Summary
Previous studies have shown that specific short-tandem-repeat (STR) and single-nucleotide-polymorphism (SNP)–based haplotypes within and among unaffected and fragile X white populations are found to be associated with specific CGG-repeat patterns. It has been hypothesized that these associations result from different mutational mechanisms, possibly influenced by the CGG structure and/or cis-acting factors. Alternatively, haplotype associations may result from the long mutational history of increasing instability. To understand the basis of the mutational process, we examined the CGG-repeat size, three flanking STR markers (DXS548-FRAXAC1-FRAXAC2), and one SNP (ATL1) spanning 150 kb around the CGG repeat in unaffected (n=637) and fragile X (n=63) African American populations and compared them with unaffected (n=721) and fragile X (n=102) white populations. Several important differences were found between the two ethnic groups. First, in contrast to that seen in the white population, no associations were observed among the African American intermediate or “predisposed” alleles (41–60 repeats). Second, two previously undescribed haplotypes accounted for the majority of the African American fragile X population. Third, a putative “protective” haplotype was not found among African Americans, whereas it was found among whites. Fourth, in contrast to that seen in whites, the SNP ATL1 was in linkage equilibrium among African Americans, and it did not add new information to the STR haplotypes. These data indicate that the STR- and SNP-based haplotype associations identified in whites probably reflect the mutational history of the expansion, rather than a mutational mechanism or pathway.
Introduction
The fragile X syndrome (MIM 309550) is an X-linked mental-retardation disorder caused by the expansion of a CGG trinucleotide repeat located in the 5′ UTR of the fragile X mental-retardation-1 gene (FMR-1) (Fu et al. 1991; Verkerk et al. 1991) The frequency of the syndrome is estimated to be about 1 in 4,000 males for a white general population (Turner et al. 1996), and there is evidence that it is at least as prevalent in the African American general population (Schwartz et al. 1988a; Crawford et al. 1999).
Among most individuals in the general population, the polymorphic CGG repeat ranges from 6 to 60 repeats and is usually interspersed every 9–10 repeats with an AGG (Fu et al. 1991; Yu et al. 1992; Snow et al. 1993; Kunst and Warren 1994; Eichler et al. 1994; Snow et al. 1994; Hirst et al. 1994; Zhong et al. 1995b). These alleles tend to be inherited in a stable manner from parent to offspring. It is not until the allele is of a larger repeat size (61–199 repeats, termed “premutation”) that the repeat becomes unstable and confers risk of expanding in the next generation. Once unstable, the probability of expanding to the full mutation (>200 repeats) in the next generation, resulting in an offspring with the fragile X syndrome, is positively correlated with maternal repeat size (Fu et al. 1991; Yu et al. 1992; Heitz et al. 1992; Snow et al. 1993; Nolin et al. 1996; Sherman et al. 1996; Ashley-Koch et al. 1998).
On expansion to the full mutation, the CpG island upstream of FMR-1 becomes hypermethylated and histone deacetylated, repressing the transcription of FMR-1 (Verkerk et al. 1991; Fu et al. 1991; Oberle et al. 1991; Sutcliffe et al. 1992; Coffee et al. 1999). Therefore, it is the lack of the fragile X mental retardation protein (FMRP), an RNA-binding protein, that gives rise to the fragile X–syndrome phenotype.
To date, little is known about the initial event(s) that causes a stable allele to begin the expansion process. Several lines of evidence suggest that one event is the conversion of the most 3′ AGG to a CGG (Eichler et al. 1994; Kunst and Warren 1994) First, pedigree analyses show that alleles in the general population with >34–37 pure CGG repeats at the 3′ end of the repeat tend to be inherited in an unstable manner (Eichler et al. 1994). Second, Kunst and Warren (1994) observed normal alleles with >24 repeats at the 3′ end of the repeat shared the same haplotype background with many of the fragile X chromosomes, suggesting a susceptibility for instability. Finally, it has been observed that most premutation alleles have either one AGG interruption, at the 5′ end of the repeat, or none at all (Eichler et al. 1994; Snow et al. 1994; Zhong et al. 1995b).
In addition to the purity of the 3′ end of the CGG repeat, results from haplotype studies of CGG flanking markers suggested possible cis-acting factors that influence CGG-repeat stability (Eichler et al. 1996). For example, among chromosomes from whites, specific short-tandem-repeat (STR) marker-based haplotypes—including DXS548, FRAXAC1, and FRAXAC2—were associated with the full mutation (for a review, see Chiurazzi et al. 1996c). Furthermore, Eichler et al. (1996) found that specific STR-based haplotypes in unaffected white chromosomes were associated with specific CGG/AGG structures. In particular, it was shown that the 2-1-3 haplotype was associated with long, interrupted CGG repeats that slowly progressed to the premutation state in a stepwise manner, whereas the 6-4-4 and 6-4-5 haplotypes were associated with the “asymmetrical” repeat patterns (i.e., 9+12+9; see fig. 1 for nomenclature) that rapidly expanded to the premutation state. It has been hypothesized that these associations result from different mutational mechanisms, possibly influenced by the CGG structure and/or cis-acting factors (Eichler et al. 1996).
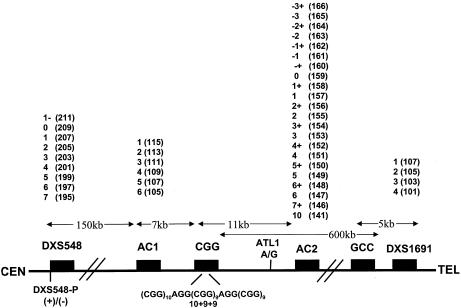
Figure 1.
Location and nomenclature of FRAXA interspersion patterns and STR- and SNP-based haplotypes. This figure was adapted from Eichler et al. (1996). We show here the position of the STR and SNP markers relative to the FRAXA CGG repeat. We also included the FRAXE GCC repeat, the STR marker DXS1691, and the recently described polymorphism within DXS548 (DXS548-P). The numbers above the marker represent the allele assignment and the numbers in parentheses represent the base-pair size, according to our protocol. The numbering systems and interspersion-pattern nomenclature are from Eichler et al. (1996) and Murray et al. (1996). We have extended the numbering system for DXS548 and FRAXAC2 to include the alleles not previously described in a white population. All STR-based haplotypes are constructed as follows: 5′-DXS548-P (if there is an insertion or deletion event)-DXS548-FRAXAC1-FRAXAC2-3′. Interspersion patterns are described as the number of consecutive CGGs followed by a plus sign (+) to designate the presence of an AGG interruption.
Further studies that used the novel single nucleotide polymorphism (SNP) ATL1, which is located about 5.6 kb distal to the CGG repeat, have also shown marked linkage disequilibrium in both the unaffected and fragile X white populations (Gunter et al. 1998). When ATL1 was combined with CGG/AGG data, strong associations between ATL1 alleles and the pattern of the 5′ end of the repeat were revealed: the ATL1 allele most frequent in the white fragile X population was associated with alleles containing an AGG in the tenth position (9+n) of the repeat, whereas the ATL1 allele most frequent in the unaffected population was associated with alleles containing an AGG in the eleventh position (10+n) of the repeat (Gunter et al. 1998). Thus, the findings of Gunter et al. (1998) suggested that the 5′ position of the AGG, independent of the length of pure 3′ repeats, might influence CGG-repeat stability.
