Summary
Evidence for genetic influences in epilepsy is strong, but reports identifying specific chromosomal origins of those influences conflict. One early study reported that human leukocyte antigen (HLA) markers were genetically linked to juvenile myoclonic epilepsy (JME); this was confirmed in a later study. Other reports did not find linkage to HLA markers. One found evidence of linkage to markers on chromosome 15, another to markers on chromosome 6, centromeric to HLA. We identified families through a patient with JME and genotyped markers throughout chromosome 6. Linkage analysis assuming equal male-female recombination probabilities showed evidence for linkage (LOD score 2.5), but at a high recombination fraction (θ), suggesting heterogeneity. When linkage analysis was redone to allow independent male-female θs, the LOD score was significantly higher (4.2) at a male-female θ of .5, .01. Although the overall pattern of LOD scores with respect to male-female θ could not be explained solely by heterogeneity, the presence of heterogeneity and predominantly maternal inheritance of JME might explain it. By analyzing loci between HLA-DP and HLA-DR and stratifying the families on the basis of evidence for or against linkage, we were able to show evidence of heterogeneity within JME and to propose a marker associated with the linked form. These data also suggest that JME may be predominantly maternally inherited and that the HLA-linked form is more likely to occur in families of European origin.
Introduction
The problems in finding genes that contribute to the expression of common, complex diseases have become more apparent as different groups report conflicting findings. One of the first common diseases for which a probable locus was identified was juvenile myoclonic epilepsy (JME [MIM 254770]), a syndrome of the group called the “idiopathic generalized epilepsies” (IGE). These epilepsies do not originate in one spot in the brain (as does “focal” epilepsy) but manifest over the whole brain simultaneously (hence, “generalized”). The causes of the IGE are uncertain but are thought to be largely genetic (Greenberg et al. 1992; Janz et al. 1992)
The genetics of these epilepsies and the relationship among them has become murkier as more loci contributing to their expression are reported. Greenberg et al. (1988) reported evidence for linkage of JME to the human leukocyte antigen (HLA) region of chromosome 6, which was later confirmed ( Durner et al. 1991; Weissbecker et al. 1991), and the locus was designated EJM1 (Greenberg et al. 1989). Sander et al. (1997a) reconfirmed strong evidence for linkage of JME to HLA. However, Whitehouse et al. (1993) detected some evidence for linkage in the HLA region (LOD score ∼1.4) at a high recombination fraction (θ), but it was not considered statistically significant. Later, Elmslie et al. (1997) found evidence of linkage of JME to chromosome 15, in an area in which an acetylcholine receptor subunit was known to exist. However, Neubauer et al. (1998) found evidence of linkage to Rolandic epilepsy—a focal, not generalized, form of epilepsy—to this same region of chromosome 15, whereas Sander et al. (1997b) and Durner et al. (in press) found no evidence supporting linkage of JME to any chromosome 15 loci. Further adding to the confusion, Liu et al. (1995) reported linkage of a large JME pedigree from Belize to markers on chromosome 6, but in a region ∼36 cM centromeric to HLA. However, both Elmslie et al. (1996) and Greenberg et al. (1998) failed to find any evidence suggesting linkage of JME to markers centromeric to HLA. Finally, Durner et al. (1992), Obeid et al. (1994), Greenberg et al. (1995), and Moen et al. (1995) found evidence of association of JME and HLA alleles.
The apparently contradictory evidence involving linkage of JME suggests heterogeneity as a major influence in these studies. Sander et al. (1997a) reported significant evidence of linkage of JME to HLA markers in a sample of German families. However, JME families they collected elsewhere in the world gave evidence against linkage, suggesting that linkage of JME to the HLA region might represent a European phenomenon. The findings of Liu et al. (1996) appear to support this hypothesis. Thus, ethnic factors may play a role in finding linkage in epilepsy.
Heterogeneity represents a major analysis problem because it significantly reduces the power to detect linkage in a data set. Measures to reduce heterogeneity at the clinical level are essential in common disease genetics. Analysis programs that take account of ``admixture'' heterogeneity are of some aid, but testing for linkage heterogeneity by using hypotheses based on clinical differences is a more powerful approach. Durner et al. (1999) showed that families separated on the basis of clinical criteria yielded significant evidence of linkage, whereas simply assuming admixture heterogeneity in an analysis including all families failed to yield a level of significance that would even qualify as interesting. Our approach in the current study was to reduce heterogeneity in JME by analysis of families ascertained through a carefully diagnosed JME case, then to test the hypothesis of linkage of JME to HLA-region markers with a new series of families. Expansive data analysis then ultimately yielded clues about heterogeneity that we could exploit to reveal the phenomena complicating the linkage results.
