999), and the light units were normalized against the β-galactosidase activity, to take into account the different transfection efficiencies. The values obtained were expressed as fold activation over that observed with Gal4 DBD alone, averaged from six independent experiments.
Results
Mutations in the AIRE Gene
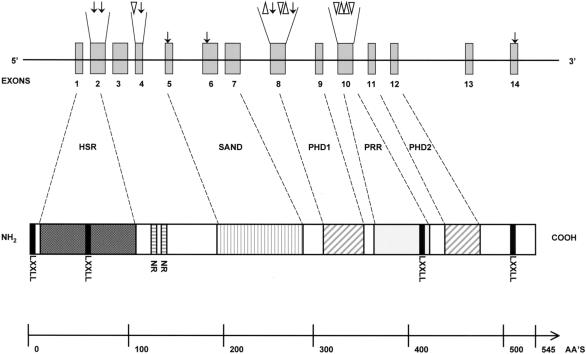
Initially, we used SSCP to screen for mutations in the AIRE gene in a series of 112 patients with APECED. Among 96 families with APECED, 16 different mutations were identified in the coding region of AIRE. All DNA samples from 23 patients with only one or no identified mutation were reanalyzed by sequencing all 14 exons from new PCR products. In a total of six patients, no mutation could be established after this effort, and in six other patients, only one disease-causing allele was found. Eight of the mutations represent new APECED mutations. The mutations, which represent single-nucleotide substitutions, small insertions, and deletions, are summarized in table 4 and in figure 1. They appear to be spread throughout the coding region of the AIRE gene. However, four evident mutational hotspots were observed: the first was located in exon 2, in the putative homodimerization domain; the second, in exon 6, between the SAND domain and the first PHD finger; the third, in exon 8, in the region coding for the first PHD finger; and the fourth, in exon 10, between the two PHD fingers (fig. 1).
Table 4.
Summary of the APECED-Causing Mutations Characterized in the Present Study
| Mutationa | Consequence | Haplotype | Origin | No. of Alleles |
| G358T | Val80Leu | 3-2-5 | Italian | 1 |
| 13-bp ins 628–629 | Frameshift, truncated 219-amino-acid protein | 5-1-4 | German | 1 |
| A374G | Tyr85Cys | 4-4-5 | Iranian Jewish | 26 |
| C637T | Gln173→STOP, truncated 172-amino-acid protein | ND | Italian | 1 |
| C727T | Arg203→STOP, truncated 202-amino-acid protein | 4-10-3 | Italian | 5 |
| C889→TFin major | Arg257→STOP, truncated 256-amino-acid protein | 4-3-5 | Finnish, Swedish | 104 |
| 2-3-5 | Finnish | 2 | ||
| 3-3-5 | Finnish | 1 | ||
| 5-3-5 | Finnish | 4 | ||
| 6-3-5 | Finnish | 1 | ||
| 4-3-6 | Finnish | 1 | ||
| 3-2-5 | Finnish | 1 | ||
| 5-2-5 | Finnish | 1 | ||
| 4-4-7 | Italian | 2 | ||
| 4-5-2 | German | 1 | ||
| 4-5-3 | German | 2 | ||
| 5-3-5 | British, Italian | 3 | ||
| 4-5-5 | Dutch | 2 | ||
| 5-4-3 | German, Swiss | 6 | ||
| T1051 deletion | Frameshift, truncated 376-amino-acid protein | 4-9-4 | French | 2 |
| G1052A | Cys311Tyr | 4-4-5 | Finnish | 2 |
| 4-bp ins 1086–1087 | Frameshift, truncated 371-amino acid protein | 5-3-5 | Italian | 4 |
| 1085–1097del | Frameshift, truncated 372-amino-acid protein | 4-5-5 | Dutch, German, British | 9 |
| 5-5-5 | British, Finnish, Canadian | 5 | ||
| 5-3-5 | Italian, Swedish | 2 | ||
| 5-10-5 | Italian, British | 3 | ||
| C1094A | Pro326Gln | 5-4-5 | Finnish | 1 |
| A ins 1283–1284 | Frameshift, truncated 422-amino-acid protein | 5-4-3 | Finnish | 2 |
| C1309 del | Frameshift, truncated 478-amino-acid protein | 5-2-5 | French | 2 |
| C1313 del | Frameshift, truncated 478-amino-acid protein | 2-10-7 | Italian | 4 |
| C ins 1364–1365 | Frameshift, truncated 422-amino-acid protein | 4-4-5 | German | 2 |
| A1758T | X546Cys, 60-amino-acid–longer protein | 5-5-7 | Finnish | 3 |
Eight of the mutations are novel. A heterozygous G→T substitution at nt 358, resulting in a Val→Leu missense mutation at amino acid 80, was observed in an Italian patient. The common Iranian Jewish mutation, an A→G substitution at nt 374 that leads to a Tyr→Cys change at amino acid 85, was detected in all of the disease chromosomes from this population. A German patient was heterozygous for a 13-bp insertion in exon 4, between nt 628 and 629. A homozygous deletion of a T at position 1051 was observed in one French patient. One Finnish patient was a compound heterozygote for a G→A transition at position 1052, resulting in a substitution of the first cysteine residue of the PHD1 with a tyrosine. A homozygous C→A substitution at nt 1094 was found in a Finnish patient. A C1309 deletion was found in a heterozygous form in an Italian patient. A German patient was homozygous for a C insertion between nt 1364–1365. The haplotypes over the APECED region were constructed from the markers JAI, D21S1912, and PFKL. For the patient denoted as “ND,” no parental samples were available; therefore, the haplotypes remain uncertain. The nucleotide positions for the cDNA are based on GenBank accession number Z97990.