Alternatively, these STR- and SNP-based haplotype associations may be specific to the white population and may be the result of low-frequency, recurrent mutations that drift through the population with selection occurring only after ⩾15 generations when they result in the fragile X syndrome. Because this interpretation remains a possibility, it is important to examine these STR- and SNP-based haplotypes in other ethnic populations. Most studies in nonwhite populations have not had the opportunity to examine haplotype associations in both unaffected and fragile X populations, and some are based on small sample sizes (Richards et al. 1994; Zhong et al. 1994; Chiurazzi et al. 1996a; Eichler and Nelson 1996; Kunst et al. 1996) Also, the STR markers used to examine the various populations were often different, making comparisons across studies and across populations difficult. Therefore, to better understand the implications of the associations identified in white populations, we have examined the overall CGG-repeat size (FRAXA), three flanking STR markers (DXS548, FRAXAC1, and FRAXAC2), and one SNP (ATL1) spanning 150 kb around the CGG repeat in large unaffected, fragile X, white, and African American populations.
Comparison of these populations revealed many important differences. First, the expected heterozygosity and number of alleles was higher for the STR loci in the unaffected African Americans when compared with the unaffected white population, a finding that was expected; however, they were lower for the FRAXA locus. Second, whereas one specific haplotype was previously associated with intermediate alleles (41–60 repeats) among whites (for review, see Chiurazzi et al. 1996c), among African American intermediate alleles no such associations were observed. Third, in fragile X populations, we observed a striking difference between the two ethnic groups: several haplotypes were significantly associated among white fragile X chromosomes, whereas only two unique haplotypes predominated among African Americans. Fourth, a putative “protective” haplotype found among whites was found to present with equal frequency among normal and fragile X African Americans. Finally, the possible risk factor based on the position of the 5′ AGG was not indicated in the African American population. These data indicate that the STR- and SNP-based haplotype associations most likely reflect the mutational and population history of the CGG-repeat expansion, rather than identifying susceptible haplotypes involved in the mechanism of instability.
Subjects and Methods
Study Population
The majority of white fragile X chromosomes were ascertained from Georgia (n=27) and from South Carolina (n=75). The majority of the African American fragile X chromosomes were ascertained from South Carolina (n=37). The remaining 25 chromosomes were drawn from Georgia (n=5), New York (n=2), Mississippi (n=1), Alabama (n=1), Texas (n=1), Oklahoma (n=1), Michigan (n=4), North Carolina (n=7), Virginia (n=1), and Maryland (n=2). An additional African American fragile X chromosome from Michigan (GM05282A) was obtained through Coriell Cell Repository as a fibroblast cell line.
The unaffected white (n=721) and African American (n=578) study populations consisted of males aged 7–10 years in special-education-needs classes from metropolitan Atlanta. Ascertainment of this population was described elsewhere (Meadows et al. 1996; Crawford et al. 1999). Because a large number of African American males with the fragile X syndrome were ascertained from South Carolina, we also studied a random set of unaffected African Americans from that same population (n=59). Neither the CGG-repeat allele distribution nor the STR-haplotype distribution differed between the two unaffected African American populations (data not shown); therefore, both populations were combined for all subsequent analyses (n=637). STR-based and SNP-based haplotype data were previously reported (Gunter et al. 1998) on a subset of the unaffected and fragile X white population described in this study.
Laboratory Methods
FRAXA CGG-repeat sizes and FRAXE GCC-repeat sizes were determined by a fluorescent sequencer method, as described elsewhere (Meadows et al. 1996). The STR markers DXS548, FRAXAC1, FRAXAC2, and DXS1691 were multiplexed, and allele sizes were determined by use of a fluorescent sequencer method, also described elsewhere (Murray et al. 1996). Allele assignments for DXS548, FRAXAC1, and FRAXAC2 were based on the numbering of Eichler et al. (1996). However, the list of allele assignments for DXS548 and FRAXAC2 was extended in this study, because of the number of unique alleles found in this African American population. Allele assignments for DXS1691 were based on the numbering of Murray et al. (1996).
In addition to the STR markers, the recently described polymorphism within the primer set of DXS548 was also examined in this study (referred to here as DXS548-P). This polymorphism is an insertion/deletion polymorphism in the (C)11 tract of a (C)4G(C)11 sequence located ∼50 bp proximal to the CA repeat. A forward primer was designed (Chiurazzi et al. 1996a, 1996b) in conjunction with the DXS548 reverse primer that amplified the CA-repeat sequence only. Comparison of the data generated by this primer set with the data generated by the original DXS548 primer set determined the allele status of the insertion/deletion polymorphism. Because insertions and deletions within the primer set of DXS548 could be determined, allele assignments were not based on the previously published numbering system (Chiurazzi et al. 1996a, 1996b). Instead, insertions of one base pair were designated (+), and deletions of one base pair were designated (−) and were placed before the allele representing DXS548 for the complete haplotype. If there was no insertion or deletion in the (C)11 sequence, nothing preceded the allele for DXS548 for the complete haplotype. The allele status at ATL1 was determined as described elsewhere (Gunter et al. 1998).
For a few African American alleles of interest, the AGG interspersion pattern was determined by sequencing the CGG-repeat array. In brief, the CGG repeat and surrounding DNA sequences were amplified from genomic DNA by Pfu polymerase (Stratagene) from the PCR protocol described elsewhere (Chong et al. 1994). The PCR products were then run on a 1.5% agarose gel at 80 volts for 1 h to check for amplification of a single allele. The PCR products were concentrated for sequencing by the Microcon YM-100 centrifugal device (Millipore). The concentrated PCR products were then subjected to sequencing by the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA Polymerase, FS. Because of the high G/C content of the template, 1 μl of dimethyl sulfoxide was added to the sequencing reaction. The primer used in the sequencing reactions was 5′-GACGGAGGCGCCGCTGCCAGG (Brown et al. 1993). The cycling conditions and subsequent purification of the sequencing products were performed as recommended by the manufacturer. All sequencing reactions were run on the ABI 373 Stretch and analyzed by ABI DNA sequencing software.
Statistical Methods
A χ2 test of independence was performed (by the software StatXact, Cytel) to compare FRAXA CGG-repeat allele distributions and STR allele distributions between ethnic groups. When the expected frequencies were <5 for any cell in any given table, associations between specific STR haplotypes and FRAXA CGG-repeat alleles were determined by the Fisher-Freeman-Halton exact test with the Monte Carlo method of repeated sampling (StatXact, Cytel). To correct for multiple testing, the P value for significance was adjusted for each population on the basis of the number of tests performed for that population. Because each comparison yielded a different number of tests performed, the significant P values and the number of tests performed are given in each table. The expected heterozygosities for the STR markers were calculated with a method described elsewhere (Nei 1978).
