Summary
Generalized epilepsy with febrile seizures plus (GEFS+) is a recently recognized but relatively common form of inherited childhood-onset epilepsy with heterogeneous epilepsy phenotypes. We genotyped 41 family members, including 21 affected individuals, to localize the gene causing epilepsy in a large family segregating an autosomal dominant form of GEFS+. A genomewide search examining 197 markers identified linkage of GEFS+ to chromosome 2, on the basis of an initial positive LOD score for marker D2S294 (Z=4.4, recombination fraction [θ] = 0). A total of 24 markers were tested on chromosome 2q, to define the smallest candidate region for GEFS+. The highest two-point LOD score (Zmax=5.29; θ=0) was obtained with marker D2S324. Critical recombination events mapped the GEFS+ gene to a 29-cM region flanked by markers D2S156 and D2S311, with the idiopathic generalized epilepsy locus thereby assigned to chromosome 2q23-q31. The existence of the heterogeneous epilepsy phenotypes in this kindred suggests that seizure predisposition determined by the GEFS+ gene on chromosome 2q could be modified by other genes and/or by environmental factors, to produce the different seizure types observed.
Epilepsies are one of the most common neurological conditions, affecting ∼2% of the population (Hauser et al. 1993). The epilepsies are broadly divided into generalized and localization-related epilepsies (Commission on Classification and Terminology of the International League against Epilepsy 1989). Although the majority of generalized epilepsies are believed to be genetically determined, a number of methodological problems, including clinical and genetic heterogeneity, unclear mode of inheritance, and the possible presence of susceptibility genes that interact to determine a single phenotype, have made it difficult to identify causative genes. We recently identified a clinical and genetic subset of the generalized epilepsies characterized by febrile seizures (FSs) (MIM 121210) in most affected family members with a variety of afebrile generalized seizure types. Inheritance of this syndrome, which is known as “generalized epilepsy with febrile seizures plus (GEFS+),” appears to be complex in some families. In other families, there is evidence of a major autosomal dominant gene (Scheffer and Berkovic 1997; Singh et al. 1999). In one large family, the locus was mapped to chromosome 19q, and a missense mutation affecting the function of the β1 subunit gene (SCN1B) of the neuronal sodium channel was found (Wallace et al. 1998). However, most families with GEFS+ do not appear to have mutations in SCN1B. In the present study, we describe mapping of a second locus for GEFS+; we studied the Victorian family in which the GEFS+ syndrome was originally described (Scheffer and Berkovic 1997). Preliminary linkage data have been presented in abstract form (Lopes-Cendes et al. 1996)
We studied a large family residing in northern Victoria, Australia, whose ancestors derived from Yorkshire, United Kingdom. Forty-one family members, including 21 affected individuals with GEFS+, were included in the linkage analysis. The mean age at seizure onset was 1.6 years. Most patients had the benign-epilepsy phenotype of FS+ characterized by generalized tonic-clonic seizures (GTCS) with or without fever, with seizure offset occurring by the second decade (Scheffer and Berkovic 1997). Neurological examination and intellect were normal in all individuals except one, who had moderate intellectual disability. Electroencephalography recordings were normal in all individuals, except for three who had generalized epileptiform activity and four who had mild or moderate diffuse background slowing. This study was approved by the appropriate institutional review boards, and informed consent was obtained from all participants.
Genomic DNA was extracted from blood samples or from lymphoblastoid cell lines, by use of standard protocols (Sambrook et al. 1989). A panel of 210 dinucleotide (CA)n–repeat polymorphic markers with high heterozygosity (75%) was chosen from the 1993–94 Généthon map (Gyapay et al. 1994). Dinucleotide markers were spaced an average of 20 cM from each other, throughout the 22 autosomes. Genotyping of microsatellite markers was accomplished by means of PCR. The reaction mixture was prepared in a total volume of 13 ml, with the use of 80 ng genomic DNA, 1.25 μl 10 × buffer with 1.5 mM MgCl2, 0.65 μl BSA (2.0 mg/ml); 100 ng each oligonucleotide primer; 200 mM each dCTP, dGTP, and dTTP; 25 mM dATP; 1.5 mCi [35S]-dATP, and 0.5 UTaq DNA polymerase (PE Biosystems). Reaction samples were transferred to 96-well plates and were subjected to the following conditions: 35 cycles of denaturation for 30 s at 94°C; annealing for 30 s at 55°C–57°C, depending on the specificity of the oligonucleotide primers; and elongation for 30 s at 72°C. PCR-reaction products were electrophoresed on 6% denaturing polyacrylamide sequencing gels.