Families and Methods
Family Data
All families were ascertained through at least one JME index case. The major inclusion criterion was the presence of myoclonic jerks, involving the arms and shoulders, that occur upon or shortly after affected individuals awaken from sleep. These jerks are experienced with full consciousness. The age at onset of seizures was >8 years and <20 years. Findings from the neurologic examination and magnetic resonance imaging (MRI) or computed tomography (CT) scans were normal (Commission on Classification 1989). To reduce instances of other epilepsy syndromes, we excluded families when the index patient had (1) myoclonic absences (absences accompanied by rhythmic [3/s] jerking) or atonic attacks, (2) focal seizures, (3) alcohol or other substance abuse, (4) mental retardation, or (5) any suggestion that the epilepsy was the result of a structural, metabolic, or degenerative disease.
We ascertained 85 JME cases (48 of which ultimately provided information for linkage at HLA). Seven patients had myoclonic jerks as their only seizure type; 78 had myoclonic jerks plus tonic-clonic seizures, and 21 also had juvenile-onset absence seizures (short staring episodes lasting a few seconds and occurring a few times a day). Informed consent was obtained for all participants.
Family Members
Although all families were ascertained through a JME patient, 28 families had an additional family member or members with JME or other forms of IGE. Family members with any form of confirmed IGE were classified as affected. All available family members were given 1-h electroencephalograms (EEGs). Fourteen family members who were clinically normal but with spike-and-wave EEG or paroxysmal bursts of generalized high-amplitude slow waves in the theta range (in the presence of a normal awake background rhythm) were also classified as affected. In previous work (Greenberg et al. 1988; Durner et al. 1991; Sander et al. 1997a), the abnormal EEGs in normal family members appear to be genetically related to the epilepsy phenotype. Thus, families are ascertained through a JME patient, but family members are counted as affected if they have any IGE or a generalized EEG abnormality of the type seen in IGE cases, making the overall phenotype broader than just JME.
Patients with Other (Non-JME) Forms of IGE
To look for association of markers with JME, we analyzed patients with forms of IGE different from JME. These patients were ascertained through the same clinics, from the same populations, and in the same manner as the JME index patients. The non-JME patients had IGE syndromes similar in many respects to JME (juvenile absence, tonic-clonic seizures, and adolescent age at onset; see Durner et al. [1999] for an extensive description) but lacked the awakening myoclonic jerks. Families with these non-JME forms of IGE yielded strong evidence against linkage with HLA (Greenberg et al. 1996). There were 38 index patients with non-JME forms of epilepsy. These forms show evidence of linkage to markers on chromosome 8 (Durner et al. 1999), whereas JME shows significant evidence against linkage to the same markers.
Genetic Markers
We typed markers along the entire length of chromosome 6 and additional markers within the HLA region. The markers we typed were (telomere) D6S344–F13A–D6S309–D6S277–D6S470–D6S260–D6S289–D6S422–D6S299–D6S258–D6S1621–D6S265–D6S306–D6S273–DRB1–DQA1–DQB1–D6S1666–D6S1610–D6S271–D6S257–D6S286–D6S460–D6S462–D6S434–D6S287–D6S262–D6S292–D6S441–D6S1581–D6S264–D6S281. The majority of these markers were from the ABI marker panel (Perkin-Elmer). Others, mostly in the HLA region, were selected from Dib et al. (1996).
Linkage Analysis
We performed both two-point and multipoint linkage analyses. LIPED (Ott 1974) was used for two-point analysis, allowing independent male and female θs (θm, θf). GENEHUNTER (Kruglyak et al. 1996) was used for multipoint analysis. We did the multipoint analyses both with and without allowing for heterogeneity.
Following previous reports of linkage of HLA markers to JME (Durner et al. 1991; Greenberg et al. 1992), we assumed dominant inheritance with 70% penetrance in the HLA region. Even moderate changes in the value of the assumed penetrance will often not greatly affect the value of the final LOD score in an analysis (Greenberg 1989). When analyzing the rest of the chromosome, we assumed both dominant and recessive inheritance, each with a penetrance value of 50%, as recommended by Hodge et al. (1997). Extensive work has demonstrated that the LOD score statistic has the most power to detect linkage in complex disease, even when any increase in type I error is taken into account (Greenberg et al. 1998; Abreu et al. 1999).
Because the HLA-DRB and HLA-DQB loci are highly polymorphic, we did not expect that inclusion of additional HLA-region markers under multipoint analysis would provide further linkage information in the HLA region beyond that provided by DR and DQ. (The potential advantage of multipoint analysis lies in extraction of more information for linkage by combination of allele information from adjacent but less-informative markers.) We typed additional markers in the HLA region to look for recombinants, thus narrowing the region containing the EJM1 locus.