Figure 1.
Schematic presentation of the APECED gene and protein structure, with the locations of all of the mutations characterized in the present study indicated. The dark gray–shaded box indicates the highly conserved N-terminal HSR domain; blackened boxes, the LXXLL nuclear receptor–binding domains; horizontally striped boxes, the putative nuclear targeting signal; the vertically striped box, the putative DNA-binding SAND domain; hatched boxes, the PHD zinc fingers; and the light gray–shaded box, the proline-rich region. The arrow indicates a single-nucleotide substitution; the triangle, a small deletion; and the inverted triangle, a small insertion.
Six of the mutations occurred in more than one family. The common Finnish mutation, R257X, was found in 114 (89%) Finnish APECED chromosomes examined, with 72% of Finnish patients being homozygous for R257X. The carrier frequency of this mutation in Finland was calculated to be 1/250 (The Finnish-German APECED Consortium 1997). This mutation was also observed in 16 disease chromosomes from non-Finnish individuals (in one allele each from 6 German, 4 Italian, 2 Dutch, 2 Swiss, 1 British, and 1 Swedish individual), combined with six different haplotypes in the APECED region. One German, one Italian, and two Swiss patients were homozygous for this mutation. A 13-bp deletion occurring at nt 1094–1106 in exon 8 was also observed in affected chromosomes from individuals of many different geoethnic origins. It was observed in 7 of 10 British disease alleles. Furthermore, it was found in two Finnish, one Swedish, two Italian, two German, and four Dutch alleles. The results of haplotype analyses suggest that this mutation has occurred independently several times, since it was found on five different APECED haplotypes.
We also analyzed samples from 13 Iranian Jewish patients with APECED. The whole coding region of the AIRE gene and all of the exon-intron boundaries were completely sequenced. The only difference from the wild-type alleles was a single-nucleotide substitution at G374A in exon 2, which resulted in the amino acid change of tyrosine to a cysteine at position 85. This Y85C mutation was found in homozygous form in all of the Iranian Jewish patients, and it was found in heterozygous form in 1 of 35 unrelated Iranian Jewish control individuals. The Y85C mutation was not detected either in patients with any other ethnic background or in the European control panel.
Subcellular Location of the Mutant APECED Proteins
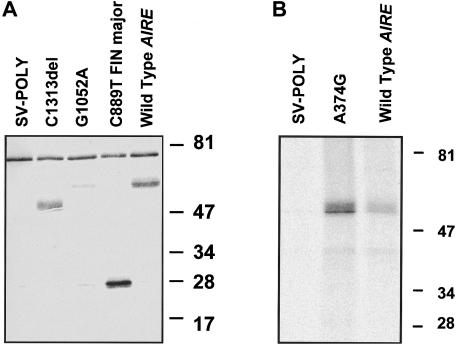
For in vitro detection of the APECED protein, we used both a polyclonal rabbit antibody raised against a synthetic 17-amino-acid peptide corresponding to amino acid residues 151–167 (Björses et al. 1999) and an antibody against the N-terminal half of the APECED polypeptide. The wild-type AIRE, as well as the four different mutant forms in the SV-poly vector, were transiently expressed in transfected COS-1 cells. The mutant forms examined included the common Finnish and Iranian Jewish mutants, a mutant with a cysteine-to-tyrosine change in the first PHD finger (G1052A), and a mutant lacking the second PHD finger (C1313del). The results of western blotting of the transfected cell lysates with use of the peptide antiserum revealed two polypeptides (fig. 2A). The molecular weight of the wild-type APECED protein, as well as that of the polypeptide produced by the mutant construct carrying the 1-bp substitution G1052A, was 58 kD. However, the signal derived from the protein with the mutation was significantly weaker than that of the wild type. The construct carrying the major Finnish mutation yielded a 28-kD polypeptide, and the construct lacking the second PHD finger yielded a 50-kD polypeptide; both values are in agreement with the calculated molecular masses of these mutated polypeptides. Surprisingly, we did not detect, by means of western blotting, any polypeptide produced by the plasmid carrying the Iranian Jewish A374G mutation. However, by immunoprecipitation assay done with the use of the antibody against the N-terminal half of the APECED protein, we were able to visualize the 58-kD polypeptide produced by this construct (fig. 2B). A 76-kD polypeptide was detected in all samples including both the nontransfected and empty vector-transfected COS-1 cells, and it was considered to be unspecific. APECED-specific immunostaining was totally abolished by preincubation of the antiserum with the synthetic APECED peptide. No specific staining was observed with preimmune serum.