Results
STR Loci in Whites and African Americans
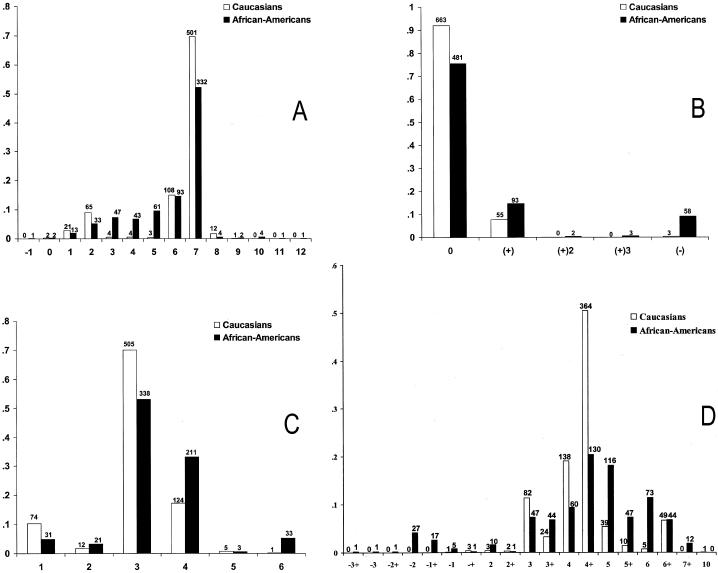
A large population of unaffected African American males from the Southeastern United States (n=637), as well as a large population of unaffected white males (n=721) were genotyped for the FRAXA CGG-repeat allele and three STR loci that surround the CGG repeat of FMR-1 (DXS548-FRAXAC1-FRAXAC2) (fig. 1). In general, the African American population at all three STR loci had a greater number of distinct alleles, a higher expected heterozygosity, and a higher variance when compared with the white population (table 1). Furthermore, the allele distributions for all three STR loci were significantly different between the two populations, as suggested by Schwartz et al. (1988b) (figs. 2A, 2C, 2D) (Fisher's exact test, P<.001), although the most frequent alleles for each of these loci in both populations are identical (alleles 7, 3, and 4+, respectively). For FRAXAC2 and DXS548, several unique alleles were identified in the African American population that were not found in this white population. Also, for FRAXAC2, there was 1 allele (allele 10) found in the white population not found in the African American population (fig. 2D).
Table 1.
FRAXA and STR Loci among Unaffected White and African American Populations
|
Locus |
||||||||
| FRAXA |
DXS548 |
FRAXAC1 |
FRAXAC2 |
|||||
| Unaffected Population | No. of Alleles | Expected Heterozygosity (Variance) | No. of Alleles | Expected Heterozygosity (Variance) | No. of Alleles | Expected Heterozygosity (Variance) | No. of Alleles | Expected Heterozygosity (Variance) |
| Whitesa | 42 | .838±.014 (44.106) | 10 | .485±.019 (3.131) | 6 | .469±.019 (.639) | 13 | .687±.017 (.856) |
| African Americansb | 34 | .808±.016 (17.863) | 14 | .685±.018 (3.355) | 6 | .603±.019 (.912) | 18 | .880±.013 (4.419) |
n=721.
n=637.
Figure 2.
A, DXS548; B, DXS548-P; C, FRAXAC1; and D, FRAXAC2 allele distributions in whites (n=721; unshaded bars) and African Americans (n=637; shaded bars). Absolute numbers are reported above each bar for each allele of each locus in both populations.
As observed for the other three STR loci, the African American population had a greater number of distinct alleles for DXS548-P (fig. 2B). Both insertion events (+) and deletion events (−) were more prevalent in the African American population when compared with the white population. For both populations, insertion events were more frequent than deletion events. Insertion events of three (3+) and of two (2+) were observed only in the African American population. All three (3+) alleles were observed on DXS548 allele 5, and the (2+) alleles were observed on DXS548 alleles 6 and 7.
In contrast to the results of the STR loci, the FRAXA locus based on overall repeat size did not reflect the increased diversity in the African American population when compared with that in the white population (table 1). Like the STR loci, the distributions between the white and African American populations differed at the FRAXA locus (P<.001). This difference was primarily due to the lack of small alleles (20 and 23 repeats) and intermediate alleles (41–60 repeats) in the African American population, as reported elsewhere (Crawford et al. 1999).
Repeat-Size Correlations among the STR Markers and FRAXA
A positive correlation between the number of repeats at FRAXA, FRAXAC1, and DXS548 has been reported among whites (Zhong et al. 1995a; Brown et al. 1996; Arrieta et al. 1999). Although there was no significant positive correlation between the number of repeats at FRAXA and that at FRAXE, a locus about 600 kb distal to FRAXA, significantly larger FRAXE alleles were found to segregate with FRAXA full-mutation alleles when compared with alleles from a control population (Brown et al. 1997). Furthermore, instabilities at the FRAXA or FRAXE locus were sometimes accompanied by instability at an independent locus (Murray et al. 1997). These findings suggested a trans-acting mechanism of instability that globally affected small repeats. An alternative explanation for this phenomenon could be that in the white population, the larger FRAXA alleles evolved on haplotype backgrounds with the larger CA-repeat sizes of FRAXAC1 and DXS548 (i.e., the 2-1-3 haplotype background) by chance. If this explanation is correct, no correlation among the STR loci and the FRAXA locus would be expected among other ethnic groups.
Our white population showed the same positive correlation between the FRAXA CGG repeat and the CA repeats of FRAXAC1, DXS548, and FRAXAC2, as previously reported (Pearson correlation r=.260, r=.286, r=.093, respectively; P<.05). In contrast, the African American population showed no correlation with nearby FRAXAC1 and FRAXAC2, although there was a significant correlation with DXS548 (Pearson correlation r=.080; P<.05). Neither the white population nor the African American population showed a significant positive correlation between FRAXA (CGG)n and FRAXE (GCC)n. Taking the data on all flanking markers together, the resulting differences among populations do not support a trans-acting mechanism of instability that globally affected small-repeat loci.
FRAXA and STR-Based Haplotype Associations
White population
The loci DXS548, FRAXAC1, and FRAXAC2 were used to create the 5′→3′ STR-based haplotypes. Both the white and African American populations were then tested for associations between FRAXA CGG-repeat size and STR-based haplotypes (see “Statistical Methods”). In the white population, 16 out of a total of 71 observed haplotypes were significantly associated with a specific FRAXA CGG-repeat size (table 2). As shown previously, the most common white haplotype, 7-3-4+, was positively associated with the most common allele of 30 CGG repeats and was negatively associated with the 29-repeat allele. Also, the 6-4-4 and 2-1-3 haplotypes found frequently in the white fragile X population were associated with different CGG-repeat sizes in an unaffected population: the 6-4-4 haplotype was associated with a repeat size of 29, and the 2-1-3 haplotype was associated with several intermediate repeat sizes (41, 42, and 43 repeats).
Table 2.
FRAXA and STR-Haplotype Associations in an Unaffected White Population[Note]
| FRAXA CGG-Repeat Size | STR Haplotype | Frequency |
| Positive association: | ||
| 20 | 6-3-4+ | .022 |
| 23 | 2-3-4 | .015 |
| 7-3-4 | .028 | |
| 29 | 1-1-3 | .015 |
| 7-3-4 | .028 | |
| (+)7-4-6+ | .026 | |
| 30 | 7-3-3+ | .025 |
| 7-3-4+ | .216 | |
| 31 | 7-3-4+ | .046 |
| 32 | 6-4-4 | .015 |
| 41 | 2-1-3 | .007 |
| 42 | 2-1-3 | .004 |
| 43 | 2-1-3 | .008 |
| Negative association: | ||
| 29 | 7-3-4+ | .008 |
| 32 | 7-3-4+ | .006 |
| 33 | 1-1-3 | .006 |
Note.—Significance level declared at P<.0002, given 229 tests of association; n=721.
Among the white fragile X chromosomes, the haplotypes 2-1-3, 6-4-4, and 6-4-5 were the three most common of 25 observed haplotypes, representing about 1/3 of the chromosomes (table 3). When compared with the unaffected white population, the haplotypes 2-1-3 and 6-4-5 were positively associated with the full mutation, whereas the haplotype 7-3-4+ was significantly underrepresented in the fragile X population. These findings are similar to other, smaller surveys (Eichler et al. 1996). We also found the same diversity of haplotypes, as noted elsewhere (Morton and Macpherson 1992), including 11 haplotypes that were rare in the fragile X population, 8 of which were not found in the unaffected white population (table 3).
