Summary
Recent studies suggest that hereditary prostate cancer (PRCA) is a complex disease, involving multiple susceptibility genes and variable phenotypic expression. Through linkage analysis, potential prostate cancer susceptibility loci have been mapped to 3 regions on chromosome 1. To investigate the reported linkage to these regions, we conducted linkage studies on 144 PRCA families by using microsatellite markers in regions 1q24-25 (HPC1) and 1q42.2-43 (PCAP). We also examined the 1p36 (CAPB) region in 13 PRCA families with at least one case of brain cancer. No significant evidence of linkage to the HPC1 or PCAP region was found when the entire data set was analyzed. However, weak evidence for linkage to HPC1 was observed in the subset of families with male-to-male transmission (n=102; maximum multipoint nonparametric linkage [NPL] 1.99, P=.03). Weak evidence for linkage with heterogeneity within this subset was also observed (HLOD 1.21, P=.02), with ∼20% of families linked. Although not statistically significant, suggestive evidence for linkage to PCAP was observed for the families (n=21) that met the three criteria of male-to-male transmission, average age of diagnosis <66 years, and ⩾5 affected individuals (maximum multipoint NPL 1.45, P=.08). There was no evidence for linkage to CAPB in the brain cancer–prostate cancer subset. These results strengthen the argument that prostate cancer is a heterogeneous disease and that multiple genetic and environmental factors may be important for its etiology.
Introduction
Prostate cancer (MIM 176807) is one of the most common human cancers, occurring in as many as 15% of men in the United States (Kosary et al. 1995). Although the majority of cases of prostate cancer are sporadic, it has long been recognized that familial clustering exists, with an increased relative risk occurring in relatives of affected men (Woolf 1960; Cannon et al. 1982; Meikle and Stanish 1982; Carter et al. 1990; Steinberg et al. 1990; Spitz et al. 1991; Goldgar et al. 1994; Whittemore et al. 1995). Segregation analysis of prostate cancer suggests the presence of at least 1 dominant susceptibility locus that may account for up to 10% of all prostate cancers (Carter et al. 1992; Schaid et al. 1998). Although genetic linkage analysis is a powerful technique for the identification of disease susceptibility loci, it is confounded by several factors in prostate cancer families. These factors include a late age of onset, a high phenocopy rate, and a lack of distinguishing features between the hereditary and sporadic forms of the disease.
In spite of these complications, three presumed prostate cancer susceptibility loci, as well as a rare prostate cancer–brain cancer susceptibility locus, have been mapped through linkage studies of high-risk prostate cancer families. Starting with a genomewide linkage screen on prostate cancer families with at least three affected first-degree relatives, Smith et al. (1996) mapped a locus for hereditary prostate cancer (HPC1 [MIM 601518]) to chromosome 1q24-25. The evidence for linkage to HPC1 was provided mostly by large pedigrees (more than five affected) with an early age of diagnosis (⩽65 years [Gronberg et al. 1997a]). Subsequently, Berthon et al. (1998) localized a second susceptibility locus, PCAP (HPC2 [MIM 602759]), distal to HPC1 at 1q42.2-43. A third locus, HPCX (MIM 300147), at Xq27-28 (Xu et al. 1998) was reported in a combined study with a total of 360 families. More recently, evidence for a prostate cancer–brain cancer susceptibility locus, CAPB at 1p36 (MIM 603688), was reported by Gibbs et al. (1999b), who used linkage studies in high-risk prostate cancer families with at least one family member with primary brain cancer.
Several confirmation studies of the HPC1 locus have been reported, with varying results. Using nonparametric methods, two groups (Cooney et al. 1997; Hsieh et al. 1997) confirmed linkage in their prostate cancer families to chromosome 1q24-25. However, the evidence in both of these studies was weak. Three other studies (McIndoe et al. 1997; Berthon et al. 1998; Eeles et al. 1998) found no evidence of linkage to this locus by use of both parametric and nonparametric methods. Neither of two recent confirmation studies of the PCAP (HPC2) locus found evidence for linkage with use of either parametric or nonparametric methods (Gibbs et al. 1999a; Whittemore et al. 1999).
