Summary
Hereditary spastic paraplegia (HSP) comprises a group of clinically and genetically heterogeneous disorders causing progressive spasticity and weakness of the lower limbs. We report a large family of French descent with autosomal dominant pure HSP. We excluded genetic linkage to the known loci causing HSP and performed a genomewide search. We found evidence for linkage of the disorder to polymorphic markers on chromosome 2q24-q34: a maximum LOD score of 3.03 was obtained for marker D2S2318. By comparison with families having linkage to the major locus of pure autosomal dominant HSP (SPG4 on chromosome 2p), there were significantly more patients without Babinski signs, with increased reflexes in the upper limbs, and with severe functional handicaps.
Hereditary spastic paraplegia (HSP) comprises a group of clinically and genetically heterogeneous disorders causing progressive spasticity and weakness of the lower limbs. The disease is characterized pathologically by axonal degeneration in the long ascending and descending tracts of the spinal cord, especially in their terminal portions (Behan and Maia 1974). The corticospinal tracts are mainly affected, but the columns of Goll and the spinocerebellar tracts are also involved. HSPs are classified according to (1) the mode of inheritance (autosomal dominant, autosomal recessive, or X linked) and (2) the neurological or nonneurological signs associated with spastic paraplegia (Harding 1981; Fink 1997). In the pure form of HSP, in addition to pyramidal signs, in the lower limbs, which dominate the clinical picture, sphincter disturbances and diminished vibration sense are frequently observed (Harding 1981; Polo et al. 1993; Dürr et al. 1994; Fontaine et al. 1995). The so-called complex forms of HSP are defined by the association of spastic paraplegia with peripheral neuropathy, epilepsy, extrapyramidal disturbances, ataxia, dementia, skin lesions, optic neuropathy, retinopathy, and deafness.
At least 11 different loci—SPG1–SPG11—have been described as being involved in HSP (MIM 182600, MIM 182601, MIM 270800, MIM 312900, MIM 312920, MIM 600363, MIM 601162 MIM 602783, and MIM 603563 (McKusick 1990; Martinez Murillo et al. 1999; Reid et al. 1999); however, only genes encoding for SPG1, SPG2, SPG4, and SPG7 have been identified. SPG1 and SPG2 have been assigned to chromosome regions Xq28 and Xq22, respectively. SPG2 is caused by mutations in the proteolipid protein and is an allelic variant of Pelizaeus-Merzbacher disease (Saugier-Veber et al. 1994). SPG1 results from mutations in the L1CAM gene and represents an incomplete form of the mental retardation, aphasia, shuffling gait, and adducted thumbs (MASA) syndrome (Jouet et al. 1994). Paraplegin is a mitochondrial metalloprotease encoded by SPG7, located on chromosome 16q. Mutations in paraplegin have been shown to cause both pure and complex forms of HSP with optic, cerebral, and cerebellar atrophy of autosomal recessive inheritance (Casari et al. 1998). Spastin is a member of the AAA family (members of which are ATPases associated with diverse cellular activities) and is encoded by SPG4, located on chromosome 2p. Mutations in spastin have been shown to cause autosomal dominant HSP (Hazan et al. 1999).
In this study, we report a large family with autosomal dominant pure spastic paraplegia. We excluded linkage to known HSP loci at the beginning of our study and performed a genomewide screen for a new HSP locus. We found significant linkage between the disorder and dinucleotide repeats in the chromosome 2q region.
The clinical inclusion criteria for the patients, prior to the linkage study, were the existence of progressive spastic paraplegia with pyramidal signs in the lower limbs and, in the family overall, a disease segregation pattern compatible with autosomal dominant inheritance (Dürr et al. 1994, 1996). Twenty-eight individuals were examined in family SAL-612, and paraclinical investigations were performed in the index case (individual III:4; see fig. 1), showing normal results for cerebral and cervical imaging, conduction velocities and electromyography of the lower limbs, fundoscopy, and evoked visual potentials. Disability was assessed on a three-point scale: 1= normal gait or very light stiffness of the legs, 2 = inability to run, and 3 = either inability to walk without help or confinement to a wheelchair (table 1). A severity score related to disease duration was obtained by dividing the disability score by disease duration (in years) and multiplying the result by 100 (table 1). To compare the clinical features of family SAL-612 and SPG4-linked families, the following statistical tests were used: t-test, for comparison of means, and χ2 test, for frequencies (with Yates's correction, if necessary).