Table 3.
White Fragile X STR-Haplotype Associations[Note]
|
Chromosomes |
||||
| Fragile X |
Unaffected |
|||
| STR Haplotype | Number | Frequency | Number | Frequency |
| 2-1-3a | 26 | .255 | 36 | .050 |
| 6-4-5a | 16 | .157 | 25 | .035 |
| 7-3-4+b | 10 | .098 | 284 | .394 |
| 6-4-4 | 8 | .078 | 24 | .033 |
| 7-1-3 | 6 | .059 | 7 | .001 |
| 6-4-6+ | 5 | .049 | 6 | .008 |
| 7-3-4 | 5 | .049 | 82 | .114 |
| 2-3-4 | 3 | .029 | 16 | .022 |
| 6-3-4 | 3 | .029 | 6 | .008 |
| 6-3-4+ | 3 | .029 | 34 | .047 |
| 7-2-4 | 3 | .029 | 2 | .003 |
| 2-4-5 | 1 | .010 | 0 | 0 |
| 3-3-3 | 1 | .010 | 0 | 0 |
| 3-4-5 | 1 | .010 | 0 | 0 |
| 5-1-5 | 1 | .010 | 0 | 0 |
| 5-4-3 | 1 | .010 | 0 | 0 |
| 6-1-3 | 1 | .010 | 1 | .001 |
| 6-4-4+ | 1 | .010 | 1 | .001 |
| 6-4-6 | 1 | .010 | 0 | 0 |
| 7-3-3+ | 1 | .010 | 21 | .029 |
| 7-4-5 | 1 | .010 | 6 | .008 |
| 8-3-4+ | 1 | .010 | 10 | .014 |
| (+)4-4-5 | 1 | .010 | 0 | 0 |
| (+)7-3-4+ | 1 | .010 | 14 | .019 |
| (−)7-3-4+ | 1 | .010 | 0 | 0 |
| Total | 102 | 1.000 | 575 | .787 |
Note.—Significance level declared at P<.002, given 25 tests of association.
Positive association.
Negative association.
African American population
In contrast to the unaffected white population, the unaffected African American population had a greater number of distinct STR-based haplotypes (202 vs. 71) and fewer significant associations (9 vs. 16; table 4). Similar to that in whites, the most common haplotype among the unaffected African American population was 7-3-4+, and it was positively associated with the 30 repeat allele and negatively associated with the 29 repeat allele.
Table 4.
FRAXA and STR-Haplotype Associations in an Unaffected African American Population[Note]
| FRAXA CGG-Repeat Size | STR Haplotype | Frequency |
| Positive association: | ||
| 29 | 3-3-4 | .014 |
| (+)7-3-6+ | .011 | |
| (+)7-4-6+ | .022 | |
| 30 | 6-3-4+ | .027 |
| 7-3-4+ | .088 | |
| 7-4-3+ | .019 | |
| 31 | 6-4-−2 | .013 |
| 39 | 7-6-5+ | .008 |
| Negative association: | ||
| 29 | 7-3-4+ | .003 |
Note.—Significance level declared at P<.00015 given 329 tests of association (n=637).
Unlike the white population, in the unaffected African American population a single haplotype was not observed among the intermediate alleles (41–60 repeats; table 4). In fact, among 22 African American chromosomes with intermediate alleles, 18 different haplotype backgrounds were observed, compared with 18 haplotype backgrounds among the 54 white intermediate alleles (data not shown). Thus, the lack of association with intermediate alleles in the African American population was due to the diversity of haplotype backgrounds among these alleles, not to a smaller sample size for this class of alleles.
We further examined the 2-1-3 haplotype background as this one was found to be associated with white intermediate alleles. Among the unaffected African American chromosomes, this haplotype was found on CGG-repeat alleles of 33 (n=1), 38 (n=1), and 43 (n=2) repeats. Thus, the 2-1-3 haplotype did not represent the majority of intermediate alleles in this population.
Striking differences were observed among the African American fragile X population when it was compared with the white fragile X population. Surprisingly, two previously undescribed haplotypes represented almost one-half of the fragile X chromosomes: (+)4-4-5 and (−)3-4-5 (table 5). When the haplotype distributions were compared, these two unique haplotypes were present at a significantly higher frequency in the African American fragile X population when compared with the unaffected population. As in the white population, the 7-3-4+ haplotype was observed, but it was not significantly underrepresented in the African American fragile X population when compared with the unaffected population (Fisher's exact, P=.700). Also, the 2-1-3 and 6-4-5 haplotypes were observed in the African American fragile X population but were not positively associated with full-mutation alleles after correction for multiple testing as observed in the white population (Fisher's exact test, P=.019 and .191, respectively; significance declared at P<.003, given 21 tests of association were performed).
Table 5.
African American Fragile X STR-Haplotype Associations[Note]
|
Chromosomes |
||||
| Fragile X |
Unaffected |
|||
| STR Haplotype | Number | Frequency | Number | Frequency |
| (+)4-4-5a | 20 | .317 | 11 | .017 |
| (−)3-4-5a | 6 | .095 | 3 | .005 |
| 7-3-4+ | 9 | .143 | 83 | .130 |
| (+)7-6-5+ | 4 | .063 | 5 | .008 |
| 2-1-3 | 3 | .048 | 4 | .006 |
| 6-4-5 | 3 | .048 | 14 | .022 |
| 7-1-3 | 2 | .032 | 8 | .013 |
| 7-4-5 | 2 | .032 | 19 | .030 |
| (+)7-6-4+ | 2 | .032 | 0 | 0 |
| 2-4-5 | 1 | .016 | 4 | .006 |
| 3-4-5 | 1 | .016 | 6 | .009 |
| 5-4-3 | 1 | .016 | 1 | .002 |
| 7-3-6 | 1 | .016 | 37 | .058 |
| (+)6-3-6+ | 1 | .016 | 0 | 0 |
| (+)6-4-3+ | 1 | .016 | 0 | 0 |
| (+)7-3-6+ | 1 | .016 | 7 | .011 |
| (+)7-4-6+ | 1 | .016 | 15 | .024 |
| (−)3-4-3+ | 1 | .016 | 1 | .002 |
| (−)4-4-5 | 1 | .016 | 1 | .002 |
| (−)5-4-3+ | 1 | .016 | 1 | .002 |
| (−)7-4-3+ | 1 | .016 | 0 | 0 |
| Total | 63 | 1.000 | 220 | .347 |
Note.—Significance level declared at P<.003, given 21 tests of association.
Significant positive association.
There was slightly less diversity of haplotypes among the African American fragile X chromosomes compared with the white chromosomes: of 21 distinct haplotypes, 7 were rare in the fragile X population and 4 were not observed in the unaffected population. However, this decreased diversity may be caused by the smaller sample size of the African American fragile X population compared with the white fragile X population reported here.
Because the two unique haplotypes were found at such a high frequency in the African American fragile X population, the current U.S. origin of each ascertained fragile X male was examined to determine whether there was a founder effect within the African American populations of South Carolina or the Southern United States. For this purpose, the African American fragile X haplotypes were examined in two ways. First, the South Carolina fragile X chromosomes (n=37) were examined by county. Of the 12 South Carolina chromosomes with the (+)4-4-5 haplotype, 4 chromosomes were ascertained within a 50-mile radius of 1 county. Although pedigree information did not reveal any recent common ancestors, additional genotyping of markers located >600 kb distal to the FRAXA locus, FRAXE, and DXS1691, revealed that all four of these chromosomes shared the same alleles for both of these loci. Two other chromosomes ascertained from different counties also had the same alleles at the distal loci and therefore may also have been distantly related. For the (−)3-4-5 haplotype, 2 of the 4 chromosomes had the same alleles at the distal loci and were ascertained from the same county in northern South Carolina.