Current data regarding the genetics of hereditary prostate cancer suggests that multiple susceptibility genes are involved and that phenotypic expression is highly variable. In an endeavor to confirm the reported linkage to the various chromosome 1 regions, we conducted linkage studies on 144 prostate cancer families by using microsatellite markers in the regions of 1q24-25 (HPC1) and 1q42.2-43 (PCAP). Additionally, we examined the 1p36 (CAPB) region for linkage in 13 prostate cancer families that also contained at least one case of brain cancer.
Methods
Family Ascertainment
All men who received a radical prostatectomy for clinically localized prostate cancer in the Department of Urology, or who received radiation therapy in the Division of Radiation Oncology at the Mayo Clinic (Rochester, MN), were sent a family cancer history survey (Schaid et al. 1998). A total of 12,675 surveys were sent on two separate occasions: March 1995 and July 1997. On the basis of family history, 196 families were identified for further follow-up. More detailed family histories were obtained over the telephone, and three- to four-generation pedigrees were constructed. From this group, a total of 144 families having a minimum of three men affected with prostate cancer were collected for linkage studies. For 72 of the families, blood was collected from as many family members as possible, including a minimum of three living affected men. For the remaining 72 families that met the selection criteria, blood was collected only on affected sib pairs, because the other affected family members were deceased. All men who contributed a blood specimen and who had prostate cancer had their cancers verified by review of medical records, particularly pathology reports. We were unable to review medical records for deceased individuals. The average age of diagnosis was 66.5 years (range, 54–77 years), with 67 pedigrees having an average age of diagnosis <66 years. There were 47 pedigrees with at least five affected men. The average number of affected men per pedigree was 4.2 (range, 3–11), the average number of affected men with blood specimens per pedigree was 2.7 (range, 2–7), and the average number of total blood specimens per pedigree was 3.4 (range, 2–12).
A subset of families was identified that contained at least one case of brain cancer in the pedigree. Although an effort was made to determine whether the brain cancer was a primary tumor versus a metastasis, this was not possible in all cases. The pedigrees that contained known metastatic brain cancer were excluded. This resulted in 11 families from the 144 pedigrees described earlier, plus two additional pedigrees that were obtained subsequently, resulting in a total of 13 prostate cancer–brain cancer families.
The research protocol and informed consent forms were approved by the Institutional Review Board at the Mayo Clinic. DNA was isolated from peripheral blood lymphocytes by use of standard methods.
Genotyping
For the HPC1 locus (1q24-25), linkage analysis was done with six polymorphic microsatellite markers: D1S452, D1S212, D1S466, D1S158, D1S422, and D1S413 (Genome Database). For the PCAP (HPC2) locus (1q42.2-43), linkage analysis was done with six additional markers: D1S235, D1S2678, D1S2785, D1S2842, D1S2850, and D1S321. Four markers, D1S1597, D1S402, D1S407, and D1S507, mapping to the CPAB region (1p36) were used for linkage analysis on the subset of 13 families with at least one case of brain cancer.
Forward primers were labeled with phosphoramidite dyes. Each 15-μl reaction contained 25 ng of genomic DNA, 200 mM dNTPs, 8 mM each primer, 0.5 U AmpliTaq Gold (PE Biosystems), and 1.5–2.5 mM MgCl2. Reactions were cycled in either a PE Biosystems GeneAmp PCR System 9600 or an MJS Tetrad Cycler as follows: 10 min at 95°C, then 35 cycles of 30 s at 95°C, 30 s at 58°C or 55°C, 30 s at 72°C; followed by an extension step of 10 min at 72°C. PCR reactions were held at 5°C until analysis. The PCR products were resolved on a 5% denaturing polyacrylamide gel and detected by use of an ABI 377 DNA sequencer. Genotypes were analyzed by use of ABI Genescan 2.1 and ABI Genotyper 2.0.
Linkage Analysis
We performed genetic linkage analyses by both parametric and model-free methods. The parametric two-point LOD scores were computed by the LINKAGE package (FASTLINK) by use of an assumed prostate cancer susceptibility allele frequency of .003 and an autosomal-dominant model. We performed two different analyses using two different models of age-dependent penetrances. Model A is essentially the same as that used by Smith et al. (1996) in the first reported linkage finding for hereditary prostate cancer. In brief, model A assumed a 15% phenocopy rate; affected men had penetrances of .001 and 1.0 for noncarriers and carriers, respectively; the lifetime penetrances for unaffected men at age ⩾75 years were 16% for noncarriers and 63% for carriers; and unaffected men at age <75 years and all women were not informative (i.e., unknown phenotype). Model B was more refined, with 11 age-dependent liability classes based on published segregation models (Carter et al. 1992) and SEER data, as implemented by Xu et al. (in press). Linkage in the presence of heterogeneity was assessed by use of Smith’s admixture test for heterogeneity (HOMOG program). Multipoint LOD scores were computed with the GENEHUNTER program. Because the inheritance of prostate cancer is complex, we also performed multipoint identical-by-descent model–free linkage analyses for affected pedigree members by use of the Z-all statistic in the GENEHUNTER program. Allele frequencies were estimated from the data set.
