Summary
In this study, which is a continuation and an extension of an earlier study, we enrolled two new families (N=31) and recruited more individuals from the previously ascertained families (N=56). The eight multiplex families (N=171) presented in this study were ascertained from a sample of adult probands whose childhood reading history is well documented through archival information. Six phenotypes were constructed to span a range of dyslexia-related cognitive processes. These phenotypes were (1) phonemic awareness (of spoken words); (2) phonological decoding (of printed nonwords); (3) rapid automatized naming (of colored squares or object drawings); (4) single-word reading (orally, of printed real words); (5) vocabulary; and (6) spelling (of dictated words). In addition, the diagnosis of lifelong dyslexia was established by clinical means. Genotyping was done with nine highly polymorphic markers from the 6p22.3–6p21.3 region. The results of two- and multipoint identity-by-descent and identity-by-state analyses supported the importance of a putative locus in the D6S464–D6S273 region for a number of dyslexia-related cognitive deficits.
Recently, a number of research groups have followed up on an initial report by Cardon et al. (1994, 1995) that suggested a putative quantitative-trait locus (QTL) involved in specific reading disability. The original article reported that the area of interest mapped to 6p21.3. This region is just distal to the HLA complex and relatively well characterized at the physical level (Feder et al. 1996). The four subsequent searches (Grigorenko et al. 1997; Field and Kaplan 1998; Fisher et al. 1999; Gayán et al. 1999) covered broader regions of chromosome 6p, differentially bracketing the putative chromosomal sector by regions as narrow as 7 cM or as wide as 43 cM.
Grigorenko et al. (1997), using a sample of six extended families (N=94) that were ascertained through adult probands who had been evaluated and identified as children, replicated and extended the initial chromosome 6 findings by (1) saturating the ∼20-cM–wide region with a panel of markers that was denser than the original set of polymorphic markers, (2) dissecting the composite phenotype of dyslexia into some of its hierarchical components (i.e., phonemic awareness, phonological decoding, rapid naming, and single-word reading), and (3) showing differential probabilities of linkage in the region to some of these phenotypic components (the most compelling evidence came from analyses by means of the phonemic-awareness phenotype, although linkage results with the phonological-decoding and single-word-reading phenotypes were also statistically significant).
Fisher et al. (1999), using a sample of 181 sib pairs from 82 British nuclear families selected on the basis of a dyslexic proband, covered the area of interest with 15 highly polymorphic markers, spanning ∼30 cM in the 6p25–6p21.3 region. The researchers used five quantitative phenotypes (single-word reading, IQ-related discrepancy, orthographic coding, phonological decoding, and an age-adjusted additive indicator of the last two phenotypes). Fisher et al. (1999) suggested the presence, in 6p21.3, of a QTL influencing several components of dyslexia, particularly in single-word-reading, phonological, and orthographic skills.
Gayán et al. (1999) performed a set of QTL analyses with a newly ascertained sample of 126 sib pairs. The area of interest in that study was covered by eight polymorphic markers spanning distance of 14.7 cM (6p22.3–6p21.3). These researchers defined 10 quantitative phenotypes, targeting the processes of phonemic awareness, phonological decoding, single-word reading, and orthographic coding, as well as intelligence. Gayán et al. (1999) located the putative QTL influencing several reading components (most notably, phonological awareness and orthographic coding), in a region, between markers D6S461 and D6S306/D6S258, reported elsewhere (Cardon et al. 1994; Grigorenko et al. 1997).
In contrast with these findings, a recent report by Field and Kaplan (1998) failed to replicate linkage to the 6p23–6p21.3 region in a sample of 79 Canadian families with at least two affected siblings. Their typing covered a region of 43 cM (6p25.3–6p21.3) with seven highly informative markers.
In the present study, which is a continuation and an extension of our earlier study (Grigorenko et al. 1997), we enrolled two new families (N=31) and recruited more individuals from the previously ascertained families (N=56). These efforts enlarged the sample size by 82%. The eight multiplex families (N=171) presented in this study were ascertained from a sample of adult probands whose childhood reading history is well documented through archival information (Felton et al. 1990). The probands in this study constitute a subset of 115 adults who were evaluated during childhood and whose childhood records were preserved for research purposes in the June Lyday and Samuel T. Orton Collection of the Columbia University Libraries. A detailed description of the sample and the administered battery can be found elsewhere (Felton et al. 1990). Probands were selected on the basis of childhood reading scores that placed these individuals’ reading achievement in the bottom 10% of the population. Selection required a deficit, below the 10th percentile of the normal population, on two tests, at least one of which was a single-word-reading test.