Figure 2.
A, Western blot analyses of cell extracts from COS-1 cells transiently expressing the wild-type and mutated forms of the APECED protein. The cells were transfected with the plasmids indicated in the figure. All of the polypeptides were resolved by means of 11% SDS-PAGE. The blot was probed with the rabbit antiserum against the APECED peptide. The molecular masses of the specific polypeptides are in conformity with their calculated values. Cells transfected with the empty SV-poly vector were used as controls. B, Metabolic labeling and immunoprecipitation of the wild-type APECED polypeptide and the polypeptide carrying the common Iranian Jewish mutation. The COS-1 cells were transfected with the constructs indicated in the figure. After 1 h of metabolic labeling with 200 μCi of [35S]-Cys, the cell-culture medium was collected and the cells were lysed. The antibody against the N-terminal half of the APECED protein with Protein A Sepharose was used to immunoprecipitate the polypeptides. The immunoprecipitated samples were electrophoresed on 11% SDS-PAGE, and the polypeptides were visualized by autoradiography. The molecular masses of the low-range marker (Bio-Rad) are indicated on the right side of both figures.
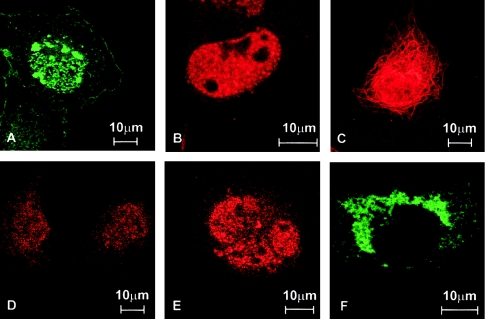
To gain information on the effects of the APECED mutations on the intracellular targeting of the protein, we performed immunohistochemical analyses of multiple mammalian cell lines transfected with either the wild-type or mutant forms of APECED. The subcellular location was investigated by means of immunofluorescence, with use of both the antibody described previously and a commercial antibody against FLAG tag cloned, in frame, to the 5′ end of the coding region of expressed cDNAs. The wild-type protein was localized mainly to the cell nucleus, where it formed discrete speckles (fig. 3A). The mutated polypeptide produced by the common Iranian Jewish mutation revealed a similar staining pattern (fig. 3C). The bulk of the mutated polypeptide carrying the disrupted first PHD finger was cytoplasmic, although some nonorganized nuclear staining was also observed. In the cytoplasm, the protein was visible as small granules, but some filamentous staining was also detected (fig. 3D). The truncated forms of APECED protein were constantly detected in the cytoplasm. The protein resulting from the most common Finnish mutation, which lacks both PHD fingers, mainly yielded a filamentous staining pattern (fig. 3B), whereas the protein lacking only the second PHD finger was observed as large granules in the cytoplasm (fig. 3E). The immunohistochemical-staining patterns obtained with the polyclonal APECED antiserum completely overlapped with the stainings obtained with the monoclonal anti-FLAG antibody (data not shown).
Figure 3.
Subcellular locations of the wild-type and mutated APECED polypeptides. The proteins were visualized by indirect immunofluorescent analysis of COS-1 cells transiently transfected with different AIRE expression constructs. The cells were fixed, permeabilized, and incubated with the polyclonal antibody raised against the APECED peptide. A, The wild-type APECED protein is localized mainly in the cell nucleus, where it forms distinct speckles. B, The protein carrying the common Iranian Jewish missense mutation was mainly transported to the nucleus, where it was observed as speckles similar to those formed by the wild-type protein. C, The bulk of the 256-amino-acid–long truncated APECED protein mimicking the common Finnish mutant was found in the cytoplasm, where the staining pattern was mainly filamentous. In addition, some large cytoplasmic granules were detected in a fraction of cells. No staining could be observed in the nucleus. D, A point mutation disrupting the first PHD finger of the APECED protein was mainly located in the cytoplasm. E, In a small fraction of the cells transfected with this mutant form, some nuclear staining could also be detected, but the mutant protein failed to form any distinct structures. F, The APECED protein lacking the second PHD finger, as a consequence of a single-nucleotide deletion, was detected, only in the cytoplasm, as small granules. A filamentous staining pattern was observed in very few transfected cells.