Results
HPC1 (1q24-25) Locus
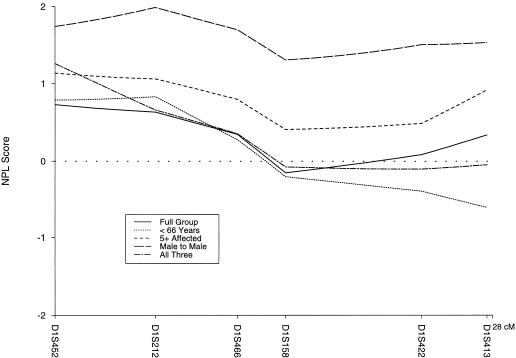
The parametric two-point LOD scores (model A) for the six markers in the HPC1 region are shown in table 1. There was no significant evidence of linkage for the entire data set with either model A or model B. The maximum cumulative two-point LOD score was 0.40 at a recombination fraction of .3 for D1S413 with model A. By using model B, we found that the peak LOD score was 0.23 at a recombination fraction of .2 for this same marker. The maximum multipoint LOD score was −40.54 for model A and −1.77 for model B. Generally, results from models A and B were consistent throughout all analyses, with model B results being less extreme (i.e., LOD scores closer to 0). When Smith’s admixture test (HOMOG) was used, there was no evidence for linkage and heterogeneity with either model. However, there were three families with maximum multipoint LOD scores >1 (model A), indicating that a few of the families may be linked to the HPC1 region (on the basis of simulations under the null hypothesis, less than one family would be expected to have a LOD score >1). The nonparametric methods also failed to show, in our data set, significant evidence of linkage to the HPC1 locus with a maximum multipoint nonparametric linkage (NPL) score 0.73 (P=.23; fig. 1).
Table 1.
Two-Point LOD Scores for Model A[Note]
|
LOD Score at Recombination Fraction = |
||||||||
| Marker | Intermarker Distance(cM) | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| HPC1 (n = 144): | ||||||||
| D1S452 | −42.21 | −29.55 | −14.13 | −7.1 | −1.76 | −.24 | .02 | |
| D1S212 | 5.5 | −53.04 | −38.22 | −19.67 | −10.65 | −3.2 | −.69 | −.02 |
| D1S466 | 5.1 | −33.43 | −24.05 | −12.22 | −6.57 | −1.97 | −.46 | −.06 |
| D1S158 | 2.4 | −68.9 | −48.44 | −24.14 | −13.08 | −4.22 | −1.16 | −.2 |
| D1S422 | 4.4 | −42.41 | −31.06 | −16.18 | 9.06 | −3.12 | −.94 | −.16 |
| D1S413 | 4.9 | −25.4 | −17.35 | −7.44 | −3.04 | −.07 | .4 | .18 |
| PCAP (n = 144): | ||||||||
| D1S235 | −51.75 | −39.28 | −22.44 | −13.68 | −5.54 | −1.97 | −.43 | |
| D1S2678 | 1.6 | −38 | −29.29 | −17.09 | −10.53 | −4.29 | −1.52 | −.33 |
| D1S2850 | 0 | −39.55 | −29.21 | −15.47 | −8.73 | −3.05 | −.94 | −.18 |
| D1S2785 | 9.4 | −64.9 | −46.75 | −24.67 | −14.21 | −5.23 | −1.7 | −.34 |
| D1S321 | 2.3 | −18.94 | −14.15 | −7.64 | −4.27 | −1.34 | −.31 | −.03 |
| D1S2842 | 5.3 | −50 | −36.82 | −19.44 | −10.75 | −3.49 | −.94 | −.15 |
| CAPB (n = 13): | ||||||||
| D1S1597 | −4.41 | −3.24 | −1.7 | −.97 | −.36 | −.12 | −.03 | |
| D1S402 | 1.19 | −9.9 | −6.95 | −3.36 | −1.73 | −.45 | −.05 | .03 |
| D1S407 | 2.73 | −2.71 | −2.17 | −1.41 | −.94 | −.43 | −.16 | −.03 |
| D1S507 | .1 | −5.25 | −3.73 | −1.82 | −.83 | −.06 | .12 | .09 |
Note.— Scores for model B were consistent in terms of conclusions but tended to be less extreme than those for model A for both negative and positive LOD scores (see text).