The inclusion criteria for the eight families in the present study required that probands (1) be married and have children and (2) have at least three first- or second-degree relatives with a documented history of specific reading problems. The exclusion criteria included (1) a history of significant neurological impairment, (2) mental retardation, or (3) a major sensory handicap. Two families of the eight selected had a bilateral family history of reading problems, which suggests some assortative mating.
Recent studies question the validity of both the global-composite-reading phenotype (e.g., see Grigorenko 1997; Grigorenko et al. 1997; Field and Kaplan 1998; Fisher et al. 1999; Gayán et al. 1999) and the IQ/reading-performance-discrepancy phenotype (e.g., see Lyon 1995; Siegel and Himel 1998). In this study, as in our earlier study (Grigorenko et al. 1997), we used a variety of reading-related cognitive processes as phenotypes and added a clinical diagnosis of lifelong dyslexia. As discussed later, this diagnosis was made if the individuals ever had met criteria for reading disability, even if they currently were reading well.
Adults were assessed with (1) the Test of Auditory Analysis Skills (Rosner 1979), a measure of syllable and phoneme segmentation; (2) the Lindamood Auditory Conceptualization Test (Lindamood and Lindamood 1971), a test of phonological awareness including phonemic discrimination and analysis skills; (3) Decoding Skills Test Part II (Richardson and DiBenedetto 1985), a measure requiring decoding of phonetically regular monosyllabic and polysyllabic real and nonreal words; (4) the Rapid Automatized Naming Test (Denckla and Rudel 1976), a test requiring rapid naming of colors, objects, digits, and letters; (5) the Woodcock-Johnson Psychoeducational Battery reading cluster (Woodcock and Johnson 1977), an achievement test with subtests for sight word identification, word attack, and passage comprehension; (6) the Wide Range Achievement Test-Revised (Jastak and Wilkinson 1984), with reading (sight word identification) and dictated spelling subtests; (7) the Peabody Picture Vocabulary Test-Revised (Dunn and Dunn 1981); and (8) the Wechsler Intelligence Scale vocabulary subtest (Wechsler 1974, 1981). Children of age 6–16 years were administered the same battery as were the adults, with appropriate age norms.
Six phenotypes were constructed to span a range of dyslexia-related cognitive processes. These phenotypes were (1) phonemic awareness (of spoken words); (2) phonological decoding (of printed nonwords); (3) rapid automatized naming (of colored squares or object drawings); (4) single-word reading (orally, of printed real words); (5) vocabulary; and (6) spelling (of dictated words). For each of the first five phenotypes, two separate tests were combined: for phonemic awareness, the tests were the Test of Auditory Analysis Skills (Rosner 1979) and the Lindamood Auditory Conceptualization Test (Lindamood and Lindamood 1971); for phonological decoding, the Woodcock Johnson Word Attack subtest (Woodcock and Johnson 1977) and the Decoding Skills Test nonword section of Part 2 (Richardson and DiBenedetto 1985); for rapid naming, the color and object naming subtests of the rapid automatized naming (RAN) test (Denckla and Rudel 1976; these subtests were judged to be less confounded by lifelong exposure to print than were the letter and number subtests); for single-word reading, the Woodcock Johnson Word Identification subtest (Woodcock and Johnson 1977) and the Decoding Skills Test real-word section of Part 2 (Richardson and DiBenedetto 1985); and, for vocabulary, the Peabody Picture Vocabulary Test (Dunn and Dunn 1981) and the vocabulary subtest of the age-appropriate Wechsler intelligence scale (Wechsler 1974, 1981). For each of these phenotypes, affected status required scoring either below the normative 10th percentile on one of the tests or below the normative 25th percentile on both tests. The test norms were developed on the basis of two independent epidemiological samples of adults and children. Only one spelling test was available in this sample; therefore, affected status required scoring below the 15th percentile on the spelling subtest of The Wide Range Achievement Test-Revised (Jastak and Wilkinson 1984). Table 1 contains pairwise correlations (φ) between the seven phenotypes.