Results
Linkage Data
As the study progressed, the analyses of the data consistently yielded maximum LOD scores at θf=.01 (the lowest tested) and θm=.5 (indicating no linkage). Although the genetic length of this region of chromosome 6 is approximately twice as great in females as in males (Cooperative Human Linkage Center database), that would not explain the observation of the difference in θm and θf at the maximum LOD score in our family data. Initially, we assumed that difference in θm and θf was due to small sample size, but, as the sample size grew and the position of the maximum did not change, we were forced to the conclusion that the phenomenon was real. Table 1 shows the grid of LOD scores for the analysis with independent male-female θs. The maximum LOD score was 4.2 at θm,f=.5,.01. (The maximum LOD score along the diagonal [θm=θf]) is 2.5 at a θ estimate of .1.) Even with correction for the increase in type I error when independent male-female θs are used, which requires subtraction of .5 from the maximum LOD score (Lander and Lincoln 1988), the LOD score is highly significant. Moreover, this is the third data set reporting linkage of JME to the HLA region. An affected-only analysis yielded a LOD score of 2.76 at θm,f=.5,.05.
Table 1.
LOD Scores for the DQB1 Locus and JME, Allowing Independent Male-Female θs
| LOD Score at θf = |
|||||||
| θm | .01 | .05 | .1 | .2 | .3 | .4 | .5 |
| .5 | 4.218 | 4.057 | 3.723 | 2.771 | 1.654 | .637 | .000 |
| .4 | 4.181 | 4.018 | 3.684 | 2.732 | 1.620 | .607 | −.022 |
| .3 | 4.041 | 3.877 | 3.543 | 2.595 | 1.490 | .491 | −.118 |
| .2 | 3.695 | 3.534 | 3.205 | 2.265 | 1.171 | .191 | −.383 |
| .1 | 2.944 | 2.797 | 2.481 | 1.559 | .479 | −.469 | −.974 |
| .05 | 2.259 | 2.126 | 1.823 | .919 | −.140 | −1.049 | −1.481 |
| .01 | 1.274 | 1.164 | .886 | .032 | −.957 | −1.753 | —2.058 |
Superficially, this result implies that there is little recombination (i.e., close linkage) in females but independent segregation of disease and marker (i.e., no linkage) in males. Such a finding might suggest maternal transmission of the disease allele— that is, that affected family members usually share a maternal marker allele. However, the expected pattern of LOD scores, with independent male and female θs under maternal transmission, would show no variation with respect to θm (e.g., the same LOD score value for all assumed θm at a given θf) and variation only with respect to θf. Our results show a great deal of variation of the LOD score with θm. One explanation for the pattern we observe is that there exists admixture of two types of families, with the linked form showing maternal inheritance and with the other form being unlinked.
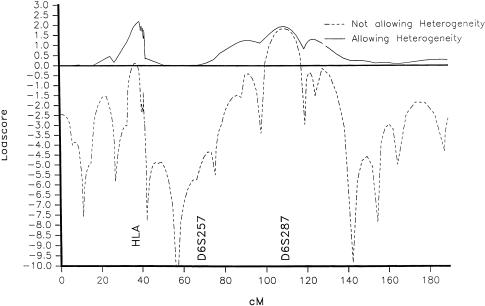
Linkage analyses reported in the literature usually assume equal male and female θs. In table 1, LOD scores for equal male-female θs are seen on the diagonal (bottom left to top right). The maximum LOD score along the diagonal is 2.5 at a θ estimate of .1. This estimate, if taken at face value, would put EJM1 beyond the HLA region. However, the fact that the maximum LOD score occurs at a high θ when previous results (and the multipoint analysis) had put the gene within the HLA region provides another suggestion of heterogeneity in the data (Ott 1991; P. C. Abreu, D. A. Greenberg, and S. E. Hodge, unpublished data). A multipoint LOD score analysis of chromosome 6, assuming heterogeneity, yielded a LOD score of 2.3 for the HLA region, with an estimated α (the percentage of families linked) of 50% (fig. 1). The multipoint LOD score assumed θm=θf. This maximum LOD score value is close to the two-point maximum LOD score when θm=θf is assumed. Thus, both the two-point analysis and the multipoint analysis at θm=θf suggest heterogeneity.
Figure 1.
Multipoint LOD scores for JME on chromosome 6, showing curves both with and without the assumption of heterogeneity.