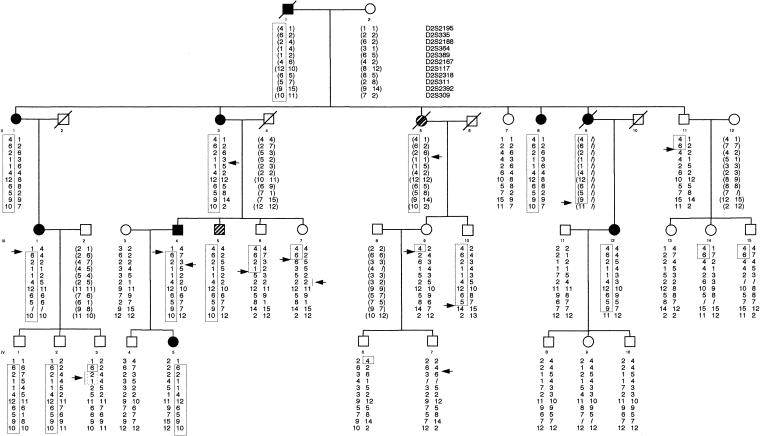
Figure 1.
Pedigree of family SAL-612. Affected individuals are indicated by blackened symbols. The haplotype segregating with the disease is boxed. Recombination events are noted by an arrow. Reconstructed genotypes are in parentheses. The hatched symbols represent individuals considered, for purposes of LOD-score calculations, to be of undetermined clinical status: patient II:5 in family SAL-612, who, on the basis of her medical history, was considered to be affected but who was never examined by one of us; and individual III:5, who was 50 years old and who had increased and spread reflexes in the lower limbs and an extensor plantar response (by Babinski sign) but no spasticity.
Table 1.
Clinical Characteristics of Family SAL-612 and Comparison with Individuals with Linkage to SPG4
| Characteristic | Family SAL-612 | Individuals with SPG4 | Pa |
| No. of patients (no. unaware of symptoms) | 9 (2) | 83 (23) | |
| Mean age at onset (years) | 39±16 (17-68) | 29±15 (2-63) | NS |
| Mean age at examination (years) | 52±18 (20-76) | 45±20 (3-85) | |
| Clinical signs present:b | |||
| Severe functional handicap | 44% (4/9) | 11% (9/83) | <.05 |
| Bilateral extensor plantar reflexes | 56% (5/9) | 88% (68/77) | <.05 |
| Increased and spread reflexes in lower limbs | 67% (6/9) | 94% (78/83) | NS |
| Increased reflexes present in upper limbs | 67% (6/9) | 22% (18/83) | <.05 |
| Decreased vibration sense | 77% (7/9) | 56% (45/81) | NS |
| Urinary urgency | 14% (1/7) | 34% (28/81) | NS |
| Pes cavus or scoliosis | 0% (0/9) | 12% (10/79) | NS |
NS = not significant.
Comparison of clinical features in family SAL-612 versus those in families linked to SPG4 was as described in the text.
Blood samples were collected from all consenting individuals, and DNA was prepared according to standard methods. Primers for dinucleotide repeats used in this study, as well as their location on genetic maps, were as described by Dib et al. (1996). In addition, the following markers were used for testing: D14S1055, D14S269, D14S1031, and D14S978, for SPG3 (Hazan et al. 1993); D2S2325, D2S2351, D2S2347, and D2S352, for SPG4 (Hazan et al. 1994); D15S1021, D15S1002, and D15S122, for SPG6 (Fink et al. 1995); and D8S1793, D8S1799, and D8S1832, for SPG8 (Hedera et al. 1999). PCRs were performed with fluorescently labeled primers (6-FAM, HEX, and TET), as recommended by the supplier. Amplification products were then pooled, loaded onto a 6% acrylamide denaturing gel on an ABI 377 DNA sequencer (PE Biosystems), and analyzed by GENESCAN version 2.1.1 and GENOTYPER version 1.1.1 softwares.