Table 1.
Pairwise Correlations for the Seven Phenotypes
|
φ for the Comparison |
|||||||
| Phenotype | Phonemic Awareness | Decoding | Rapid Naming | Single-Word Reading | Vocabulary | Spelling | Lifelong Diagnosis |
| Phonemic awareness | |||||||
| Decoding | .46 (.00) | ||||||
| Rapid naming | .25 (.00) | .29 (.00) | |||||
| Single-word reading | .37 (.00) | .48 (.00) | .37 (.00) | ||||
| Vocabulary | .26 (.00) | .27 (.00) | .18 (.05) | .45 (.00) | |||
| Spelling | .30 (.00) | .46 (.00) | .32 (.00) | .68 (.00) | .44 (.00) | ||
| Lifelong diagnosis | .49 (.00) | .63 (.00) | .45 (.00) | .50 (.00) | .32 (.00) | .54 (.00) | |
The lifelong diagnosis of dyslexia was established by means of clinical information. A diagnosis was made if an individual, either adult or child, was reported to have had difficulty acquiring initial reading skills, had been certified as learning disabled in reading, and/or required tutoring or special reading classes as a child. Of the 68 judged to have some degree of dyslexia, 33 were classified as clearly impaired, with adults continuing to show impairment in reading and with children needing ongoing specialized reading instruction; the remaining 35 were classified as “borderline,” with adults having obtained literacy level (at least eighth grade on most tests) and with children no longer requiring ongoing reading help. The affected group was composed of both the impaired and the borderline group. Individuals were classified as “normal” if they had no reported history of difficulty with reading acquisition and if they were deficient in no more than one aspect of reading (e.g., only phonological decoding or only phonemic awareness).
DNA was extracted from whole blood and inner-cheek tissue. Blood was drawn from 94 family members; 60 ml of blood were drawn from each consenting adult, but only 20 ml were collected from each participant of age <18 years. Children's consent forms were signed by their parents. Genomic DNA was prepared from EDTA-preserved whole blood, according to standard techniques, except that salting out was substituted for phenol extraction (Miller et al. 1988). From the remaining 77 family members, four cytological brushings were collected (two from each cheek). Genomic DNA was prepared from the collected tissue, with the BIORAD biomatrix extraction technique (T. Webb, personal communication).
Genotyping was done with nine highly polymorphic markers from the 6p22.3(D6S285)–6p21.31(D6S273) region. The relative chromosomal location and heterozygosity of each marker are shown in figure 1. The most probable order of markers and intermarker distances was derived from current linkage and physical maps of 6p. PCR primers were labeled with 6-FAM, HEX, or TET phosphoramidite; PCR reactions were done in 96 well plates on a PE Biosystems thermocycler. Products of appropriate sizes were pooled together and were run on a 377 sequencer (Applied Biosystems), and the results were analyzed by means of GENESCAN (version 2.0) and GENOTYPER (version 1.1) software.
Figure 1.
Map of the markers studied (heterozygosity is shown in parentheses)
Two sets of marker-allele frequencies were used: (1) the published frequencies (Genome Database) and (2) the frequencies obtained by counting alleles in the parents and married individuals (see Field and Kaplan 1998). Analyses were done twice—once with the published estimates and once with the estimates obtained from counting alleles.
The raw ABI data were imported into EXCEL and were processed with SAS and SPSS statistical software macros. Model-based linkage analyses were done with version 5.2 of the LINKAGE program (Lathrop et al. 1984). Model-free analyses were done by use of the computer programs SIMWALK (Sobel and Lange 1996), SIMIBD (Davis et al. 1996), and APM (Weeks and Lange 1988). SIMIBD and APM offer three weighting schemes (1, 1/sqr(p), and 1/p), of which the preferred is 1/sqr(p). In this report, only the results obtained with the preferred weighting scheme (1/sqr[p]), recommended by the authors of the software, are presented. Thus, the analyses were done by the methods used in our earlier research (Grigorenko et al. 1997) and by more recent methods developed for large-extended-pedigree analyses.