Analysis of Recombinants
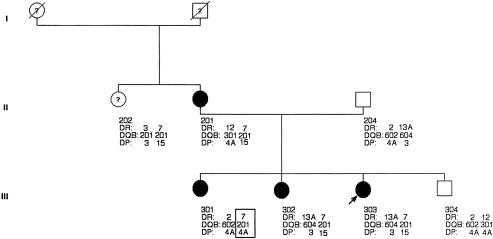
Analysis of recombinations seen in the families we studied showed a clear recombinant in one family between DQ and DP (fig. 2). Sander et al. (1997a) had reported a recombinant centromeric to DQ in one of the families in their study. Taken together, these two recombinant events mean that the EJM1 locus is likely to be centromeric of DQ and telomeric of DP. The known expressed loci in this region are DNA, RING3, DMA, DMB, LMP2, TAP1, LMP7, TAP2, and DOB.
Figure 2.
Pedigree of a family with epilepsy, showing a recombinant (box) between DQ and DP, suggesting that EJM1 lies distal to DP.
The existence of heterogeneity complicates the determination of which of the possible candidates may be EJM1. Since it is essential to try to separate the hypothesized linked from the unlinked form, we decided to see whether an allele of a marker locus in the region between DQ and DP was associated with JME probands from families showing positive evidence of linkage compared with “controls” (probands from non-JME families). If a particular allele occurred more frequently among the JME patients showing linkage than among controls, then the distribution of alleles among the non-EJM1 form of JME might be similar to the distribution found in non-JMEs. This might lead to a marker for the two different forms of JME.
Use of Association Data to Resolve the Heterogeneity
Having shown strong evidence for linkage and, simultaneously, for heterogeneity, we sought a means to distinguish the EJM1-influenced JME from the non-EJM1 form. Previous reports suggested an association of JME with HLA alleles. Durner et al. (1992), Obeid et al. (1994), and Greenberg et al. (1996) reported an association of JME with DR alleles. In the Greenberg et al. (1996) report, DQ alleles showed a somewhat stronger association than did DR alleles.
With the increased data, we again looked at the DR and DQ association with JME. We no longer detect an association with DR13. Taking DQB1*603 and 604 together, we observe a nonsignificant χ2 of 3.1 (P<.078). However, we know that we also now have substantial heterogeneity in the data set, making an association more difficult to detect.
We analyzed the 18 JME families showing the highest positive LOD scores (range 0.1–1.4) for linkage with the DRB1 marker. We then compared the distribution of alleles in the families showing positive linkage with the distribution in probands from the non-JME families. The distribution of DRB1 alleles was not different in the index cases from the linked-only families and the non-JME families. There was a significant difference in the distribution of DQB603 and 604, with a .17 frequency of 603 and 604 in the linked-only probands compared with .04 in the non-JME cases (χ2=6.04; P<.014) (table 2). However, despite the continued evidence for association, the number of linked-only JME cases with the 603 and 604 alleles was too small to use them as the basis for a subset for linkage.
Table 2.
Differences in Allele Frequencies between Probands in JME Families with Positive LOD Scores versus Probands from Families with Other Forms of IGE for the DQB1 and RING3 Loci
| Locus and Allele | JME(n) | non-JME(n) | JME(%) | non-JME(%) | χ2 |
| DQB1 | |||||
| 603+604 | 6 | 4 | 16.8 | 4.1 | |
| Other | 30 | 94 | 83.3 | 96.7 | 6.03 (P < .02, 1 df) |
| Total | 36 | 98 | 100 | 100 | |
| RING3 | |||||
| 3 | 18 | 18 | 50.00 | 23.68 | |
| 4 | 16 | 50 | 44.5 | 65.79 | |
| Other | 2 | 8 | 5.5 | 10.3 | 8.7 (P < .025, 2 df) |
| Total | 36 | 76 | 100 | 100 |
We searched for other polymorphic marker loci in the region. We discovered a CA repeat located in the middle of the RING3 locus, referred to below as CA.RING3 (GenBank accession number AF107699). Amplification of this microsatellite region revealed six alleles, two of which comprised 91% of the total (table 2). We examined the distribution of these alleles in the linked-JME versus non-JME patients and noted that the frequency of allele 3 (CA.RING3.3) was .5 in the patients from families showing linkage and .24 in the non-JME cases. Although the difference in the allele distribution between the two groups was significant (χ2=8.7; P<.025, 2 df), our main purpose was not to demonstrate a statistically significant association but to find a means to divide the families in a way that could help unravel the heterogeneity.