Figure 1 .
Multipoint NPL scores for the whole data set (n=144) and four subsets, on a six-marker map of the HPC1 region. The subsets are average age of diagnosis <66 years (n=67), at least five affected individuals (n=47), male-to-male transmission (n=102), and a combination (n=21) of <66 years, at least five affected, and male-to-male transmission.
Families were stratified by the presence (n=102) or absence (n=42) of male-to-male transmission, average age at diagnosis (<66 years, n=67, vs. ⩾66 years, n=77), and number of affected individuals (fewer than five, n=97, vs. at least five, n=47) and then reexamined for linkage. The subset of pedigrees that showed male-to-male transmission, consistent with autosomal inheritance, had a maximum multipoint NPL score of 1.99 (P=.03) at D1S212. Using parametric analysis, we did not detect any evidence for linkage when homogeneity was assumed, but there was weak evidence for linkage after allowing for heterogeneity (HLOD score 1.21, P=.02). If we assume heterogeneity, an estimated 20% of the male-to-male transmission families may show linkage to the HPC1 region. The subset of families (n=21) that met all three criteria (the presence of male-to-male transmission, average age at diagnosis <66 years, and at least five affected individuals) had a peak multipoint NPL score of 1.26 (P=.11). In addition, evidence for linkage and heterogeneity in this subset was observed with an HLOD score of 1.39 (P=.01), with ∼44% of these families linked. Stratification by average age of diagnosis did not result in significantly increased LOD scores for any of the HPC1 markers. When stratified by number of affected individuals per family, a maximum multipoint NPL score of 1.14 (P=.13) at marker D1S452 was attained for the subset of families with at least five affected individuals. Figure 1 shows the multipoint NPL graphs for the various subsets.
PCAP (HPC2 [1q42.2-43]) Locus
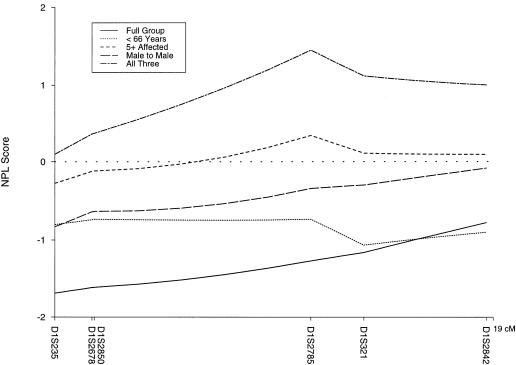
The studies on the PCAP locus also failed to show evidence for linkage for the entire data set. There were no positive cumulative two-point LOD scores with use of either model (table 1), nor was there evidence for heterogeneity by use of HOMOG. The multipoint LOD scores were negative for both models, with a maximum of −53.33 for model A and −5.16 for model B. Nonparametric methods also failed to show evidence of linkage for the whole group, with a maximum NPL score of −0.76 (P=.78; fig. 2).
Figure 2 .
Multipoint NPL scores for the whole data set (n=144) and for four subsets, on a six-marker map of the PCAP region. The subsets are average age of diagnosis <66 years (n=67), at least five affected individuals (n=47), male-to-male transmission (n=102), and a combination (n=21) of <66 years, at least five affected, and male-to-male transmission.
When stratified, data for the 21 families that met all three criteria (the presence of male-to-male transmission, average age at diagnosis <66 years, and at least five affected individuals) had a peak multipoint NPL score of 1.45 (P=.08). Stratification of the families by age of diagnosis did not result in higher LOD scores for the PCAP locus. Positive, but not significant, multipoint NPL scores (maximum 0.34, P=.36) were observed in the group of 47 families with at least five affected members. When stratified by the occurrence of brain cancer, the families with at least one case of brain cancer had a multipoint NPL score of 1.29 (P=.10) at D1S2785. The multipoint NPL graphs for the PCAP region for these various subsets are shown in figure 2.