Second, the haplotypes among the fragile X chromosomes ascertained from the Southern United States (other than South Carolina) and those outside of this region (Michigan, Oklahoma, New York, and Maryland) were examined. For the Southern fragile X chromosomes (n=16), the (+)4-4-5 haplotype was found in North Carolina, Georgia, Alabama, and Mississippi, whereas the (−)3-4-5 was found only in Georgia (table 6). For the non-Southern fragile X chromosomes (n=11), the (+)4-4-5 haplotype was identified in Michigan and the (−)3-4-5 haplotype was identified in Maryland. These haplotypes observed in northern cities are consistent with historical records that show a substantial migration of southern African Americans to the north after World War I (Johnson and Campbell 1981; Tanner 1995). Unlike the South Carolina chromosomes, there was no association with specific FRAXE and DXS1691 alleles among the northern cases. Thus, it seems that the South Carolina fragile X chromosomes with these unique haplotypes may be derived from a recent single founder. However, not all such chromosomes can easily be traced to a recent single founder.
Table 6.
Current United States Origin of Fragile X Chromosomes in the African American Population
| STR Haplotype | SC | NC | GA | MI | MD | NY | AL | OK | MS | TX | VA |
| (+) 4-4-5 | 12 | 4 | 1 | 1 | … | … | 1 | … | 1 | … | … |
| (−) 3-4-5 | 4 | … | 1 | … | 1 | … | … | … | … | … | … |
| 7-3-4+ | 3 | 1 | 1 | 2 | 1 | … | … | … | … | … | 1 |
| 2-1-3 | 3 | … | … | … | … | … | … | … | … | … | … |
| 6-4-5 | 2 | … | 1 | … | … | … | … | … | … | … | … |
| (+) 7-6-4+ | 2 | … | … | … | … | … | … | … | … | … | … |
| (+) 7-6-5+ | 2 | 1 | 1 | … | … | … | … | … | … | … | … |
| 2-4-5 | … | … | … | … | … | 1 | … | … | … | … | … |
| 3-4-5 | 1 | … | … | … | … | … | … | … | … | … | … |
| 5-4-3 | 1 | … | … | … | … | … | … | … | … | … | … |
| 7-1-3 | 1 | … | … | … | … | … | … | … | … | 1 | … |
| 7-3-6 | … | 1 | … | … | … | … | … | … | … | … | … |
| 7-4-5 | 1 | … | … | … | … | 1 | … | … | … | … | … |
| (+) 6-3-6+ | … | … | … | … | … | … | … | 1 | … | … | … |
| (+) 6-4-3+ | 1 | … | … | … | … | … | … | … | … | … | … |
| (+) 7-3-6+ | 1 | … | … | … | … | … | … | … | … | … | … |
| (+) 7-4-6+ | 1 | … | … | … | … | … | … | … | … | … | … |
| (−) 3-4-3+ | … | … | … | 1 | … | … | … | … | … | … | … |
| (−) 4-4-5 | 1 | … | … | … | … | … | … | … | … | … | … |
| (−) 5-4-3+ | 1 | … | … | … | … | … | … | … | … | … | … |
| (−) 7-4-3+ | … | … | … | 1 | … | … | … | … | … | … | … |
| Total | 37 | 7 | 5 | 5 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
To further examine the two unique haplotypes found among the African American fragile X chromosomes, we sequenced the CGG-repeat array of the unaffected chromosomes with these haplotype backgrounds. Among the unaffected African American chromosomes with the (+)4-4-5 haplotype background, we observed common repeat sizes either with no AGG interruptions or with one AGG interruption at the 3′ end of the repeat (table 7). In contrast, the unaffected chromosomes on the (−)3-4-5 haplotype background all contained a single 5′ AGG interruption at the 10th position and displayed a more diverse range of repeat sizes and structures (table 7).
Table 7.
CGG-Repeat Structures of Unaffected Chromosomes from African American Populations with Haplotypes Associated with the Fragile X Syndrome
| STR Haplotype | CGG-Repeat Structures |
| (+) 4-4-5 | 23 pure repeats (n = 1) |
| 19+9 (n = 1) | |
| 20+9 (n = 4) | |
| 22+9 (n = 1) | |
| (−) 3-4-5 | 9+9+9 (n = 1) |
| 9+10+10 (n = 1) | |
| 9+28 (n = 1) |
ATL1 in the African American Population
The ATL1 SNP marker was also typed to further characterize the haplotype associations in the African American population and findings were compared with the results from the white population (Gunter et al. 1998). Among whites, ATL1-G was strongly associated with both the intermediate and the full-mutation alleles (table 8). This strong association may indicate that the intermediate alleles represent a pool of alleles susceptible to expansion to the full mutation (Gunter et al. 1998). In addition, it was suggested that ATL1-G represented the ancestral state, as chromosomes with this allele had a greater diversity of STR-based haplotypes when compared with ATL1-A. Further evidence supporting this suggestion stemmed from the fact that ATL1-G was the more frequent allele among a small sample size of African, African American, and nonhuman primate samples which were used to represent genetically older populations (Gunter et al. 1998).
Table 8.
Comparison of the Frequency of ATL1 Allele G between a White and an African American Population
|
Fragile X Status |
|||
| Ethnicity | Unaffected (6–60 repeats) | Intermediate (41–60 repeats) | Full Mutation (>200 repeats) |
| Whitea | 40% (n=564) | 83%b (n=24) | 83%b (n=152) |
| African American | 74% (n=468) | 87% (n=15) | 88%b (n = 56) |
From Gunter et al. (1998).
Significant difference between this allelic form and the normal form at P<.05.
In contrast to that seen in the white population, ATL1-G in the African American population was more frequent among unaffected X chromosomes (74% vs. 40% in the white population) (table 8). Although the frequency of ATL1-G in the unaffected African American population was significantly different than the frequency in fragile X population (88%; Fisher's exact test, P<.05), it did not differ in frequency from the population of X chromosomes with intermediate alleles (87%; Fisher's exact test, P>.05) (table 8). Therefore, ATL1 allele status does not identify a pool of susceptible alleles in the African American unaffected population, as it does in the white population. ATL1-G was found among 28 distinct FRAXA CGG-repeat sizes while ATL1-A was found among 17 distinct repeat sizes. Similarly, ATL-G was found on 145 STR-based haplotype backgrounds, compared with ATL1-A found on 30 STR-based haplotype backgrounds. Both the high frequency of ATL1-G in the African American population and the greater diversity of FRAXA alleles and STR-based haplotypes observed with this allele support the idea that ATL1-G is the ancestral allele (Gunter et al. 1998; Hacia et al. 1999).
Discussion
To date, there have been many reports describing different white populations for the STR loci FRAXAC1, FRAXAC2, and DXS548 (Chiurazzi et al. 1996c). Comparing across studies, however, has proved difficult in the past because of differences in genotyping methods among the laboratories involved (Chiurazzi et al. 1996c; Chiurazzi et al. 1999). Thus, we characterized two large populations of different ethnic backgrounds to determine susceptibility factors for instability. Our primary goal was to determine whether haplotype background (i.e., cis-acting factors) influences stability of the CGG repeat.