Combining Linkage and Association Data
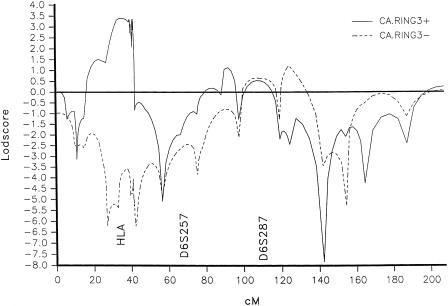
We then applied the partitioned linkage-association (PAL) test (Greenberg and Doneshka 1996). This test requires division of the families into two groups, on the basis of the presence or absence of the associated allele in the index patient, and performance of separate linkage analyses in the two groups. We divided the JME families on the basis of the presence (CA.RING3.3+) or absence (CA.RING3.3−) of the CA.RING3.3 allele. We then performed linkage analysis separately on the two groups. We observed, first, that the θs at which the maximum LOD score occurred in the CA.RING3.3+ group changed from θmθf=.5,.01 to .01,.01 and, second, that the LOD score rose from 1.27 to 3.97 at those θ values (table 3). Recall that analysis of the entire sample showed a maximum LOD score of 1.3 at θm,f=.01,.01 (table 1). The CA.RING3.3− group (table 4) became negative (−2.43; multipoint LOD score −7 [fig. 3]). Also, the CA.RING3.3+ group showed little variation with respect to θm, in contrast to the analysis of the undivided group (table 1). If inheritance is usually maternal, this is what one expects to see, whether the cause is biological or due to ascertainment bias. Removal of the heterogeneous families resolved the apparent anomaly that the maximum LOD score occurred at θm,f=.5,.01. The multipoint maximum LOD score was 3.4 for CA.RING3.3+ families [fig. 3]). When we performed a statistical test for heterogeneity (Morton 1955) between the RING3+ and RING3− groups, based on the LOD scores assuming equal male-female θs, χ2 was 6.77 (P<.01, 1 df).
Table 3.
LOD Scores for the DQB1 Locus Allowing Independent Male-Female θ in CA.RING3.3+ Families
| LOD Score at θf = |
|||||||
| θm | .01 | .05 | .1 | .2 | .3 | .4 | .5 |
| .5 | 3.4314 | 3.1727 | 2.7983 | 1.9565 | 1.1073 | .4019 | .0000 |
| .4 | 3.4708 | 3.2124 | 2.8400 | 2.0031 | 1.1605 | .4617 | .0648 |
| .3 | 3.6024 | 3.3431 | 2.9714 | 2.1417 | 1.3104 | .6251 | .2397 |
| .2 | 3.7720 | 3.5134 | 3.1447 | 2.3254 | 1.5075 | .8373 | .4664 |
| .1 | 3.9182 | 3.6648 | 3.3027 | 2.4967 | 1.6916 | 1.0344 | .6782 |
| .05 | 3.9616 | 3.7125 | 3.3557 | 2.5574 | 1.7586 | 1.1055 | .7562 |
| .01 | 3.9718 | 3.7278 | 3.3758 | 2.5851 | 1.7900 | 1.1397 | .7950 |
Table 4.
LOD Scores for the DQB1 Locus Allowing Independent Male-Female θs in CA.RING3.3− Families
| LOD Score at θf = |
|||||||
| θm | .01 | .05 | .1 | .2 | .3 | .4 | .5 |
| .5 | .8106 | .9045 | .9406 | .8231 | .5511 | .2360 | .0000 |
| .4 | .7423 | .8339 | .8681 | .7457 | .4704 | .1540 | −.0797 |
| .3 | .4950 | .5863 | .6189 | .4929 | .2136 | −.1028 | −.3274 |
| .2 | .0229 | .1167 | .1505 | .0217 | −.2609 | −.5743 | −.7777 |
| .1 | −.8041 | −.7031 | −.6636 | −.7894 | −1.0709 | −1.3661 | −1.5160 |
| .05 | −1.4842 | −1.3744 | −1.3273 | −1.4435 | −1.7113 | −1.9719 | −2.0566 |
| .01 | −2.4313 | −2.3038 | −2.2373 | −2.3129 | −2.5159 | −2.6667 | −2.6286 |
Figure 3.
Multipoint linkage analysis for families in which the index patient was CA.RING3.3+ or CA.RING3.3−
These results may also explain the findings of Whitehouse et al. (1993), who found a LOD score of 1.4 in the HLA region, at a high θ value. Those authors did not test independent male-female θs. Had they done so, they might have found a pattern similar to that reported here.
Other Findings on Chromosome 6
Region identified by Liu et al. (1995).—We paid particular attention to markers D6S271, D6S257, and D6S460, ∼36 cM centromeric of HLA. Liu et al (1995) reported significant evidence of linkage in a JME family from Belize in this area. Two-point LOD scores for these markers ranged from −6 (D6S271) to −1.5 (D6S460). Multipoint LOD scores ranged from −7 to −4 (fig. 1). LOD scores assuming heterogeneity were ∼0 (fig. 1). Division of the families on the basis of whether absence seizures were present in the family (Liu et al. 1996) also did not yield any positive evidence for linkage (multipoint LOD score −2 to −4).
A possible locus on 6q.—We noted a small positive LOD score for JME in the region of D6S287, on the long arm of chromosome. 6. The LOD score value for all JME families was 1.9 and was essentially the same when we allowed for heterogeneity (2.1, α=.87).