All analyses were conducted for all seven diagnostic schemes. As suggested by Elston (1997, 1998), the precise P values are presented, rather than those adjusted for multiple comparisons. The underlying logic here is that the adjustment assumes that the tests in question were independent; this assumption does not hold in our case because (1) all phenotypes in this study correlate with each other and (2) the genetic markers are located in close proximity to each other. Therefore, a traditional correction for multiple comparisons in our case would have most likely been overly conservative.
Parametric linkage analyses were completed by use of three models of transmission (dominant, additive, and recessive; for details, see E. L. Grigorenko, F. B. Wood, M. S. Meyer, J. E. D. Pauls, L. A. Hart, D. L. Pauls, unpublished data). The analyses were done with phenotype-specific sets of parameters obtained from segregation analyses of the patterns of family transmission of corresponding phenotypes. The segregation analyses were conducted with POINTER (Lalouel et al. 1983), with the assumptions of a prevalence of 13% and of a 1:1 male:female ratio. Although there were some weakly positive results (e.g., LOD = .25 for θ=.00 at D6S464, under recessive-model parameters with published allele frequencies for the phenotype of single-word reading), none of the pairwise LOD scores were statistically significant. In general, parametric analyses were uninformative, with the majority of LOD scores between −2 and +1.
The results of model-free two-point identity-by-descent (IBD) (done with SIMIBD) and identity-by-state (IBS) (done with APM) analyses are shown in table 2. Field and Kaplan (1998), commenting on the results that Grigorenko et al. (1997) found by using IBS, pointed out that IBS analyses may have a tendency toward false-positive results, because correction for allele frequencies, when done on published data, may sometimes be inadequate. As a consequence, we conducted IBS analyses by using both published and counted allele frequencies. The IBS analyses revealed a consistent pattern of significant P values for all seven phenotypes (both for published and for counted allele frequencies) in the region D6S464–D6S306. The IBD analyses showed a consistent pattern in this region, but for only three phenotypes: single-word reading, vocabulary, and spelling (both for published and counted allele frequencies).
Table 2.
Model-Free Pairwise Analyses
|
Statistic (P) for |
|||||||||
| Phenotype | D6S285 | D6S109 | D6S461 | D6S299 | D6S464 | D6S105 | D6S306 | D6S258 | D6S273 |
| IBD: | |||||||||
| Published: | |||||||||
| Phonemic awareness | 39.8 (.25) | 29.6 (.91) | 42.1 (.86) | 41.4 (.84) | 77.3 (.20) | 47.8 (.92) | 36.3 (.75) | 55.8 (.54) | 60.9 (.19) |
| Decoding | 41.2 (.29) | 31.1 (.72) | 40.6 (.92) | 55.2 (.50) | 78.2 (.44) | 52.9 (.34) | 35.3 (.77) | 55.5 (.21) | 32.9 (.79) |
| Rapid naming | 56.4 (.02) | 29.0 (.89) | 51.3 (.48) | 36.9 (.45) | 70.0 (.32) | 30.7 (.88) | 35.6 (.39) | 33.2 (.50) | 53.2 (.54) |
| Single-word readinga | 21.4 (.26) | 13.7 (.57) | 22.1 (.22) | 33.0 (.08) | 38.7 (.05) | 42.3 (.01) | 20.1 (.07) | 19.3 (.06) | 28.0 (.60) |
| Vocabularyb | 10.7 (.48) | 12.2 (.43) | 17.5 (.17) | 20.7 (.27) | 20.7 (.30) | 25.8 (.02) | 9.0 (.59) | 20.7 (.07) | 17.1 (.55) |