Transcriptional-Activation Properties of the APECED Protein
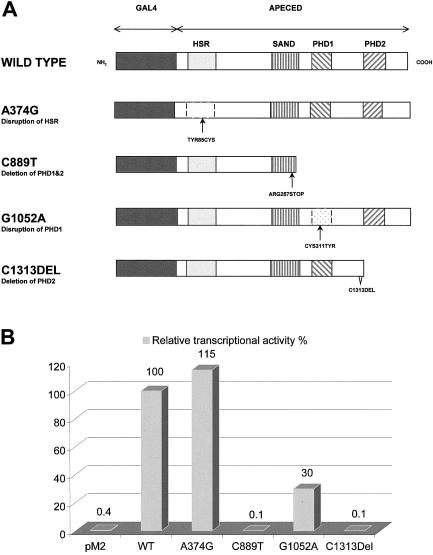
Several predicted domains of the APECED protein, together with its location in discrete speckles in the cell nucleus, suggested that this protein may play a role in the regulation of gene transcription. To test this hypothesis, we used a eukaryotic transactivation assay. cDNAs encoding for wild-type and for mutated APECED proteins were fused, in frame, to the yeast transcription factor Gal4 DNA-binding domain (DBD) in the pM2 expression vector (fig. 4A) and were cotransfected into COS-1 cells with luciferase reporter plasmids. Two reporter constructs with different promoters controlling luciferase transcription were used: (1) the herpes simplex virus thymidine kinase (tk) promoter with four Gal4 binding sites (pUAS4tk-LUC) and (2) a minimal promoter containing adenovirus E1b promoter TATA sequence with five upstream Gal-4 response elements (pG5LUC). The luciferase activity obtained for each fusion construct was compared with that produced by the pM2 vector expressing the Gal4 DBD domain alone.
Figure 4.
A, Schematic diagram of the wild-type and different mutant Gal4 DBD-APECED constructs. The dark gray–shaded boxes indicate the GAL4 DBD domains; the light gray–shaded boxes, the HSR domains; the striped boxes, the SAND domains; and the hatched boxes, PHD zinc fingers 1 and 2. B, Comparison of the transcription-activation function of the constructs shown in panel A. The wild-type APECED tethered to the GAL4 DBD strongly activates the pG5LUC reporter target, and this activation level is taken as 100% transcription activation. The mutation A374G, which is the major Iranian Jewish mutation, disrupts the HSR domain of the protein. This disruption has no major effect on the transcription-activation level, which is 115% of the wild-type activity. The Finnish major C889T mutation, which deletes both PHD zinc fingers, has no transactivation function. Similarly, the mutant protein carrying the C1313 deletion and, thus, lacking the second PHD zinc finger fails to cause any transcription activation. The G1052A substitution disrupts the first PHD finger, which in turn decreases the transactivation function of the protein to 30%.
The results of six independent transfection experiments revealed that the wild-type APECED protein stimulates the E1b promoter 250-fold and the UAS4tk promoter 100-fold. Surprisingly, the protein harboring the Iranian Jewish APECED mutation also activated these promoters 300-fold and 110-fold, respectively. Experiments with the mutant construct expressing a polypeptide with a mutation in the first PHD finger produced transcription levels that were 77-fold and 23-fold higher, respectively. The Gal4 DBD fusion proteins lacking one or both PHD fingers did not activate either of the tested promoters (fig. 4B). Interestingly, the wild-type AIRE in the pcDNA3.1(+) vector that produces APECED protein lacking the Gal4 DBD also slightly activated transcription from both of the promoters (∼15-fold). In control experiments with an empty pcDNA3.1(+) vector, no transcriptional activation was detected.
Discussion
Mutations in the AIRE Gene
Mutations in genes encoding transcriptional regulators have been described in a number of human disorders. A high proportion of these mutations lead to loss of function of the protein, and the disease is the result of haploinsufficiency of the gene product (Engelkamp and van Heyningen 1996). The AIRE gene encodes a novel putative transcription coactivator that is found to be mutated in an autoimmune disease APECED. Twenty-one disease-causing mutations in AIRE had previously been identified (The Finnish-German APECED Consortium 1997; Nagamine et al. 1997; Myhre et al. 1998; Pearce et al. 1998; Rosatelli et al. 1998; Scott et al. 1998; Wang et al. 1998; Heino et al. 1999b). This report describes a mutational analysis of an extended series of 112 affected individuals, among whom 16 different mutations (small insertions, deletions, or 1-bp substitutions), all occurring in the coding region of the gene, were identified. Eight of these mutations are novel, bringing the total number of the identified mutations causing APECED to 29. Each mutation is predicted to result either in a truncated form of the APECED protein or a change in the amino acid sequence. None of the mutations was found in homozygosity in any of the control samples. By sequencing the complete coding region and the exon-intron boundaries of the AIRE gene, we failed to identify any mutations in 6 of the 112 patients analyzed. However, the clinical findings and the linkage data, taken together, indicate that these patients do have APECED disease. In addition, in six patients, only one mutant allele of AIRE was found. Because our sequence analyses did not cover the promoter region or the intronic sequences, the missing mutations most probably are located in these sequences.