Pairwise and multipoint LOD scores were calculated by use of the MLINK and LINKMAP programs of the LINKAGE package. A disease-allele frequency of .0001 was assumed. Only affected individuals who had been clinically tested by one of us were considered as affected for purposes of the LOD-score calculations. Patient II:5 in family SAL-612, who, on the basis of her medical history, was considered to be affected was, for purposes of the LOD-score calculations, considered as being of undetermined clinical status, since she was never examined by one of us. Individual III:5, who was 50 years old and had increased and spread reflexes in the lower limbs and an extensor plantar response (by Babinski sign) but no spasticity, also was, for purposes of the LOD-score calculations, considered as being of undetermined clinical status (fig. 1). Since penetrance of autosomal dominant HSP is a function of age, liability classes as described elsewhere (Dürr et al. 1996), as well as a penetrance of .90, were used. No significant differences between the two models were noted. LOD scores were calculated both with the assumption of equal allele frequency and with allele frequencies determined in the Centre d'Étude du Polymorphisme Humain pedigrees. Both methods gave similar results.
At the beginning of our study for autosomal dominant HSP, we tested linkage between the disease segregating in family SAL-612 and dinucleotide repeats at four known loci (SPG3, SPG4, SPG6, and SPG8; reported by Hazan et al. [1993, 1994], Fink et al. [1995], and Hedera et al. [1999], respectively), and we observed negative LOD scores, as well as obligate recombinants in patients, at all four of these loci (data not shown). These results indicated that the disease was not linked to any of these loci in family SAL-612. We then performed a genomewide search, using 318 dinucleotide repeats evenly spaced every 10–15 cM on the autosomes.
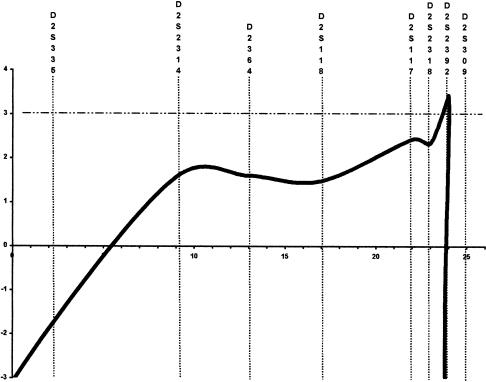
A maximum LOD score (Zmax) of 3.03 was observed for marker D2S2318, at a recombination fraction of 0. To test whether this positive LOD score resulted from random association in a relatively small kindred or from true linkage, 25 markers spaced every 1–5 cM were further tested, and the haplotype segregating with the disease was constructed. A selection of 11 markers is shown in figure 1. Multipoint linkage analysis using markers D2S335, D2S2314, D2S364, D2S118, D2S117, D2S2318, D2S2392, and D2S309 gave a Zmax of 3.43 at microsatellite D2S2392 (fig. 2). Construction of the haplotype segregating with the disease showed that all markers flanked by D2S2195 and D2S309 cosegregated with the disease (fig. 1). In family SAL-612, individuals who carried the disease haplotype but who were normal on the basis of neurological examination were noted: unaffected individuals III:10, IV:1, and IV:2 (32, 31, and 28 years old, respectively) were haplotype carriers (fig. 1). To precisely delimit the locus by means of the analysis of recombinants, we chose the most conservative method—that is, we considered recombinations only in affected individuals. The centromeric boundary at D2S2195 was defined by the recombination event detected in patients III:1 and III:4 and transmitted to affected offspring IV:5, whereas the telomeric boundary at D2S309 was defined by the recombination event observed in patient III:12 (fig. 1). These results demonstrated the existence of a new HSP locus, which maps to a 30-cM interval flanked by markers D2S2195 and D2S309 on chromosome 2q24-q34.
Figure 2.
Genetic linkage to chromosome 2q24-q34 in family SAL-612. Multipoint LOD scores between chromosome 2q24-q34 markers and HSP.