| Spellingc | 14.0 (.91) | 19.9 (.36) | 27.2 (.26) | 33.0 (.25) | 49.8 (.16) | 37.0 (.05) | 32.0 (.48) | 40.7 (.04) | 30.7 (.09) |
| Lifelong diagnosis | 36.7 (.85) | 43.3 (.74) | 49.4 (.93) | 68.2 (.68) | 122.1 (.2) | 73.0 (.17) | 51.2 (.25) | 69.1 (.46) | 57.0 (.77) |
| Counted: | |||||||||
| Phonemic awareness | 29.0 (.66) | 29.5 (.92) | 36.4 (.80) | 34.8 (.91) | 62.4 (.18) | 39.0 (.92) | 27.6 (.91) | 29.2 (.91) | 42.0 (.45) |
| Decoding | 30.0 (.62) | 31.8 (.74) | 38.8 (.76) | 46.2 (.57) | 53.9 (.32) | 46.7 (.56) | 29.3 (.82) | 31.3 (.84) | 37.2 (.71) |
| Rapid naming | 56.4 (.02) | 26.3 (.88) | 36.9 (.45) | 38.3 (.72) | 38.2 (.70) | 44.1 (.49) | 33.3 (.44) | 37.5 (.73) | 41.1 (.66) |
| Single-word reading | 17.1 (.33) | 21.3 (.23) | 19.4 (.17) | 26.0 (.09) | 39.4 (.03) | 29.4 (.01) | 19.3 (.08) | 18.5 (.50) | 21.8 (.69) |
| Vocabulary | 12.0 (.45) | 13.5 (.33) | 17.1 (.16) | 16.8 (.36) | 5.7 (.59) | 19.8 (.00) | 8.8 (.61) | 14.2 (.38) | 14.5 (.52) |
| Spelling | 14.0 (.95) | 49.6 (.49) | 56.0 (.59) | 27.7 (.31) | 42.6 (.23) | 30.3 (.02) | 17.9 (.24) | 21.0 (.50) | 25.1 (.18) |
| Lifelong diagnosis | 37.4 (.74) | 43.5 (.72) | 45.6 (.84) | 54.6 (.84) | 122.4 (.0) | 58.4 (.32) | 43.1 (.45) | 41.1 (.95) | 49.0 (.77) |
| IBS: | |||||||||
| Published: | |||||||||
| Phonemic awareness | 1.8 (.03) | 1.7 (.05) | 7.2 (.00) | 2.1 (.02) | 14.8 (.00) | .8 (.21) | 2.9 (.00) | 5.9 (.00) | 5.1 (.00) |
| Decoding | 2.7 (.00) | 3.6 (.00) | 3.6 (.00) | 2.8 (.00) | 15.3 (.00) | 2.8 (.00) | 2.6 (.00) | 7.4 (.00) | 2.1 (.02) |
| Rapid naming | 5.0 (.00) | 4.4 (.00) | 9.1 (.00) | 3.1 (.00) | 15.1 (.00) | 3.5 (.00) | 3.7 (.00) | 5.5 (.00) | 4.1 (.00) |
| Single-word reading | 1.7 (.04) | 3.1 (.00) | 2.4 (.01) | 2.5 (.01) | 10.7 (.00) | 5.1 (.00) | 1.8 (.05) | 4.5 (.00) | 3.3 (.00) |
| Vocabulary | .4 (.33) | 2.2 (.01) | 4.1 (.00) | 3.2 (.00) | 14.7 (.00) | 7.7 (.00) | 2.3 (.01) | 6.3 (.00) | 1.0 (.15) |
| Spelling | .2 (.43) | 3.4 (.00) | 3.2 (.00) | 2.5 (.01) | 12.6 (.00) | 5.8 (.00) | 3.0 (.00) | 5.8 (.00) | 2.6 (.00) |
| Lifelong diagnosis | 1.3 (.10) | 3.9 (.00) | 5.4 (.00) | 2.3 (.01) | 14.4 (.00) | 3.7 (.00) | 4.1 (.00) | 6.2 (.00) | 3.0 (.00) |
| Counted: | |||||||||
| Phonemic awareness | −.1 (.55) | 1.8 (.05) | 4.1 (.00) | .5 (.30) | 4.4 (.00) | 1.0 (.16) | 1.7 (.05) | .7 (.24) | 2.2 (.01) |
| Decoding | .7 (.23) | 3.6 (.00) | 2.9 (.00) | 1.0 (.16) | 4.7 (.00) | 1.2 (.12) | 1.6 (.05) | .9 (.17) | 1.4 (.08) |
| Rapid naming | 2.2 (.01) | 4.5 (.00) | 4.8 (.00) | 1.4 (.08) | 2.7 (.00) | 2.3 (.01) | 3.1 (.00) | .6 (.28) | 2.7 (.00) |
| Single-word reading | .7 (.25) | 3.4 (.00) | 1.6 (.05) | .7 (.23) | 6.1 (.00) | 3.0 (.00) | 1.5 (.07) | .7 (.24) | 1.2 (.12) |
| Vocabulary | −.3 (.51) | 2.9 (.00) | 3.4 (.00) | 1.8 (.04) | 1.7 (.04) | 6.7 (.00) | 2.0 (.02) | 2.8 (.00) | .6 (.27) |
| Spelling | −.4 (.65) | 3.7 (.00) | 2.4 (.01) | 1.1 (.15) | 4.2 (.00) | 5.2 (.00) | 2.1 (.02) | .7 (.24) | 1.1 (.13) |
| Lifelong diagnosis | .3 (.38) | 4.0 (.00) | 3.5 (.00) | .4 (.34) | 7.0 (.00) | 2.3 (.01) | 2.6 (.00) | .1 (.46) | 1.9 (.03) |
The analyses for the single-word-reading phenotype are done on seven of the eight pedigrees. In the eighth pedigree there were only two affected individuals who were unrelated to each other.