Eleven of the 16 APECED mutations observed in this study are predicted to result in a severely truncated, most likely nonfunctional, protein. Three different single-nucleotide substitutions in the N-terminal half of AIRE, which change an amino acid into a premature STOP codon, lead to a polypeptide without either of the predicted PHD fingers. In addition, a 13-bp insertion in exon 4 results in an even shorter polypeptide lacking the putative DNA-binding SAND domain (nt 687–960). Moreover, a single nt deletion, a 4-bp insertion, and a 13-bp deletion, all occurring in the region encoding the first PHD finger, also result in a protein without any intact PHD fingers. Four different one-nucleotide insertions or deletions occurring between the two PHD fingers are predicted to eliminate the second PHD finger.
In our patient series, we observed five missense mutations in the AIRE gene. These mutations are of particular interest, because they may reveal functionally critical regions of the APECED protein. Two of them, including the common Iranian Jewish mutation, occur in the amino-terminal conserved region of the gene encoded by the first two exons, and they show significant homology to the HSR domain of Sp100 protein (Sternsdorf et al. 1997; Gibson et al. 1998; Sternsdorf et al. 1999). The HSR domain found in the amino termini of the human and mouse Sp100 and Sp140 proteins, predicted to have a function in the regulation of gene expression, has recently been shown to function as a homodimerization domain and, also, to be responsible for the targeting of these proteins to the nuclear bodies (Sternsdorf et al. 1999). The clustering of the missense mutations in this domain, reported elsewhere (Pearce et al. 1998), affords further evidence for the functional significance of the motif in APECED protein.
The first PHD domain of the APECED protein harbors two different point mutations. The C→A transition at nt 1052 changes one of the proline residues in the PHD1 into glutamine and may have the potential to disrupt the structure of the zinc finger. The other point mutation changes the first conserved cysteine residue of PHD1 into tyrosine, which most likely prevents the coordination of the zinc atom. The binding of zinc ions has been shown to be critical for maintainance of the native three-dimensional structure of similar zinc fingers (Borden et al. 1995a, 1995b). The growing number of pathological conditions caused by mutations in PHD fingers emphasizes the true functional significance of this particular motif.
In our patients, two evident recurrent mutations, R257X and 1085–1097del, were together responsible for 67% of the disease alleles. The common Finnish mutation R257X, observed in 89% of the Finnish and in 33% of the non-Finnish affected chromosomes analyzed in this study, is also reported to be the common north Italian mutation (Scott et al. 1998). R257X was the second-most-common mutation among both the American and British patients with APECED. In those populations, the most common mutation was shown to be the 1085–1097del, which was responsible for 56.3% and 71% of the disease alleles, respectively (Pearce et al. 1998; Wang et al. 1998). The R257X mutation was observed on six different haplotype backgrounds, and the 1085–1097del was observed on five different haplotype backgrounds. These data, together with the population of origin of the patients with APECED, provide strong evidence for the independent origin of these mutations occurring in the mutation-sensitive CpG sequence.
Mutations That Alter the Subcellular Location of APECED Protein
The correct function of any protein depends on its proper subcellular location and its ability to maintain its macromolecular interactions. We and others have earlier shown that the wild-type APECED protein is mainly located in the nucleus, where it forms distinct speckled domains throughout the nucleoplasm, excluding the nucleoli (Björses et al. 1999; Heino et al. 1999a; Rinderle et al. 1999). To initiate the analysis of the molecular and cellular effects of the disease-causing mutations in AIRE, we have analyzed the subcellular distribution of selected mutated forms of the protein, when transiently expressed in mammalian cell lines. Two of the mutations studied—the major Finnish mutation and the C1313 deletion—were shown to produce truncated proteins of 28 kD and 47 kD, respectively, and were lacking either both or the second PHD zinc-finger domains. Despite the intact nuclear targeting sequence, these mutated APECED proteins were predominantly retained in the cytoplasm. We observed that the protein lacking both the PHD fingers mainly produces a cytoplasmic filamentous staining pattern, which was also occasionally seen with the wild-type protein. This would support the previous suggestion that the domain responsible for the vimentin or microtubule binding may be located in the N-terminal half of the protein (Rinderle et al. 1999). Nevertheless, the protein lacking only the second PHD finger (C1313del) failed to reveal any filamentous cytoplasmic staining and was observed only as large cytoplasmic aggregates. This may have been the result of an altered conformation that compromised the vimentin-binding properties of the protein. The mutated form of APECED protein, with the disrupted first PHD finger, was also detected mainly in the cytoplasm, where it produced small granules. In ∼10% of the cells transiently expressing this form of the protein, some nuclear staining was also observed, but no distinct structures were visualized. Both a nuclear and a cytoplasmic location for the 226- and 308-amino-acid–long N-terminal APECED fragments were also reported by Rinderle et al. (1999). They also found that the shorter truncated protein was associated with vimentin or microtubuli and that the longer one formed aggregates in the cytoplasm.