There are, in fact, several possible explanations for the patterns of associations previously identified in white populations. First, all full mutations may descend from one, or a few, chromosomes, creating a founder effect. This explanation was invoked after the cloning of FMR-1 as it was observed that all full mutation alleles were the result of a multistep process occurring over many generations and that, in specific white populations, the majority of the full mutation alleles were found on a few haplotype backgrounds (for a review, see Chiurazzi et al. 1996c).
An alternative explanation to founder effects is the existence of “at risk” and “protective” chromosomal backgrounds involving cis-acting factors influencing repeat instability. Eichler et al. (1996) suggested at least two different mutational pathways, as indicated by two different haplotype backgrounds and repeat structures. It was postulated that regularly interspersed CGG repeats found on one haplotype background (2-1-3) are resistant to loss of AGG interruptions, thereby progressing more slowly to the premutation by the addition of 3′ repeats. The second haplotype background (6-4-4/5) found in association with asymmetrical CGG/AGG structures was postulated to be more susceptible to loss of interruptions, causing the allele to rapidly expand to the premutation. These distinct pathways may be the result of yet-unidentified cis-acting factors or the structure of the repeat itself.
In an effort to more clearly define the mutational history of the fragile X syndrome, Gunter et al. (1998) combined the STR-based haplotypes with a novel SNP (ATL1) and refined the definition of “at risk” and “protective” haplotypes in an unaffected white population. In particular, the 7-3-4+/A chromosomal background was significantly underrepresented in the unaffected white population when compared with the white fragile X population. Furthermore, the 10+ structures were highly associated with this “protective” haplotype while the 9+ structures were highly associated with the “at risk” haplotypes of 2-1-x and 6-4-x (Gunter et al. 1998). Thus, Gunter et al. (1998) concluded that the position of the 5′ AGG in the repeat might be yet another factor that impacts CGG-repeat stability.
A third possible explanation for these haplotype associations could be that the initial mutational event independently recurs in different ethnic groups at a relatively low frequency on random haplotype backgrounds. This possibility is not necessarily mutually exclusive from the first two explanations; that is, of a founder effect and causal cis-acting factors.
Much data have been amassed concerning STR-based haplotype associations in genetically younger populations such as the white population for the FRAXA locus. However, little has been done to identify these associations in genetically older populations that could help confirm or refute these possible interpretations. We used an admixed African American population to represent a genetically older population. Expectations for an older population were observed (e.g., greater haplotype diversity and fewer associations with the CGG repeat) and, for the one reported allele distribution for FRAXAC1, the allele distribution was not significantly different from those published for two African populations (Kunst et al. 1996; Chiurazzi et al. 1996a). These observations suggest that the degree of admixture is not great enough to obscure the results. On the basis of the proportion of African American STR-based haplotypes among whites, we crudely estimate the degree of admixture to be around 19%, which is consistent with previous estimates of relatively low European admixture among the African American population (Chakraborty et al. 1992; Clark et al. 1998; Parra et al. 1998; Destro-Bisol et al. 1999).
In general, the data on whites presented here for the unaffected population were similar with those previously reported in the literature. The unaffected white population had a total of 71 distinct STR-based haplotypes, 16 of which were significantly associated with a specific FRAXA CGG-repeat size (table 2). Although CGG/AGG structure information was not included in these analyses, the associations identified were similar to those of Eichler et al. (1996), who used AGG interspersion-pattern data. To briefly summarize their findings, only the 2-1-3 haplotype was associated with the intermediate alleles, and these alleles tended to maintain two AGG interruptions. The haplotype 6-4-4 was associated with the more asymmetrical CGG/AGG structures such as the 9+12+9 structure. The 7-3-4+ haplotype, in contrast to both the 2-1-3 and 6-4-4 haplotype, was associated with the 10+9+9 CGG/AGG structure.
The distribution of STR-based haplotypes for a white fragile X population and the significant associations with the full mutation reported here also were similar to others in the literature (Chiurazzi et al. 1996c; Eichler et al. 1996). Specifically, the 2-1-3 and 6-4-5/4 haplotypes were positively associated with the full mutation, and the haplotype 7-3-4+ was negatively associated with the full mutation.
For the unaffected African American population, 202 distinct STR-based haplotypes were observed, 39 of which were also found in the white population. In contrast with the white population, only 9 of the 202 STR-based haplotypes were significantly associated with a CGG-repeat allele (table 4). This would be expected for an older population, as recombination over time would have abolished linkage disequilibrium over larger distances. Of the 9 associations identified, few were common between the 2 populations: allele 30 was positively associated with the STR-based haplotype 7-3-4+, while allele 29 was negatively associated with this haplotype (table 4).
A striking difference between the white and African American haplotype patterns was the lack of associations with intermediate alleles in the African American population (table 4). This was also noted by Kunst et al. (1996), who observed no linkage disequilibrium between normal CGG repeat–length classes and FRAXAC1 alleles in a small sample of African chromosomes. This lack of association among African American intermediate alleles supports the idea that the 2-1-3 haplotype association with intermediate alleles in the white population is of recent origin (Kunst and Warren 1994; Kunst et al. 1996). This also suggests that there is no pool of susceptible alleles as identified by an STR-based haplotype in the African American population. Further support for this lack of a susceptible-allele pool as identified by repeat size alone comes from the comparison of the frequency of intermediate alleles (i.e., “susceptibility” alleles) and the prevalence of the fragile X syndrome. Previously, we found a decreased frequency of intermediate alleles among the African American population when compared with that in the white population; however, we found the fragile X syndrome to be at least as prevalent in African Americans as in whites (Crawford et al. 1999).
Because most STR loci have a higher mutation rate than SNP loci, white STR associations not found among other ethnic groups might be preserved as SNP associations. Recently, a subset of the white population described here was genotyped for the SNP ATL1 (Gunter et al. 1998). ATL1-G was associated with intermediate, premutation, and full mutation alleles in the white population, suggesting that FRAXA alleles on this background were predisposed to expansion. In contrast, ATL1 in the African American population did not seem to distinguish “predisposed” from “nonpredisposed” alleles in the unaffected population.
In fact, the majority of normal, intermediate, and full mutation chromosomes surveyed here were ATL1-G, supporting the idea that this is the ancestral allele (table 8). Indeed, smaller surveys of African and African American populations showed that the predominant CGG/AGG structure is the 9+ structure, which is tightly linked to ATL1-G in whites with 9+ structures (Eichler and Nelson 1996; Kunst et al. 1996; Gunter et al. 1998). Taken together, these results suggest that the different ATL1 alleles are associated with the 5′ end of the CGG/AGG structure and do not help identify cis-acting factors other than the structure of the repeat itself.
Another prediction stemming from the random and recurrent interpretation would be that the full-mutation allele would be found on many different STR-based haplotypes in the African American and white populations. In fact, there were many haplotype backgrounds observed on fragile X chromosomes in both African Americans and whites, some in strong linkage disequilibrium, some in linkage equilibrium, and others rare or not seen in the unaffected population. There were four and eight haplotype backgrounds on African American and white fragile X chromosomes, respectively, that could be considered “private” backgrounds. However, there is the strong possibility that these backgrounds are derived from other haplotypes and show differences because of the high mutation rate of STRs and/or because of recombination. Thus, the fragile X mutation on these “private” backgrounds may be related to other mutations on backgrounds that differ by one or two STR alleles. In fact, all 12 “private” haplotypes had closely related haplotypes among the unaffected chromosomes that differed either at the most distant marker, DXS548, or the FRAXAC2 marker, which is a complex polymorphism with at least two variable regions (Zhong et al. 1993). Further work is needed to determine whether these are true “private” mutations.