The analyses for the vocabulary phenotype are done on three of the eight pedigrees. Five smallest pedigrees were eliminated from the analyses because of the absence of nonparent-child pairs of affected relatives.
The analyses for the spelling phenotype are performed on seven of the eight pedigrees. The eighth pedigree had no affected individuals.
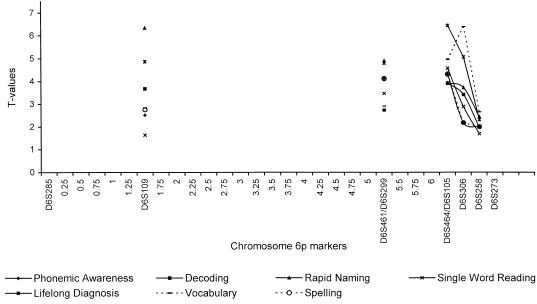
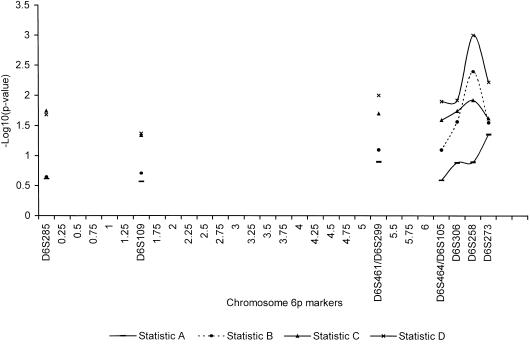
Multipoint model-free analyses were done with SIMWALK (fig. 2) and APM (fig. 3). Because of the CPU-demanding nature of SIMWALK, multipoint analyses were done only for counted allele frequencies. As was anticipated on the basis of the pairwise results, multipoint analyses supported a putative locus in the D6S464–D6S273 region, with peaks at D6S258 (fig. 2), for SIMWALK-produced results, and at D6S105/306 (fig. 3), for APM-produced results. Together, the 1.5-LOD-support interval established through the analysis of the single-word reading phenotype and the 1-LOD-support interval established through the analysis of the vocabulary phenotype span ∼2 cM in the D6S461/D6S299–D6S273 interval (fig. 2). The most striking aspect of this finding is that this location is notably consistent with that shown, first, by the work on at least three independent samples (Cardon et al. 1994; Fisher et al. 1999; Gayán et al. 1999) and, second, by our own work on a smaller subsample (Grigorenko et al. 1997).
Figure 2.
Statistic C’s –Log10(P value) terms for the seven phenotypes. The values of statistic C were the highest among the four statistics.
Figure 3.
Three-point APM linkage analyses of chromosome 6 markers and the reading-related phenotypes. All graphed points are statistically significant at P<.05.