CAPB (1p36) Locus
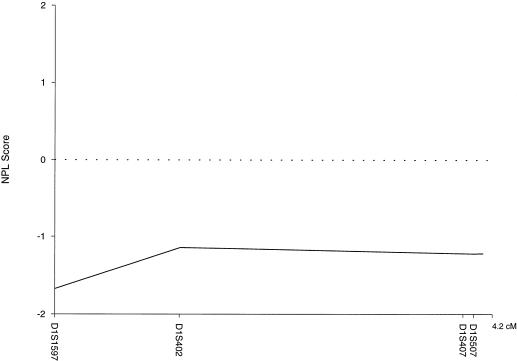
For the 13 families with prostate cancer and brain cancer, there was no evidence of linkage to the 1p36 locus by use of either parametric or nonparametric methods. The two-point LOD scores (model A) for each marker are shown in table 1. The multipoint LOD scores were all negative for both models (model A maximum −8.35, model B maximum −0.35). The multipoint NPL scores were also negative for the full group, with a maximum of −1.08 (P=.88; fig. 3).
Figure 3 .
Multipoint NPL scores for the brain cancer–prostate cancer subset (n=13) on a 4-marker map of the CAPB region
Discussion
In the present study, no significant evidence for linkage of familial prostate cancer to HPC1 (1q24-25) was observed for the whole data set. There may be several reasons for this: disease locus heterogeneity or the use of incorrect analysis models, both of which diminish statistical power. It may be that a small fraction of families are linked to HPC1 but were not detected because of low statistical power. That this might be the case is supported by the results of a meta-analysis of 772 families for linkage to the HPC1 region (Xu et al., in press). This analysis revealed that the proportion of families linked to the HPC1 locus is likely to be considerably less (as low as 6%) than the 34% originally reported. Also, the current study contained a relatively small number of pedigrees that appear to provide the greatest evidence of linkage to HPC1 (Gronberg et al. 1997a, 1997b; Xu et al., in press): large pedigrees (at least five affected sampled in two generations) with an early age of onset (mean <66 years) and male-to-male transmission. There were three families in our study that had multipoint LOD scores ⩾1 in the HPC1 region, although only two of these satisfied all three criteria.
When the data set was stratified into several groups, however, the subset with male-to-male transmission showed weak evidence for linkage to HPC1, with a maximum NPL score of 1.99 (P=.03). This is consistent with the meta-analysis for linkage to the HPC1 region that also showed the strongest evidence for linkage in the male-to-male–transmission families (Xu et al., in press). Evidence for linkage to HPC1 in the subset of families with male-to-male transmission was not observed by use of parametric methods with homogeneity, although models that allowed for heterogeneity did show some evidence for linkage in this subset, with an estimated 20% of families linked. However, the estimate of the proportion of families linked may be incorrect because the true model is unknown. Of interest, the only two confirmatory studies that have supported linkage of familial prostate cancer to the HPC1 locus have done so by use of nonparametric methods similar to those used in this study (Cooney et al. 1997; Hsieh et al. 1997). Our two-point LOD score results for the parametric models A and B were consistent in terms of conclusions, but model B results tended to be less extreme than those of model A for both negative and positive LOD scores. The biggest discrepancy was between the multipoint LOD scores and the multipoint NPL statistic. It is well known that parametric multipoint LOD scores tend to be spuriously negative when the parameters used for analysis (e.g., allele frequency, mode of inheritance, penetrance) are not correct (Risch and Giuiffra 1992). This is because nonrecombinant offspring tend to be misclassified as double recombinants, a rare event, resulting in the disease locus being “pushed off” the multipoint marker map. In contrast, the parametric two-point analyses tend to be more robust to model misspecification. The NPL statistic is ideal for complex traits (Ott 1996). The advantage of the NPL statistic is that it is not based on unknown, yet assumed, genetic models, but rather on the comparison of the observed versus expected sharing of chromosomal regions identical by descent among affected relatives. For these reasons, we have relied on the parametric two-point LOD scores and the multipoint NPL statistics for our main conclusions.