Interestingly, 2 of the 21 STR-based haplotypes found among the African American fragile X chromosomes were present on almost one half of these chromosomes. In the unaffected population, these two haplotypes were rare: the (+)4-4-5 haplotype (n=11) was found on repeat sizes in a range of 17–35 repeats and the (−)3-4-5 haplotype (n=3) was found on repeat sizes in a range of 29–38 repeats.
The high frequency of these two unusual haplotypes could be caused by a founder effect in our African American fragile X samples. Historical records show that Charleston, SC, was a major port of entry into the United States for slave trade and that the distribution of members of various African tribes brought into this country varied among the different ports (Curtin 1969; Franklin and Moss 1994). Whereas the majority of our samples are from South Carolina, we could be observing the founder allele that settled in the Southeastern United States from Africa. It would be interesting to ascertain west central African males with the fragile X syndrome to determine whether the full-mutation allele on this particular background originated in Africa and was introduced to the Americas through the slave trade.
Another possible explanation for the high frequency of these unique haplotypes is that they represent a recent mutation that has not come into equilibrium. As the record for African slaves in the United States dates as early as 1526 (Piersen 1996), the mutation could have occurred as recently as 20–25 generations ago. It may be that the CGG/AGG repeat structures on these unique haplotype backgrounds are particularly susceptible to repeat expansion, making this allele rare in the unaffected population and soon to be extinct in the fragile X population.
Alternatively, the high frequency of the (+)4-4-5 and (−)3-4-5 haplotype backgrounds in the African American fragile X population may be caused by a mechanism and origin similar to those of the 6-4-4/5 haplotype background. This haplotype has been proposed to progress rapidly to the full mutation, on the basis of its association pattern in whites (Eichler et al. 1996). The unique African American haplotypes, in fact, differ from the 6-4-4/5 haplotype only by the distant DXS548 locus, which could have changed over time because of a mutation or recombination. As proposed by Eichler et al. (1996), the asymmetrical CGG-repeat structures associated with the 6-4-4/5 haplotype may be more prone to loss of the 3′ AGG, leading to an allele susceptible to rapid expansion. Sequencing of the CGG-repeat array for the African American alleles with the (+)4-4-5 and (−)3-4-5 haplotype backgrounds (table 7), however, did not reveal the structures as observed in whites with the 6-4-4/5 haplotype backgrounds (e.g., 9+10+9 and 9+12+9). However, there is a possibility that the structures observed in the (+)4-4-5 background are the subsequent mutational step in this mutational pathway. That is, the 20+9 and 22+9 structures may have evolved from the loss of the 5′ AGG of the original asymmetrical 9+10+9 and 9+12+9 structures, respectively. In contrast to the CGG-repeat structures on the (+)4-4-5 background, the CGG-repeat structures on the (−)3-4-5 background do not seem susceptible to loss of the 5′ AGG. However, the sample size for these sequenced alleles is quite small and will require further investigation for these findings to be confirmed and interpreted.
Conclusions
We report here, for the first time, a comprehensive survey of the FRAXA CGG repeat and the surrounding STRs and SNP in a large African American unaffected and fragile X population. In comparing these data to the already established white population for these loci, we present evidence that the haplotype associations first identified in white populations are not necessarily caused by cis-acting sequences as identified by these haplotypes. This large survey in African Americans is, in a sense, preliminary, as further work must be performed in other ethnic populations before we can accurately and comprehensively identify all the factors involved in CGG-repeat stability that lead to the fragile X syndrome.
Acknowledgments
We would like to thank Dr. Allison E. Ashley-Koch, Elizabeth F. Hinkle, Mary L. Stanfield, Lisa Shubek, Dr. Don Bailey, Dr. Annette K. Taylor, and Dr. Michele M. M. Mazzocco for ascertaining fragile X male samples. We would also like to thank Dorothy L. Pettay for her technical assistance, Lorri Griffin for the establishment of lymphoblastoid cell lines (M01-RR-00039), and JoAnn Babb (Greenwood Genetic Center) for ascertainment information on the South Carolina fragile X samples. We would also like to thank Dr. Lynn Jorde for insightful comments on these data. Finally, we would like to thank the patients and their families, whose participation made this work possible. S.T.W. is an investigator of the Howard Hughes Medical Institute. This work was supported by NIH grants HD35576, HD29909, and HD08443. Partial support was also provided by the South Carolina Department of Disabilities and Special Needs.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for fragile X syndrome [MIM 309550])
References
- Arrieta I, Gil A, Nunez T, Elez M, Artinez B, Criado B, Stao C (1999) Stability of the FMR1 CGG repeat in a Basque sample. Hum Biol 71:55–68 [PubMed]
- Ashley-Koch AE, Robinson H, Glicksman AE, Nolin SL, Schwartz CE, Brown WT, Turner G, et al (1998) Examination of factors associated with instability of the FMR1 CGG repeat. Am J Hum Genet 63:776–785 [DOI] [PMC free article] [PubMed]
- Brown TC, Tarleton JC, Go RC, Longshore JW, Descartes M (1997) Instability of the FMR2 trinucleotide repeat region associated with expanded FMR1 alleles. Am J Med Genet 73:447–455 [DOI] [PubMed]
- Brown WT, Houck GEJ, Jeziorowska A, Levinson FN, Ding X, Dobkin C, Zhong N, et al (1993) Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. JAMA 270:1569–1575 [published erratum appears in JAMA 271:28] [PubMed]
- Brown WT, Zhong N, Dobkin C (1996) Positive fragile X microsatellite associations point to a common mechanism of dynamic mutation evolution. Am J Hum Genet 58:641–643 [PMC free article] [PubMed]
- Chakraborty R, Kamboh M, Nwankwo M, Ferrell R (1992) Caucasian genes in American Blacks: new data. Am J Hum Genet 50:145–155 [PMC free article] [PubMed]
- Chiurazzi P, Destro-Bisol G, Genuardi M, Oostra BA, Spedini G, Neri G (1996a) Extended gene diversity at the FMR1 locus and neighboring CA repeats in a sub-Saharan population. Am J Med Genet 64:216–219 [DOI] [PubMed]
- Chiurazzi P, Genuardi M, Kozak L, Giovannucci-Uzielli ML, Bussani C, Dagna-Bricarelli F, Grasso M, et al (1996b) Fragile X founder chromosomes in Italy: a few initial events and possible explanation for their heterogeneity. Am J Med Genet 64:209–215 [DOI] [PubMed]
- Chiurazzi P, Macpherson J, Sherman S, Neri G (1996c) Significance of linkage disequilibrium between the fragile X locus and its flanking markers. Am J Med Genet 64:203–208 [DOI] [PubMed]
- Chiurazzi P, Pomponi MG, Sharrock A, Macpherson J, Lormeau S, Morel ML, Rousseau F (1999) DNA panel for interlaboratory standardization of haplotype studies on the fragile X syndrome and proposal for a new allele nomenclature. Am J Med Genet 83:347–349 [PubMed]