Discussion
Our results yield three major findings: (1) We have been able to replicate linkage of JME to HLA-region markers, making this the third independent data set showing strong evidence for linkage. (2) We demonstrated heterogeneity in the data and were able to propose a marker that could separate the HLA-linked and unlinked forms of JME. The CA.RING3 marker had an allele occurring in 76% of probands from families showing positive evidence for linkage but only 47% among probands with other forms of IGE and 31% among JME patients from families not showing evidence for linkage. (3) The linkage pattern implied the existence not only of heterogeneity but also of transmission predominantly from mothers; this observation was strengthened when we separated the families on the basis of the presence of CA.RING3.3. This finding suggests that the maternally inherited EJM1 either is more penetrant or is preferentially transmitted by the mother in those families.
The observation of maternal transmission of epilepsy has long been noted in the literature (Ottman et al. 1985, 1988). However, although the linkage data and epidemiologic data suggest a biological basis for the maternal transmission, ascertainment bias cannot be ruled out. For ascertainment bias to account for our observations, mothers with epilepsy in their families would have to have been three times as likely to participate in the study as were fathers with epilepsy in their families. We found that, in families with epilepsy in two or more generations or in which IGE was present in relatives (enabling us to assign the origin to one side of the family or the other), there were 25 instances in which IGE was likely to come from the mothers' family but only 7 such instances on fathers' sides (Klotz et al. 1999). This does not rule out ascertainment bias but makes it less plausible.
Of interest, Weissbecker et al. (1991) also performed linkage analysis assuming independent male-female θs. Their LOD score maximized at a high female and low male θ (θm,f=.001,.2). However, the difference between that maximum LOD score and the one obtained for θf=θm was not statistically significant, whereas the difference we obtained for the two maxima in the current study was highly significant (P<.005).
We noted the existence of a small peak in both the two-point and the multipoint LOD scores (maximum LOD score ∼2) on the long arm of chromosome 6, at D6S287. Almost half the families yield LOD scores of 0 at this marker, in part because two alleles make up ∼60% of the total frequency. Further studies will continue to explore this region.
Another observation concerns ethnicity. Sander et al. (1997a) noted that JME families from non-European ethnic groups showed evidence against linkage to HLA, suggesting that EJM1 may influence JME primarily in European-origin families. We examined families on the basis of ethnicity. The problem in a “melting pot” area such as New York City is that ethnicity may be difficult to determine. We had classified families broadly into white (that is, European origin), black (based on skin color and features), Hispanic (based on Spanish language of the parents and South or Central American origin), and Asian (Chinese, Japanese, or Indian origin). The nonwhite families produced mostly negative LOD scores (12/16 families had negative LOD scores, 4/16 had positive LOD scores), and the LOD score at all θ was negative and was −1.8 at θm=θf=.01. Although we have limited data, our results, like those of Sander et al. (1997a), suggest that the EJM1 form of JME occurs mostly in families of European origin. The findings of Liu et al. (1995) support this hypothesis.
These new results on chromosome 6 in JME must be put into the perspective of all the genetic influences on IGE and JME. Results from our genome scan in IGE (M. Durner, M. A. Meddache, L. Tomasini, S. Shinnar, S. R. Resor, S. L. Moshe, D. Rosenbaum, J. Cohen, C. Harden, H. Kang, S. Wallace, D. Luciano, K. Ballaban-Gil, I. Klotz, E. Dicker, D. A. Greenberg, unpublished data) show that other genetic loci influence the expression of JME, perhaps as much as, or more strongly than, the locus on chromosome 6. The demonstration of heterogeneity within the JME syndrome means that the epilepsy syndrome we call JME is not a single genetic entity. The genome scan and the existence of heterogeneity also suggest that other loci besides EJM1 may produce the symptom of myoclonic jerks, given that the locus or loci predisposing to IGE are present. Results from our genome scan support the idea that EJM1 may act to determine the type of seizures that will be manifested in a patient who is genetically susceptible to IGE, rather than being responsible for the IGE itself. This hypothesis is supported by our observation of loci that appear to be linked to absence seizures in both JME and non-JME families (M. Durner, M. A. Meddache, L. Tomasini, S. Shinnar, S. R. Resor, S. L. Moshe, D. Rosenbaum, J. Cohen, C. Harden, H. Kang, S. Wallace, D. Luciano, K. Ballaban-Gil, I. Klotz, E. Dicker, D. A. Greenberg, unpublished data).