Two-point linkage analysis was performed with the use of the MLINK program, version 5.1, from the LINKAGE computer package (Lathrop et al. 1984). Precise values for Zmax were calculated with use of the ILINK program from the same computer package. LOD scores were generated on the basis of an autosomal dominant mode of inheritance, 80% penetrance (calculated from the pedigree used in the analysis), disease-allele frequency of 1:500, and allele frequencies for all allele markers calculated from the pedigree with use of the computer program ILINK (Lathrop et al. 1984). For this kindred, the maximum LOD score attainable, by use of the aforementioned parameters, is 6.5.
A genomewide search examining 190 markers identified three LOD scores >2: D2S294 (Zmax=4.4, θ = 0), D5S421 (Zmax=2.7, θ=0), and D7S495 (Zmax=2.2, θ=0.1). Linkage of GEFS+ to chromosome 2q was pursued, and a total of 24 markers were subsequently tested, to define the smallest GEFS+ candidate region. Table 1 shows the two-point LOD scores for 17 markers spanning the GEFS+ candidate region. The highest LOD score (Zmax=5.29; θ=0) was obtained with marker D2S324, assigning the GEFS+ locus to chromosome 2q23-q31.
Table 1.
Two-Point LOD Scores for 17 Markers Localized on Chromosome 2q23-q31
|
Recombination Fraction at θ = |
|||||||||
| Locus | 0 | .05 | .1 | .15 | .2 | .3 | .4 | Zmax | θmax |
| D2S142 | .99 | 1.94 | 1.97 | 1.85 | 1.68 | 1.22 | .66 | 1.98 | .078 |
| D2S284 | 1.3 | 1.18 | 1.06 | .94 | .82 | .57 | .3 | 1.3 | 0 |
| D2S306 | 1.9 | 2.82 | 2.74 | 2.52 | 2.25 | 1.6 | .85 | 2.82 | .057 |
| D2S156 | 2.15 | 3.05 | 2.96 | 2.73 | 2.43 | 1.73 | .93 | 3.05 | .056 |
| D2S354 | 4.72 | 4.26 | 3.82 | 3.4 | 2.97 | 2.1 | 1.13 | 4.72 | 0 |
| D2S111 | 5.15 | 4.71 | 4.26 | 3.78 | 3.29 | 2.26 | 1.17 | 5.15 | 0 |
| D2S124 | 3.5 | 3.2 | 2.89 | 2.58 | 2.26 | 1.58 | .84 | 3.5 | 0 |
| D2S382 | 4.31 | 3.93 | 3.54 | 3.14 | 2.74 | 1.91 | 1.02 | 4.31 | 0 |
| D2S399 | .48 | .4 | .33 | .27 | .22 | .14 | .08 | .48 | 0 |
| D2S294 | 4.4 | 4.04 | 3.65 | 3.25 | 2.84 | 2 | 1.07 | 4.4 | 0 |
| D2S335 | 4.76 | 4.32 | 3.91 | 3.51 | 3.1 | 2.22 | 1.21 | 4.76 | 0 |
| D2S333 | 1.42 | 1.23 | 1.04 | .87 | .72 | .45 | .22 | 1.4 | 0 |
| D2S324 | 5.29 | 4.72 | 4.16 | 3.63 | 3.13 | 2.15 | 1.14 | 5.29 | 0 |
| D2S384 | 3.85 | 3.52 | 3.17 | 2.82 | 2.45 | 1.69 | .89 | 3.85 | 0 |
| D2S152 | 1.9 | 1.7 | 1.52 | 1.36 | 1.2 | .87 | .48 | 1.9 | 0 |
| D2S311 | −.81 | 1.62 | 1.66 | 1.58 | 1.46 | 1.11 | .63 | 1.66 | .085 |
| D2S155 | −5.21 | .57 | 1.12 | 1.29 | 1.29 | 1.04 | .59 | 1.3 | .17 |
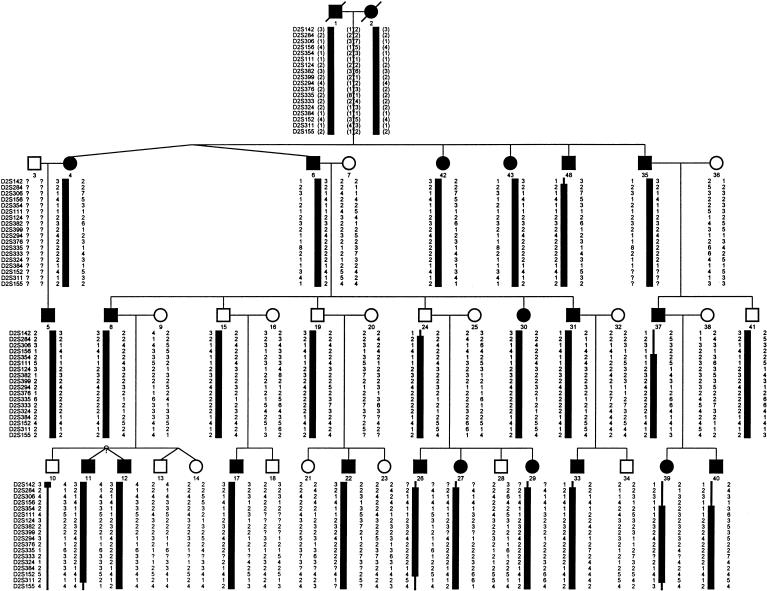
We constructed haplotypes by use of 17 markers spanning the GEFS+ candidate region (fig. 1). The centromeric boundary was defined by a recombination event between the markers D2S156 and D2S354, whereas a recombination event between markers D2S152 and D2S311 set the telomeric boundary. These critical recombination events localized the GEFS+ gene to a 29-cM region flanked by markers D2S156 and D2S311.