So far, the mechanism regulating the distribution of APECED protein between the cytoplasm and the nucleus is not known. Our results imply that the PHD zinc finger domains, possibly together with the nuclear targeting signal, may play an important role in determining the subcellular distribution of the protein and its association with specific structures in the nucleus. Similar findings defining the involvement of zinc-finger domains in the cellular location of proteins have previously been reported. For example, mutations in the RING-finger domain of the ret finger protein (rfp) result in rfp remaining in the cytoplasm of the transfected HeLa cells, whereas the wild-type protein is localized in the nucleus (Cao et al. 1997). The data presented here further suggest that the pathological consequences of the mutations disturbing the PHD domains may exert their effects by altering the correct intracellular targeting of the APECED protein. That the premature termination codons created by the nonsense mutations subject the APECED mRNAs to rapid degradation via the nonsense-mediated mRNA decay (Hentze and Kulozik 1999) is another alternative that cannot be tested by in vitro expression studies. To find evidence for this mechanism, transcripts or polypeptides should be studied in appropriate patient samples. This approach is, however, complicated by the restricted tissue expression of the AIRE gene.
Activation, by APECED Protein, of In Vitro Transcription
Both the nuclear localization and the structural features of the APECED protein suggest that it has a function in the regulation of transcription. We explored the transactivation potential of this protein, by means of cotransfection experiments with Gal4 DBD-APECED fusion protein and the luciferase reporter genes under either the tk promoter or the adenovirus E1b TATA sequence with upstream Gal4 response elements. Our results demonstrate that the APECED protein is indeed capable of activating the transcription, since GAL4 DBD-APECED fusion protein strongly stimulated both reporter genes, providing, for the first time, experimental evidence for the APECED protein being a transcriptional regulator. Moreover, the truncated proteins encoded by the cDNAs carrying APECED mutations, which in native form were inefficiently transported into the nucleus, showed no activity in the Gal4 DBD-based transactivation assay. Interestingly, a protein with a disrupted first PHD finger but an intact second PHD finger showed about one-third of the wild-type activity. Since a small proportion of the native mutant protein was found in the nuclei of the transfected cells, it is possible that this mutant polypeptide is not totally inactive.
The Iranian Jewish APECED Mutation
APECED is enriched in three isolated populations: the Finnish, the Sardinian, and the Iranian Jewish. In each of these populations, the disease is associated with one predominant haplotype, indicating a founder effect. The common mutations in Finland and in Sardinia have been described elsewhere (The Finnish-German APECED Consortium 1997; Nagamine et al. 1997; Rosatelli et al. 1998). They are both null mutations leading to severe truncation of the polypeptide. The coding region of the AIRE gene of the Iranian Jewish patients was sequenced in this work. The only alteration found in this sequence was a single-nucleotide substitution, A374G, that exchanges a tyrosine residue for a cysteine in the C-terminal part of the HSR domain. This substitution was detected as homozygosity in all Iranian Jewish patients and as heterozygosity in 1 of the 35 healthy Iranian Jewish controls, thereby implying a high carrier frequency of the major mutation in this population. It was not detected in any of the European samples. Somewhat surprisingly, in the transactivation assay done here, the Iranian Jewish mutant protein stimulated both of the promoters that were tested as effectively as did the wild-type APECED protein. The HSR domain has been shown to be a homodimerization domain of several Sp100-related proteins. The dimerization of the APECED protein may be essential for DNA-binding or for binding a currently unidentified partner, and mutations in this domain could disrupt these interactions. The transactivation assay used in this study may fail to detect the effects of the mutations on specific mechanisms of this kind. The defective biological activity resulting in APECED in Iranian Jewish patients may also be the result of rapid intracellular degradation of the mutated polypeptide. The disease-causing nature of the Y85C substitution needs to be verified by further studies. However, the clustering of three different point mutations in the region coding for six amino acids in the C-terminus of the putative dimerization domain supports the hypothesis that these sequence alterations would cause APECED disease.
Implications for Genotype-Phenotype Correlations
APECED is a disease with considerable phenotypic variation. The differences observed in the clinical components of the Finnish patients with APECED who were known to be homozygous for the common Finnish mutation and even the differences in symptoms between siblings with APECED indicate that the allelic variation in the AIRE gene cannot be the major determinant of the clinical picture of the disease. The genetic background of the individual and the environmental factors most probably play important roles in the selection of target organs for tissue destruction. However, distinct differences in the phenotype can be observed between patients from two populations showing enrichment of APECED disease. In contrast to the Finnish patients with APECED, the Iranian Jewish patients rarely have chronic mucocutaneous candidiasis, and the ectodermal dystrophies are much less common in patients in this group (Zlotogora and Shapiro 1992). Furthermore, hypoparathyroidism often is the only organ-specific autoimmune symptom. This seems to be in accord with our finding that the major Iranian Jewish mutation is a missense mutation, whereas the major Finnish mutation presumably leads to a totally nonfunctional truncated protein. The Iranian Jewish mutation did not seem to have any effect on either the subcellular location or the transactivation properties assayed in this study. Even if the bulk of the mutant polypeptide rapidly degraded, the remaining fraction of the protein may contain some biological activity. Whether this is also the main cause of the milder clinical symptoms needs to be verified. We also recognize the possibility that there may be additional nucleotide changes in the noncoding regions of AIRE that may influence the Iranian Jewish phenotype.