Another observation is that, in common disease studies, a parochial analysis of linkage data can miss evidence for linkage (Hodge et al. 1993). Whether the difference in θm and θf is due to biologic factors or to ascertainment bias, the fact that the linkage could have been missed without the data exploration afforded by the broader analysis suggests that one must take a more expansive view of the methods used to find linkage. Such an approach may be necessary to resolve inevitable contradictory results from different studies.
Acknowledgments
This work was supported in part by NIH grants NS27941, DK31775, NS37466, MH48858 and by a grant from the Epilepsy Foundation of America to (M.D.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Cooperative Human Linkage Center (CHLC), http://www.chlc.org/ [DOI] [PubMed]
- GenBank, http://www.ncbi.nlm.nih.gov/ (for CA.RING3 [accession number AF107699])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for JME [MIM 254770]) [PubMed]
References
- Abreu PC, Greenberg DA, Hodge SE (1999) Direct power comparisons between single LOD scores and NPL scores for linkage analysis in complex traits. Am J Hum Genet 65:847–857 [DOI] [PMC free article] [PubMed]
- Commission on Classification and Terminology of the International League Against Epilepsy. (1989) Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia 30:389–399 [DOI] [PubMed]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Durner M, Janz D, Zingsem J, Greenberg DA (1992) Possible association of juvenile myoclonic epilepsy with HLA-DRw6. Epilepsia 33:814–816 [DOI] [PubMed]
- Durner M, Sander T, Greenberg DA, Johnson K, Beck-Mannagetta G, Janz D (1991) Localization of idiopathic generalized epilepsy on chromosome 6p in families of juvenile myoclonic epilepsy patients. Neurology 41:1651–1655 [DOI] [PubMed]
- Durner M, Shinnar S, Resor SR, Moshe SL, Rosenbaum R, Cohen J, Harden C, et al. No evidence for a major susceptibility locus for juvenile myoclonic epilepsy on chromosome 15q. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Durner M, Zhou G, Fu D, Abreu P, Shinnar S, Resor SR, Moshe SL, et al (1999) Evidence for linkage of adolescent-onset idiopathic generalized epilepsies to chromosome 8 and genetic heterogeneity. Am J Hum Genet 64:1411–1419 [DOI] [PMC free article] [PubMed]
- Elmslie FV, Rees M, Williamson MP, Kerr M, Kjeldsen MJ, Pang KA, Sundqvist A, et al (1997) Genetic mapping of a major susceptibility locus for juvenile myoclonic epilepsy on chromosome 15q. Hum Mol Genet 6:1329–1334 [DOI] [PubMed]
- Elmslie FV, Williamson MP, Rees M, Kerr M, Kjeldsen MJ, Pang KA, Sundqvist A, et al (1996) Linkage analysis of juvenile myoclonic epilepsy and microsatellite loci spanning 61 cM of human chromosome 6p in 19 nuclear pedigrees provides no evidence for a susceptibility locus in this region. Am J Hum Genet 59:653–663 [PMC free article] [PubMed]
- Greenberg DA (1989) Inferring mode of inheritance by comparison of lod scores. Am J Med Genet 34:480–486 [DOI] [PubMed]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD score analysis. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed]
- Greenberg DA, Delgado-Escueta AV, Widelitz H, Abad P, Park MS (1989) Strengthened evidence for linkage of juvenile myoclonic epilepsy to HLA and BF: Human Gene Mapping 10. Cytogenet Cell Genet 51:1008 [Google Scholar]
- Greenberg DA, Delgado-Escueta AV, Widelitz H, Sparkes RS, Treiman L, Maldonado HM, Terasaki PI, et al (1988) Juvenile myoclonic epilepsy (JME) may be linked to the BF and HLA loci on human chromosome 6. Am J Med Genet 31:185–192 [DOI] [PubMed]
- Greenberg DA, Doneshka P (1996) The partitioned association-linkage (PAL) test: distinguishing `necessary' from `susceptibility' loci. Genet Epidemiol 13:243–252 [DOI] [PubMed]
- Greenberg DA, Durner M, Delgado-Escueta AV (1992) The evidence for several genetic influences in the expression of generalized epilepsy. Neurology 42(suppl):56–62 [PubMed]
- Greenberg DA, Durner M, Resor S, Rosenbaum D, Shinnar S (1995) The genetics of idiopathic generalized epilepsies of adolescent onset: differences between juvenile myoclonic epilepsy and epilepsy with random grand mal and with awakening grand mal. Neurology 45:942–946 [DOI] [PubMed]