- Chong SS, Eichler EE, Hughes MR, Nelson DL (1994) Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am J Med Genet 51:522–526 [DOI] [PubMed]
- Clark A, Weiss K, Nickerson D, Taylor S, Buchanan A, Stengard J, Salomaa V, et al (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed]
- Coffee B, Zhang F, Warren ST, Reines D (1999) Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet 22:98–101 [DOI] [PubMed]
- Crawford D, Meadows K, Newman J, Taft L, Pettay D, Gold L, Hersey J, et al (1999) Prevalence and phenotype consequence of FRAXA and FRAXE alleles in a large, ethnically diverse, special education-needs population. Am J Hum Genet 64:495–507 [DOI] [PMC free article] [PubMed]
- Curtin P (1969) The Atlantic slave trade. University of Wisconsin Press, Madison [Google Scholar]
- Destro-Bisol G, Maviglia R, Caglia A, Boschi, Spedini G, Pascali V, Clark A, et al (1999) Estimating European admixture in African Americans by using microsatellites and a microsatellite haplotype (CD4/Alu). Hum Genet 104:149–157 [DOI] [PubMed]
- Eichler EE, Holden JA, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, et al (1994) Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet 8:88–94 [DOI] [PubMed]
- Eichler EE, Macpherson JN, Murray A, Jacobs PA, Chakravarti A, Nelson DL (1996) Haplotype and interspersion analysis of the FMR1 CGG repeat identifies two different mutational pathways for the origin of the fragile X syndrome. Hum Mol Genet 5:319–330 [DOI] [PubMed]
- Eichler EE, Nelson DL (1996) Genetic variation and evolutionary stability of the FMR1 CGG repeat in six closed human populations. Am J Med Genet 64:220–225 [DOI] [PubMed]
- Franklin JH, Moss AA (1994) The slave trade and the New World. In: Labella P, Greiner B (eds) From slavery to freedom: a history of African-Americans. McGraw-Hill, New York, pp 27–55 [Google Scholar]
- Fu YH, Kuhl DPA, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJMH, et al (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67:1047–1058 [DOI] [PubMed]
- Gunter C, Paradee W, Crawford DC, Meadows KL, Newman J, Kunst CB, Nelson DL, et al (1998) Re-examination of factors associated with expansion of CGG repeats using a single nucleotide polymorphism in FMR1. Hum Mol Genet 7:1935–1946 [DOI] [PubMed]
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, et al (1999) Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet 22:164–167 [DOI] [PubMed]
- Heitz D, Devys D, Imbert G, Kretz C, Mandel JL (1992) Inheritance of the fragile X syndrome: size of the fragile X premutation is a major determinant of the transition to full mutation. J Med Genet 29:794–801 [DOI] [PMC free article] [PubMed]
- Hirst MC, Grewal PK, Davies KE (1994) Precursor arrays for triplet repeat expansion at the fragile X locus. Hum Mol Genet 3:1553–1560 [DOI] [PubMed]
- Johnson DM, Campbell RR (1981) Black migration in America: a social demographic history. Duke University Press, Durham [Google Scholar]
- Kunst CB, Warren ST (1994) Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell 77:853–861 [DOI] [PubMed]
- Kunst CB, Zerylnick C, Karickhoff L, Eichler E, Bullard J, Chalifoux M, Holden JJ, et al (1996) FMR1 in global populations. Am J Hum Genet 58:513–522 [PMC free article] [PubMed]
- Meadows KL, Pettay D, Newman J, Hersey J, Ashley AE, Sherman SL (1996) Survey of the fragile X syndrome and the fragile X E syndrome in a special education needs population. Am J Med Genet 64:428–433 [DOI] [PubMed]
- Morton NE, Macpherson JN (1992) Population genetics of the fragile-X syndrome: multiallelic model for the FMR1 locus. Proc Natl Acad Sci USA 89:4215–4217 [DOI] [PMC free article] [PubMed]
- Murray A, Macpherson JN, Pound MC, Sharrock A, Youings SA, Dennis NR, McKechnie N, et al (1997) The role of size, sequence and haplotype in the stability of FRAXA and FRAXE alleles during transmission. Hum Mol Genet 6:173–184 [DOI] [PubMed]
- Murray A, Youings S, Dennis N, Latsky L, Linehan P, McKechnie N, Macpherson J, et al (1996) Population screening at the FRAXA and FRAXE loci: molecular analyses of boys with learning difficulties and their mothers. Hum Mol Genet 5:727–735 [DOI] [PubMed]
- Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolin SL, Lewis FA, Ye LL, Houck GEJ, Glicksman AE, Limprasert P, Li SY, et al (1996) Familial transmission of the FMR1 CGG repeat. Am J Hum Genet 59:1252–1261 [PMC free article] [PubMed]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, et al (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252:1097–1102 [DOI] [PubMed]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, et al (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed]
- Piersen WD (1996) From Africa to America. Twayne Publishers, New York [Google Scholar]
- Richards RI, Kondo I, Holman K, Yamauchi M, Seki N, Kishi K, Staples A, et al (1994) Haplotype analysis at the FRAXA locus in the Japanese population. Am J Med Genet 51:412–416 [DOI] [PubMed]
- Schwartz CE, Phelan MC, Brightharp C, Pancoast I, Howard-Peebles PN, Thibodeau S, Brown WT, et al (1988a) Fragile X syndrome: linkage analysis in black and white populations. Am J Med Genet 30:531–542 [DOI] [PubMed]
- Schwartz CE, Phelan MC, Pulliam LH, Wilkes G, Vanner LV, Albiez KL, Potts WA, et al (1988b) Fragile X syndrome: incidence, clinical and cytogenetic findings in the black and white populations of South Carolina. Am J Med Genet 30:641–654 [DOI] [PubMed]
- Sherman SL, Meadows KL, Ashley AE (1996) Examination of factors that influence the expansion of the fragile X mutation in a sample of conceptuses from known carrier females. Am J Med Genet 64:256–260 [DOI] [PubMed]
- Snow K, Doud LK, Hagerman R, Pergolizzi RG, Erster SH, Thibodeau SN (1993) Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet 53:1217–1228 [PMC free article] [PubMed]
- Snow K, Tester DJ, Kruckeberg KE, Schaid DJ, Thibodeau SN (1994) Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet 3:1543–1551 [DOI] [PubMed]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST (1992) DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet 1:397–400 [DOI] [PubMed]
- Tanner HH (1995) The settling of North America. Macmillan, New York [Google Scholar]
- Turner G, Webb T, Wake S, Robinson H (1996) Prevalence of fragile X syndrome. Am J Med Genet 64:196–197 [DOI] [PubMed]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu Y-H, Kuhl DPA, Pizzuti A, Reiner O, et al (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 [DOI] [PubMed]
- Yu S, Mulley J, Loesch D, Turner G, Donnelly A, Gedeon A, Hillen D, et al (1992) Fragile-X syndrome: unique genetics of the heritable unstable element. Am J Hum Genet 50:968–980 [PMC free article] [PubMed]
- Zhong N, Dobkin C, Brown WT (1993) A complex mutable polymorphism located within the fragile X gene. Nat Genet 5:248–253 [DOI] [PubMed]
- Zhong N, Ju W, Pietrofesa J, Wang D, Dobkin C, Brown WT (1995a) Fragile X “gray zone” alleles: AGG patterns, expansion risks, and associated haplotypes. Am J Med Genet 64:261–265 [DOI] [PubMed]
- Zhong N, Liu X, Gou S, Houck GEJ, Li S, Dobkin C, Brown WT (1994) Distribution of FMR-1 and associated microsatellite alleles in a normal Chinese population. Am J Med Genet 51:417–422 [DOI] [PubMed]
- Zhong N, Yang W, Dobkin C, Brown WT (1995b) Fragile X gene instability: anchoring AGGs and linked microsatellites. Am J Hum Genet 57:351–361 [PMC free article] [PubMed]