Conclusion
Results of previous studies have shown that mutations in the AIRE gene are responsible for the development of APECED. In this study, we have identified eight novel mutations in the AIRE gene in patients of different ethnic backgrounds who have APECED. On the basis of its structural characteristics and subcellular location, the APECED protein is suggested to be a transcriptional regulator. For the first time, we have provided experimental evidence that APECED protein acts as a powerful transcriptional coactivator. For most of the APECED mutants analyzed in the present study, the intracellular location was altered, and the stimulating effect on transcription, if any, was reduced. Comparison of the functional properties of the mutant APECED proteins carrying the Finnish and Iranian Jewish common mutations suggests that they may partially explain differences in the clinical phenotype of these patients. Identification of the target genes regulated by the APECED protein and the molecular interactions essential for its regulatory activity remains a major challenge and is required for definition of the mechanisms that trigger the destruction of specific tissues characteristic for APECED. The information on the interacting partners and target genes for the APECED protein, supported by systematic functional analysis of its domains and different mutations, will extend our understanding of the role of APECED protein in development of a normal immune response. Furthermore, it will broaden our knowledge of the pathogenesis of autoimmune diseases in general.
Acknowledgments
We are grateful to all patients with APECED and their families, for collaboration and for donation of DNA samples. T. Airaksinen, A. Vikman, L. Puhakka, and P. Sillanpää are appreciated, for excellent technical assistance. Drs. A. Jalanko and A.-C. Syvänen gave expert methodological advice, and Juha Korhonen prepared the GST-fusion protein construct. Dr. J. Seeler is thanked for valuable discussion. Drs. J. Zlotogora, G. Ben-Zion, G. Chiumello, N. Dahl, P. Heideman, J. J. G. Hoorweg-Nijman, L. Mathivon, P. E. Mullis, M. Pohl, M. Ritzen, G. Romeo, M. S. Shapiro, C. S. Smith, and J. Soloym kindly provided the non-Finnish patient samples. We thank Drs. T. Pearlmann and I. Sadowski, for the plasmids for the transactivation assay. This study was financially supported by grants from the Academy of Finland, the Ulla Hjelt Fond of the Foundation for Pediatric Research, and the Emil Aaltonen Foundation, and by resources from the Helsinki Biomedical Graduate School M.D./Ph.D. program.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CEPH, http://www.cephb.fr/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for AIRE cDNA [accession number Z97990] and the AIRE genomic sequence [accession number AJ009610])
- European Molecular Biology Laboratory Database, http://www.ebi.ac.uk/embl/index.html (for the AIRE genomic sequence [accession number AJ009610])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for APECED [MIM 240300])
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20:56–59 [DOI] [PubMed]
- Ahonen P, Myllarniemi S, Sipila I, Perheentupa J (1990) Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med 322:1829–1836 [DOI] [PubMed]
- Björses P, Aaltonen J, Horelli-Kuitunen N, Yaspo ML, Peltonen L (1998) Gene defect behind APECED: a new clue to autoimmunity. Hum Mol Genet 7:1547–1553 [DOI] [PubMed]
- Björses P, Aaltonen J, Vikman A, Perheentupa J, Ben-Zion G, Chiumello G, Dahl N, et al (1996) Genetic homogeneity of autoimmune polyglandular disease type I. Am J Hum Genet 59:879–886 [PMC free article] [PubMed]
- Björses P, Pelto-Huikko M, Kaukonen J, Aaltonen J, Peltonen L, Ulmanen I (1999) Localization of the APECED protein in distinct nuclear structures. Hum Mol Genet 8:259–266 [DOI] [PubMed]
- Blechschmidt K, Schweiger M, Wertz K, Poulson R, Christensen HM, Rosenthal A, Lehrach H, et al (1999) The mouse Aire gene: comparative genomic sequencing, gene organization, and expression. Genome Res 9:158–166 [PMC free article] [PubMed]
- Borden KL, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, et al (1995a) The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J 14:1532–1541 [DOI] [PMC free article] [PubMed]
- Borden KL, Lally JM, Martin SR, O'Reilly NJ, Etkin LD, Freemont PS (1995b) Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J 14:5947–5956 [DOI] [PMC free article] [PubMed]
- Cao T, Borden KL, Freemont PS, Etkin LD (1997) Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J Cell Sci 110:1563–1571 [DOI] [PubMed]
- Engelkamp D, van Heyningen V (1996) Transcription factors in disease. Curr Opin Genet Dev 6:334–342 [DOI] [PubMed]