- Greenberg DA, Durner D, Shinnar S, Resor S, Cohen J, Zhou G, Tomasini L, et al (1998) Evidence from linkage analysis of heterogeneity within the JME syndrome. Epilepsia 39 (suppl 6):144 [Google Scholar]
- Greenberg DA, Durner M, Shinnar S, Resor S, Rosenbaum D, Klotz I, Dicker E, et al (1996) Association of HLA class II alleles in patients with juvenile myoclonic epilepsy compared to patients with other forms of adolescent onset generalized epilepsy. Neurology 47:750–755 [DOI] [PubMed]
- Hodge SE, Abreu P, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed]
- Hodge SE, Durner M, Vieland VJ, Greenberg DA (1993) Better data analysis through data exploration. Am J Hum Genet 53:775–776 [PMC free article] [PubMed]
- Janz D, Beck MG, Sander T (1992) Do idiopathic generalized epilepsies share a common susceptibility gene? Neurology 42:48–55 [PubMed]
- Klotz R, Hardin C, Durner M, Greenberg DA (1999) Maternal and paternal transmission of juvenile myoclonic epilepsy and non-JME idiopathic generalized epilepsy. Epilepsia 40, suppl 7:235 [Google Scholar]
- Kruglyak L, Daly MJ, Reeve DM, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed]
- Lander ES, Lincoln SE (1988) The appropriate threshold for declaring linkage when allowing sex-specific recombination rates. Am J Hum Genet 43:396–400 [PMC free article] [PubMed]
- Liu AW, Delgado-Escueta AV, Gee MN, Serratosa JM, Zhang QW, Alonso ME, Medina MT, et al (1996) Juvenile myoclonic epilepsy in chromosome 6p12-p11: locus heterogeneity and recombinations. Am J Med Genet 63:438–446 [DOI] [PubMed]
- Liu AW, Delgado-Escueta AV, Serratosa JM, Alonso ME, Medina MT, Gee MN, Cordova S, et al (1995) Juvenile myoclonic epilepsy locus in chromosome 6p21.2-p11: linkage to convulsions and electroencephalography trait. Am J Hum Genet 57:368–381 [PMC free article] [PubMed]
- Moen T, Brodtkorb E, Michler RP, Holst A (1995) Juvenile myoclonic epilepsy and human leukocyte antigens. Seizure 4:119–122 [DOI] [PubMed]
- Morton NE (1955) Sequential tests for the detection of linkage. Am J Hum Genet 7:277–318 [PMC free article] [PubMed] [Google Scholar]
- Neubauer BA, Fiedler B, Himmelein B, Kampfer F, Lassker U, Schwabe G, Spanier I, et al (1998) Centrotemporal spikes in families with rolandic epilepsy: linkage to chromosome 15q14. Neurology 51:1608–1612 [DOI] [PubMed]
- Obeid T, el Rab MO, Daif AK, Panayiotopoulos CP, Halim K, Bahakim H, Bamgboye E (1994) Is HLA-DRW13 (W6) associated with juvenile myoclonic epilepsy in Arab patients? Epilepsia 35:319–321 [DOI] [PubMed]
- Ott J (1974) A computer program for linkage analysis of general human pedigrees. Am J Hum Genet 28:528–529 [PMC free article] [PubMed] [Google Scholar]
- ——— (1991) Analysis of human genetic linkage. Revised ed. Johns Hopkins, Baltimore, pp 212–213 [Google Scholar]
- Ottman R, Annegers JF, Hauser WA, Kurland LT (1988) Higher risk of seizures in offspring of mothers than of fathers with epilepsy. Am J Hum Genet 43:257–264 [PMC free article] [PubMed]
- Ottman R, Hauser WA, Susser M (1985) Genetic and maternal influences on susceptibility to seizures: an analytic review. Am J Epidemiol 122:923–939 [DOI] [PubMed]
- Sander T, Bockenkamp B, Hildmann T, Blasczyk R, Kretz R, Wienker TF, Volz A, et al (1997a) Refined mapping of the epilepsy susceptibility locus EJM1 on chromosome 6. Neurology 49:842–847 [DOI] [PubMed]
- Sander T, Kretz R, Williamson MP, Elmslie FV, Rees M, Hildmann T, Bianchi A, et al (1997b) Linkage analysis between idiopathic generalized epilepsies and the GABA(A) receptor alpha5, beta3 and gamma3 subunit gene cluster on chromosome 15. Acta Neurol Scand 96:1–7 [PubMed]
- Weissbecker KA, Durner M, Janz D, Scaramelli A, Spence MA (1991) Confirmation of linkage between juvenile myoclonic epilepsy and the HLA-region on chromosome 6. Am J Med Genet 38:32–36 [DOI] [PubMed]
- Whitehouse WP, Rees M, Curtis D, Sundqvist A, Parker K, Chung E, Baralle D, et al (1993) Linkage analysis of idiopathic generalized epilepsy (IGE) and marker loci on chromosome 6p in families of patients with juvenile myoclonic epilepsy: no evidence for an epilepsy locus in the HLA region. Am J Hum Genet 53:652–662 [PMC free article] [PubMed]