Figure 1.
Pedigree structure and haplotypes in the linked region of chromosome 2q in the family with GEFS+. In the first generation shown, individuals 1 and 2 are uncle and niece. The identity of the markers included in the haplotypes are shown (left) and are ordered from centromere (top) to telomere (bottom). The thick vertical lines below the affected individuals denote the disease-bearing chromosome; the recombinations are shown as thinner lines. Parentheses denote inferred genotype, and a question mark (?) denotes missing data.
To date, at least 18 putative loci have been mapped for idiopathic seizure disorders (Berkovic and Scheffer 1999); however, only four genes have been identified. All four genes are ion-channel subunits from either ligand-gated (Steinlein et al. 1995) or voltage-gated ion channels, including the SCN1B mutations described in one family with GEFS+ (Bievert et al. 1998; Singh et. al. 1998; Wallace et al. 1998). Although the candidate interval identified in our kindred remains large, a number of interesting genes map to the region. These include a cluster of sodium (Na+)-channel genes and potassium (Ka+)-channel genes (identified through an electronic-database search), as well as the GAD1 gene, which encodes for glutamate decarboxylase, an enzyme involved in the syntheses of γ-aminobutyric acid (GABA) (Bu and Tobin 1994). GABA is one of the major neurotransmitters involved in synaptic inhibition in the CNS (Blair et al. 1988). The large size of the candidate interval will require further refinement of the locus prior to the identification of the gene responsible for GEFS+ in our kindred.
The seizure patterns in GEFS+ are surprisingly heterogeneous, both in families with evidence for complex inheritance and in families like the one described here with a major autosomal dominant gene (Scheffer and Berkovic 1997; Singh et al. 1999). This may indicate that, although the predisposition for GEFS+ in this family is determined by a single gene localized on chromosome 2q23-q31, the different types of generalized seizures occurring in the family may have resulted from interactions with genetic and/or environmental modifiers.
During the review of this paper, three reports of GEFS+ kindreds with linkage to the same interval of chromosome 2 have been published (Baulac et al. 1999; Moulard et al. 1999; Peiffer et al. 1999). The phenotypes in two families (Baulac et al. 1999; Moulard et al. 1999) were reported as “GEFS+” and were similar to those of our Australian kindred. The phenotype in the third family (Peiffer et al. 1999) was reported as “FS,” but, in our opinion, the phenotype was typical of GEFS+. Thus, it appears that four unrelated families with this newly recognized but relatively common generalized epilepsy syndrome may share a single locus.