We observed no significant evidence of linkage to PCAP (HPC2 [1q42.2-43]) for the entire data set, with either parametric or model-free analyses. Neither this study nor that of Gibbs et al. (1999a) has confirmed linkage to the 1q42.2-43 (PCAP) locus. Therefore, the proportion of families linked to this locus is likely to be considerably <50%, as was estimated by Berthon et al. (1998) in the original report. We did find suggestive evidence for linkage to the PCAP region (maximum NPL score 1.45; P=.08) in the subset of families that satisfied all three criteria of male-to-male transmission, large size (at least five affected sampled in two generations), and early age of onset (mean <66 years). Gibbs et al. (1999a) also found suggestive, but not significant, evidence for linkage in their subset of families with at least five affected individuals (maximum NPL score 1.2, P=.1).
There were 13 families with both prostate cancer and brain cancer present. However, there was no evidence for linkage to CAPB (1p36) in this subset. Of interest is the observation that the prostate cancer–brain cancer subset had multipoint NPL scores suggestive of linkage to the PCAP region, which may imply that a general tumor-suppressor gene is involved in this region. However, although the known cases of brain metastases were excluded, not all of the remaining cases were confirmed to be primary brain tumors. Therefore, these results are not definitive.
It is well recognized that familial prostate cancer is heterogeneous and that several loci, as well as possibly other genetic and environmental factors, are very likely to play a role in its etiology. The heterogeneity of this disease is underscored by our data set, which failed to show linkage at two of the chromosome 1 loci but did show linkage to the Xq27-28 region (with stronger evidence in a combined data set) with an estimated 16% of families linked (Xu et al. 1998). Also, a subset of our families showed weak evidence of linkage to the HPC1 region. Population differences reflecting different involved loci may further contribute to the heterogeneity of this disease. For example, two black pedigrees demonstrated positive LOD scores in the original HPC1 report (Smith et al. 1996). The present study population was entirely white, with one Hispanic and no black families. Other studies failing to report linkage to the HPC1 locus also tended to be less diverse. However, the two studies (Cooney et al. 1997; Hsieh et al. 1997) that supported linkage to the HPC1 locus had more ethnically diverse study populations, which included 6/59 and 6/79 black families, respectively.
In conclusion, our results suggest that a small subset of families, characterized by male-to-male transmission, may be linked to the HPC1 region. In addition, these results strengthen the argument that prostate cancer is a complex, heterogeneous disease with multiple genes and factors contributing to its development. Future meta-analyses of the PCAP and CAPB regions, as well as regions identified in the future, will be necessary to gain an accurate estimate of their involvement in familial prostate cancer.
Acknowledgments
We acknowledge the technical assistance of Mr. Isaac Amundson, and we would also like to thank Marcia Brumm for her study coordination efforts. This study was supported by NIH grants R01CA72818 and CA 72818 and the Mayo Clinic Cancer Center.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genome Database, http://gdbwww.gdb.org (for markers D1S452, D1S212, D1S466, D1S158, D1S422, D1S413, D1S235, D1S2678, D1S2850, D1S2785, D1S321, D1S2842, D1S1597, D1S402, D1S407, and D1S507)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nim.nih.gov/omim (for prostate cancer [MIM 176807], HPC1 [MIM 601518], HPC2 [MIM 602759], HPCX [MIM 300147], and CAPB [MIM 603688])
References
- Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, Wohr G, Latil A, et al (1998) Predisposing gene for early-onset prostate cancer, localized on chromosome 1q42.2-43. Am J Hum Genet 62:1416–1424 [DOI] [PMC free article] [PubMed]
- Cannon L, Bishop DT, Skolnick M, Hunt S, Lyon JL, Smart CR (1982) Genetic epidemiology of prostate cancer in the Utah Mormon geneology. Cancer Surv 1:47–69 [Google Scholar]
- Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC (1992) Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci USA 89:3367–3371 [DOI] [PMC free article] [PubMed]