- Finnish-German APECED Consortium, The (1997) An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains: autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Nat Genet 17:399–403 [DOI] [PubMed]
- Gibson TJ, Ramu C, Gemund C, Aasland R (1998) The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem Sci 23:242–244 [DOI] [PubMed]
- Heery DM, Kalkhoven E, Hoare S, Parker MG (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed]
- Heino M, Peterson P, Kudoh J, Nagamine K, Lagerstedt A, Ovod V, Ranki A, et al (1999a) Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun 257:821–825 [DOI] [PubMed]
- Heino M, Scott HS, Chen Q, Peterson P, Maebpaa U, Papasavvas MP, Mittaz L, et al (1999b) Mutation analyses of North American APS-1 patients. Hum Mutat 13:69–74 [DOI] [PubMed]
- Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310 [DOI] [PubMed]
- Mittaz L, Rossier C, Heino M, Peterson P, Krohn KJ, Gos A, Morris MA, et al (1999) Isolation and characterization of the mouse Aire gene. Biochem Biophys Res Commun 255:483–490 [DOI] [PubMed]
- Moilanen A-M, Karvonen U, Poukka H, Yan W, Toppari J, Jänne O, Palvimo JJ (1999) A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J Biol Chem 274:3700–3704 [DOI] [PubMed]
- Myhre AG, Björses P, Dalen A, Husebye ES (1998) Three sisters with Addison's disease. J Clin Endocrinol Metab 83:4204–4206 [DOI] [PubMed]
- Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, et al (1997) Positional cloning of the APECED gene. Nat Genet 17:393–398 [DOI] [PubMed]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770 [DOI] [PMC free article] [PubMed]
- Pearce SH, Cheetham T, Imrie H, Vaidya B, Barnes ND, Bilous RW, Carr D, et al (1998) A common and recurrent 13-bp deletion in the autoimmune regulator gene in British kindreds with autoimmune polyendocrinopathy type 1. Am J Hum Genet 63:1675–1684 [DOI] [PMC free article] [PubMed]
- Rinderle C, Christensen HM, Schweiger S, Lehrach H, Yaspo ML (1999) AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered sub-cellular distribution of mutants lacking the PHD zinc fingers. Hum Mol Genet 8:277–290 [DOI] [PubMed]
- Rosatelli MC, Meloni A, Meloni A, Devoto M, Cao A, Scott HS, Peterson P, et al (1998) A common mutation in Sardinian autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients. Hum Genet 103:428–434 [DOI] [PubMed]
- Scott HS, Heino M, Peterson P, Mittaz L, Lalioti MD, Betterle C, Cohen A, et al (1998) Common mutations in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients of different origins. Mol Endocrinol 12:1112–1119 [DOI] [PubMed]
- Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A (1998) Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA 95:7316–7321 [DOI] [PMC free article] [PubMed]
- Silver PA, Keegan LP, Ptashine M (1984) Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci USA 81:5951–5955 [DOI] [PMC free article] [PubMed]
- Stacey A, Schnieke A (1990) SVpoly: a versatile mammalian expression vector. Nucleic Acids Res 18:2829 [DOI] [PMC free article] [PubMed]
- Sternsdorf T, Grotzinger T, Jensen K, Will H (1997) Nuclear dots: actors on many stages. Immunobiology 198:307–331 [DOI] [PubMed]
- Sternsdorf T, Jensen K, Reich B, Will H (1999) The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J Biol Chem 274:12555–12566 [DOI] [PubMed]
- Syvanen AC, Aalto-Setala K, Kontula K, Soderlund H (1989) Direct sequencing of affinity-captured amplified human DNA application to the detection of apolipoprotein E polymorphism. FEBS Lett 258:71–74 [DOI] [PubMed]
- Syvanen AC, Sajantila A, Lukka M (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am J Hum Genet 52:46–59 [PMC free article] [PubMed]
- Vandenplas S, Wiid I, Grobler-Rabie A, Brebner K, Ricketts M, Wallis G, Bester A, et al (1984) Blot hybridisation analysis of genomic DNA. J Med Genet 21:164–172 [DOI] [PMC free article] [PubMed]
- Wang CY, Davoodi-Semiromi A, Huang W, Connor E, Shi JD, She JX (1998) Characterization of mutations in patients with autoimmune polyglandular syndrome type 1 (APS1). Hum Genet 103:681–685 [DOI] [PubMed]
- Wang CY, Shi JD, Davoodi-Semiromi A, She JX (1999) Cloning of Aire, the mouse homologue of the autoimmune regulator (AIRE) gene responsible for autoimmune polyglandular syndrome type 1 (APS1). Genomics 55:322–326 [DOI] [PubMed]
- Werle E, Schneider C, Renner M, Volker M, Fiehn W (1994) Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res 22:4354–4355 [DOI] [PMC free article] [PubMed]
- Zakeri H, Amparo G, Chen SM, Spurgeon S, Kwok PY (1998) Peak height pattern in dichloro-rhodamine and energy transfer dye terminator sequencing. Biotechniques 25:406–410, 412–414 [DOI] [PubMed]
- Zlotogora J, Shapiro MS (1992) Polyglandular autoimmune syndrome type I among Iranian Jews. J Med Genet 29:824–826 [DOI] [PMC free article] [PubMed]