Acknowledgments
We wish to thank the members of the kindred for their cooperation in this study. The work was supported by the Medical Research Council of Canada. Dr. I.L.-C. was supported by a fellowship from the Savoy Foundation for Epilepsy Research, Montreal.
Electronic-Database Information
Accession number and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FSs [MIM 121210])
- Généthon, http://www.genethon.fr/ (for polymorphic markers)
References
- Blair LA, Levitan ES, Marshall J, Dionne VE, Barnard EA (1988) Single sub-units of the GABAA receptor form ion channels with properties of the native receptor. Science 242:577–579 [DOI] [PubMed]
- Baulac S, Gourfinkel-An I, Picard F, Rosenberg-Bourgin M, Prud'homme J-F, Baulac M, Brice A, et al (1999) A second locus for familial generalized epilepsy with febrile seizures plus maps to chromosome 2q21-q33. Am J Hum Genet 65:1078–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovic SF, Scheffer IE (1999) Genetics of the epilepsies. Curr Opin Neurol 12:177–182 [DOI] [PubMed]
- Bievert C, Schoeder BC, Kubisch C, Berkovic SF, Propping P, Jentsch TJ, Steinlein OK (1998) A potassium channel mutation in neonatal human epilepsy. Science 279:403–406 [DOI] [PubMed]
- Bu DF, Tobin AJ (1994) The exon-intron organization of the genes (GAD1 and GAD2) encoding two human glutamate decarboxylases (GAD67 and GAD65) suggests that they derive from a common ancestral GAD. Genomics 21:222–228 [DOI] [PubMed]
- Commission on Classification and Terminology of the International League against Epilepsy (1981) Proposal for revised clinical and electroencephalographic classification of epileptic seizures. Epilepsia 22:489–501 [DOI] [PubMed]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, et al (1994) The 1993–94 Généthon human genetic linkage map. Nat Genet 7:246–339 [DOI] [PubMed]
- Hauser WA, Annegers JF, Kurland LT (1993) Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935–1984. Epilepsia 34:453–468 [DOI] [PubMed]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed]
- Lopes-Cendes I, Scheffer IE, Berkovic SF, Rousseau M, Andermann E, Rouleau GA (1996) Mapping a locus for idiopathic generalized epilepsy in a large multiplex family. Epilepsia 37 Suppl 5:127 [Google Scholar]
- Moulard B, Guipponi M, Chaigne D, Mouthon D, Buresi C, Malafosse A (1999) Identification of a new locus for generalized epilepsy with febrile seizures plus (GEFS+) on chromosome 2q24-q33. Am J Hum Genet 65:1396–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer A, Thompson J, Charlier C, Otterud B, Varvil T, Pappas C, Barnitz C, et al (1999) A locus for febrile seizures (FEB3) maps to chromosome 2q23-24. Ann Neurol 46:671–678 [DOI] [PubMed]
- Sambrook J, Fritsch EF, Maniatis T (eds) (1989) Molecular cloning: a laboratory manual, 2d ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp E.3–E.4 [Google Scholar]
- Scheffer IE, Berkovic SF (1997) Generalized epilepsy with febrile seizures plus: a genetic disorder with heterogeneous clinical phenotypes. Brain 120:479–490 [DOI] [PubMed]
- Singh NA, Charlier C, Stauffer D, DuPont BR, Leach RJ, Melis R, Ronen GM, et al (1998) A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat Genet 18:25–29 [DOI] [PubMed]
- Singh R, Scheffer IE, Crossland K, Berkovic SF (1999) Generalized epilepsy with febrile seizures plus (GEFS+): a common, childhood-onset, genetic epilepsy syndrome. Ann Neurol 45:75–81 [DOI] [PubMed]
- Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, Scheffer IE, et al (1995) A missense mutation in the neuronal nicotinic acetylcholine receptor α4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 11:201–203 [DOI] [PubMed]
- Wallace RH, Wang DW, Sing R, Scheffer IE, George AL Jr , Phillips HA, Saar K, et al (1998) Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel β1 subunit gene SCN1B. Nat Genet 19:366–370 [DOI] [PubMed]