Clinical data on family SAL-612 were used for comparison with clinical features of patients with SPG4 who had been described elsewhere (Dürr et al. 1996). There was no significant difference in the mean age at onset, the presence of increased reflexes in the lower limbs, decreased vibration sense, urinary urgency, or pes cavus (table 1); however, Babinski signs were significantly less frequent in family SAL-612 than in families with SPG4 (56% vs. 88%; P<.05), but increased reflexes in the upper limbs (67% vs. 22%; P<.05) and severe functional handicap (44% vs. 11%; P<.05) were more frequent.
We have described a new locus for autosomal dominant pure spastic paraplegia, a locus that maps to chromosome 2q24-q34. In family SAL-612, the whole genome was screened, and only markers in the chromosome 2q24-q34 region generated LOD scores over the threshold of 3.00, which was confirmed by multipoint linkage analysis. Moreover, the haplotype of this chromosomal region was constructed and shown to segregate with the disease. LOD scores were calculated conservatively; only clinically affected individuals examined by one of us were considered as affected. These results strongly support the hypothesis of true genetic linkage and demonstrate the existence of a new locus for autosomal dominant pure spastic paraplegia, on chromosome 2q24-q34.
It is now well established that the penetrance of autosomal dominant HSP is age dependant and incomplete. In families with linkage to SPG4, 30% of the gene carriers are unaware of symptoms but are clinically affected (Dürr et al. 1996). Three individuals in family SAL-612 who were normal on the basis of neurological examination but whose age at onset of the disease is younger than the mean carry the disease haplotype. This observation has important clinical implications for genetic counseling in HSP: normal clinical examination is not sufficient to establish a diagnosis; despite being normal on the basis of neurological examination, a person may carry the disease haplotype—and thus may transmit the disease.
Genotype-phenotype correlations are difficult to establish for autosomal dominant HSP, since most loci have been identified in only a small number of families. Elsewhere, we have suggested that the absence of sensory signs might be a distinctive features of SPG3 (Hazan et al. 1993; Fontaine et al. 1995). SPG4 is the only locus for which clinical data could be collected from a large number of families (Dürr et al. 1996; Fink et al. 1996). We compared the clinical features of family SAL-612 to those of 12 families with linkage to SPG4 (Dürr et al. 1996). The only significant differences were lack of Babinski signs, increased reflexes in the upper limbs, and a more severe handicap in family SAL-612. More-detailed phenotype-genotype correlations will require collaborative studies, however, in order to assemble a sufficient number of families linked to the same locus.
The gene for autosomal recessive symmetrical spastic cerebral palsy has recently been mapped to chromosome 2q24-25 (McHale et al. 1999). Although the diseases differ both in their clinical presentation and in their mode of transmission, we cannot exclude the possibility that they might represent allelic mutations of the same gene. For example, both myotonia congenita of autosomal recessive inheritance and that of autosomal dominant inheritance have been shown to be caused by mutations in the same gene encoding the chloride channel CLCN1 (Koch et al. 1992). Both autosomal recessive symmetrical spastic cerebral palsy and autosomal dominant pure spastic paraplegia mapping to chromosome 2q24-q34 affect the pyramidal tracts, and the two loci overlap; indeed, considering the recombinations in family SAL-612, we have shown that both loci overlap in the 5-cM region flanked by D2S294 and D2S2195.