- Carter BS, Carter HB, Isaacs JT (1990) Epidemiologic evidence regarding predisposing factors to prostate cancer. Prostate 16:187–197 [DOI] [PubMed]
- Cooney KA, McCarthy JD, Lange E, Huang L, Miesfeldt S, Montie JE, Oesterling JE, et al (1997) Prostate cancer susceptibility locus on chromosome 1q: a confirmatory study. J Natl Cancer Inst 89:955–959 [DOI] [PubMed]
- Eeles RA, Durocher F, Edwards S, Teare D, Badzioch M, Hamoudi R, Gill S, et al (1998) Linkage analyses of chromosome 1q markers in 136 prostate cancer families. Am J Hum Genet 62:653–658 [DOI] [PMC free article] [PubMed]
- Gibbs M, Chakrabarti L, Stanford JL, Goode EL, Kolb S, Schuster EF, Buckley VA, et al (1999a) Analysis of chromosome 1q42.2-43 in 152 families with high risk of prostate cancer. Am J Hum Genet 64:1087–1095 [DOI] [PMC free article] [PubMed]
- Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, et al (1999b) Evidence for a rare prostate cancer-susceptibility locus at chromosome 1p36. Am J Hum Genet 64:776–787 [DOI] [PMC free article] [PubMed]
- Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH (1994) Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst 86:1600–1608 [DOI] [PubMed]
- Gronberg H, Isaacs SD, Smith JR, Carpten JD, Bova GS, Freije D, Xu J, et al (1997a) Characteristics of prostate cancer in families potentially linked to the hereditary prostate cancer 1 (HPC1) locus. JAMA 278:1251–1255 [DOI] [PubMed]
- Gronberg H, Xu J, Smith JR, Carpten JD, Isaacs SD, Freiji D, Bova GS, et al (1997b) Early age at diagnosis in families provides evidence of linkage to the hereditary prostate cancer locus (HPC1) on chromosome 1. Cancer Res 57:4707–4709 [PubMed]
- Hsieh C-L, Oakley-Girvan I, Gallagher RP, Wu AH, Kolonel LN, Teh C-Z, Halpern J, et al (1997) Re: Prostate cancer susceptibility locus on chromosome 1q: a confirmatory study. J Natl Cancer Inst 89(letter):1893–1894 [DOI] [PubMed]
- Kosary CL, Ries LAG, Miller BA, Hankey BF, Harras A, Edwards BK (eds) (1995) SEER cancer statistics review, 1973–1991: tables and graphs. NIH Publ. No. 96-2789. National Cancer Institute, Bethesda. [Google Scholar]
- McIndoe RA, Stanford JL, Gibbs M, Jarvik GP, Brandzel S, Neal CL, Li S, et al (1997) Linkage analysis of 49 high-risk families does not support a common familial prostate cancer-susceptibility gene at 1q24–25. Am J Hum Genet 61:347–353 [DOI] [PMC free article] [PubMed]
- Meikle AW, Stanish WM (1982) Familial prostatic cancer risk and low testosterone. J Clin Endocrinol Metab 54:1104–1108 [DOI] [PubMed]
- Ott J (1996) Complex traits on the map. Nature 379:772–773 [DOI] [PubMed]
- Risch N, Giuiffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed]
- Schaid DJ, McDonnell SK, Blute ML, Thibodeau SN (1998) Evidence for autosomal dominant inheritance of prostate cancer. Am J Hum Genet 62:1425–1438 [DOI] [PMC free article] [PubMed]
- Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, et al (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genomewide search. Science 274:1371–1374 [DOI] [PubMed]
- Spitz MR, Currier RD, Fueger JJ, Babaian RJ, Newell GR (1991) Familial patterns of prostate cancer: a case-control analysis. J Urol 146:1305–1307 [DOI] [PubMed]
- Steinberg GD, Carter BS, Beaty TH, Childs B, Walsh PC (1990) Family history and the risk of prostate cancer. Prostate 17:337–347 [DOI] [PubMed]
- Whittemore AS, Lin IG, Oakley-Girvan I, Gallagher RP, Halper J, Kolonel LN, Wu AH, et al (1999) No evidence of linkage for chromosome 1q42.2–43 in prostate cancer. Am J Hum Genet 65:254–256 [DOI] [PMC free article] [PubMed]
- Whittemore AS, Wu AH, Kolonel LN, John EM, Gallagher RP, Howe GR, West DW, et al (1995) Family history and prostate cancer risk in black, white and Asian men in the United States and Canada. Am J Epidemiol 141:732–740 [DOI] [PubMed]
- Woolf CM (1960) An investigation of the familial aspects of carcinoma of the prostate. Cancer 13:739–744 [DOI] [PubMed]
- Xu J, International Consortium for Prostate Cancer Genetics. Combined analysis of hereditary prostate cancer linkage to 1q24-25: results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meyers D, Freije D, Isaacs S, Wiley K, Nusskern D, Ewing C, et al (Group 1) (1998) Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet 20:175–179 [DOI] [PubMed]