Acknowledgments
This work was financially supported by the Association Française contre les Myopathies, INSERM, Assistance Publique-Hôpitaux de Paris (Délégation à la Recherche Clinique), and the French Ministry of Research (ACCSV-2). We thank the patients and their families, as well as the Association Strümpell-Lorrain, for help and encouragement. We thank Sylvana Pavek, from Généthon, for help with the genotyping.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for HSP [MIM 182600, MIM 182601, MIM 270800, MIM 312900, MIM 312920, MIM 600363, MIM 601162 MIM 602783, and MIM 603563])
References
- Behan WMH, Maia M (1974) Strümpell's familial spastic paraplegia: genetics and neuropathology. J Neurol Neurosurg Psychiatry 37:8–20 [DOI] [PMC free article] [PubMed]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, et al (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93:973–983 [DOI] [PubMed]
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Dürr A, Brice A, Serdaru M, Rancurel G, Derouesné C, Lyon-Caen O, Agid Y, et al (1994) The phenotype of “pure” autosomal dominant spastic paraplegia. Neurology 44:1274–1277 [DOI] [PubMed]
- Dürr A, Davoine C-S, Paternotte C, von Fellenberg J, Cogilnicean S, Coutinho P, Lamy C, et al (1996) Phenotype of autosomal dominant spastic paraplegia linked to chromosome 2. Brain 119:1487–1496 [DOI] [PubMed]
- Fink JK (1997) Advances in hereditary spastic paraplegia. Curr Opin Neurol 10:313–318 [DOI] [PubMed]
- Fink JK, Wu CT, Jones SM, Sharp GB, Lange BM, Lesicki A, Reinglass T, et al (1995) Autosomal dominant familial spastic paraplegia: tight linkage to chromosome 15q. Am J Hum Genet 56:188–192 [PMC free article] [PubMed]
- Fink JK, Heiman-Patterson T, Bird T, Cambi F, Dubé M-P, Figlewicz DA, Haines JL, et al (1996) Hereditary spastic paraplegia: advances in genetic research. Neurology 46:1507–1514 [DOI] [PubMed]
- Fontaine B, Rime C-S, Hazan J, Dürr A, Stevanin G, Penet C, Reboul J, et al (1995) Exclusion of the candidate locus FSP1 in six families with late-onset autosomal dominant spastic paraplegia. Neuromusc Disord 5:11–17 [DOI] [PubMed]
- Harding AE (1981) Hereditary “pure” spastic paraplegia: a clinical and genetic study of 22 families. J Neurol Neurosurg Psychiatry 44:871–883 [DOI] [PMC free article] [PubMed]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine C-S, et al (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 [DOI] [PubMed]
- Hazan J, Fontaine B, Bruyn RPM, Lamy C, van Deutekom JCT, Rime C-S, Dürr A, et al (1994) Linkage of a new locus for autosomal dominant familial spastic paraplegia to chromosome 2p. Hum Mol Genet 3:1569–1573 [DOI] [PubMed]
- Hazan J, Lamy C, Melki J, Munnich A, de Recondo J, Weissenbach J (1993) Autosomal dominant familial spastic paraplegia is genetically heterogeneous and one locus maps to chromosome 14q. Nat Genet 5:163–167 [DOI] [PubMed]
- Hedera P, Rainier S, Alvarado D, Zhao X, Williamson J, Otterud B, Leppert M, et al (1999) Novel locus for autosomal hereditary spastic paraplegia, on chromosome 8q. Am J Hum Genet 64:563–569 [DOI] [PMC free article] [PubMed]
- Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, et al (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7:402–407 [DOI] [PubMed]
- Koch MC, Steinmeyer K, Lorenz C, Ricker K, Wolf F, Otto M, Zoll B, et al (1992) The skeletal muscle chloride channel in dominant and recessive human myotonia. Science 257:797–800 [DOI] [PubMed]
- MartinezMurillo F, Kobayashi H, Pegoraro E, Galluzzi G, Creel G, Mariani C, Farina E, et al (1999) Genetic localization of a new locus for recessive familial spastic paraparesis to 15q13-15. Neurology 53:50–56 [DOI] [PubMed]
- McHale DP, Mitchell S, Bundey S, Moynihan L, Campbell DA, Woods CG, Lench NJ, et al (1999) A gene for autosomal recessive symmetrical cerebral palsy maps to chromosome 2q24-25. Am J Hum Genet 64:526–532 [DOI] [PMC free article] [PubMed]
- McKusick VA (1990) Mendelian inheritance in man. 9th ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Polo JM, Calleja J, Combarros O, Berciano J (1993) Hereditary “pure” spastic paraplegia: a study of nine families. J Neurol Neurosurg Psychiatry 56:175–181 [DOI] [PMC free article] [PubMed]
- Reid E, Dearlove AM, Rhodes M, Rubinsztein DC (1999) A new locus for autosomal dominant “pure” hereditary spastic paraplegia mapping to chromosome 12q13, and evidence for further genetic heterogeneity. Am J Hum Genet 65:757–763 [DOI] [PMC free article] [PubMed]
- Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, Le Merrer M, Gil R, Boespflug-Tanguy O (1994) X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic disorders at the proteolipid protein locus. Nat Genet 6:257–262 [DOI] [PubMed]