To highlight the details of the single-word-reading phenotype finding, the distribution of the four statistics calculated by SIMWALK (Sobel and Lange 1996) for the single-word-reading phenotype is shown in figure 4. The four statistics (A–D) indicate the degree of clustering of the marker alleles descending from the pedigree founder, for affected individuals only (Sobel and Lange 1996). In particular, statistic A shows the number of different founder genes (or the total number of different trees) contributing to the marker genes appearing among the affected individuals. This statistic was significant only at the D6S273 location. Statistic B indicates the maximum number of genes, among affected individuals, attributable to any one founder gene (i.e., to any one descent tree). This statistic was significant throughout the interval between D6S461/D6S299 and D6S464/D6S105. Statistic C shows the degree of entropy among affected individuals, taking into account the number of founders, the number of affected individuals, and the number of marker genes, among affected individuals, attributable to a given descent tree. Finally, statistic D indicates the extent of allele sharing among pairs of affected individuals, as measured by their IBD kinship coefficient. Both statistic C and statistic D are generic statistics, indicating whether there are a few descent trees that are overly represented among the marker genes of affected individuals. Both these statistics (C and D) reached significance (P<.05) at D6S285 and remained significant throughout the region.
Figure 4.
−Log10(P value) terms for statistics A, B, C, and D, calculated with the phenotype of single-word reading
In summary, this extension of our previous work contributes to the pool of converging evidence, from three other independent studies, that a region on 6p21.3 influences various dyslexia-spectrum processes. This convergence was challenged by Field and Kaplan's (1998) study of Canadian dyslexic families. There are two characteristics of that study that might explain its failure to replicate other groups’ findings. First, Field and Kaplan used a rather sparse marker map covering 43 cM with only seven markers, of which only three (D6S299, D6S105, and TNFB, spanning 6 cM) were immediately adjacent to or located within the region to which the linkage was reported by other groups. Second, unlike other groups, they used a single clinically derived phenotype, which, even though partially based on the data from the specific neuropsychological tests, was heavily dependent on a conglomerate of dyslexia indicators that other groups have used as separate reading-related processes. Specifically, in defining “phonological coding dyslexia,” they relied on tests that other groups used for separate and differentially strong linkages between the region of interest and categories of phonemic awareness and phonological decoding (Grigorenko et al. 1997; Fisher et al. 1999; Gayán et al. 1999).
The results of this study have, once again, attested to the complexity of the mechanism underlying the behavioral manifestation of dyslexia. Of particular interest is that the breadth of potentially affected phenotypes may even include vocabulary, which was not assessed by any reading operations. Thus, this study and other recent molecular genetic studies provide a parallel to recent behavioral studies (see Blachman 1997), which suggest that, although phonological processes are a substantial substrate of reading disability, they do not alone encompass all the cognitive manifestations of dyslexia.
Despite converging evidence implicating the role of 6p in dyslexia-related cognitive deficits, however, we agree with Fisher et al. (1999) that, at this point, a precise estimate of the relative contributions of this locus to specific processes of the spectrum is rather difficult to obtain. The relative strength of the evidence for linkage of any one cognitive process is dependent on many factors, including ascertainment method, applied analytical tools, methods of phenotype definition, and frequency of the phenotype of interest in a given sample. Whereas IBS methods in this study continue to suggest linkage to a wider variety of phenotypes, the IBD analyses put a special emphasis on the phenotype of single-word reading, with spelling and vocabulary phenotypes defining the confidence interval of the deficit linked to 6p. The discrepancy between the IBS and IBD results can be explained by either APM’s higher sensitivity to misspecification of allele frequencies, APM’s higher rate of false-positive results, or both. Yet, it is possible to presume that IBS is a more sensitive test (if it is assumed that allele frequencies are specified correctly), especially for detection of small-effect genes involved in QTL.
Clearly, the challenge posed by this increased breadth of phenotypic expression requires still further careful studies of the various ways in which the genetic risk is expressed. It is noteworthy, nonetheless, that broadly defined phenotypes, whether discrepancy based or inclusive of multiple aspects of dyslexia (e.g., our lifelong-diagnosis phenotype or the Field-Kaplan [1998] phonological-disorder phenotype), do not seem to be as informative as are specific-processes–based phenotypes. Thus, whereas three independent studies converge to implicate various discrete cognitive phenotypes, they do not tend to support composites that merge several of these phenotypes. It is also notable that these phenotypes are related to each other significantly—and sometimes strongly—but hardly at a level strong enough to imply a factorial unity across these phenotypes. Thus, Gayán et al. (1999) reported highly significant correlations between their variables, in the range of .41–.90. In our study, all bivariate phenotypic correlations were statistically significant, but the observed magnitudes were smaller, in the range of .18–.68. Genetic modeling done on our data showed the presence of both genetic and environmental correlations between different phenotypes (Grigorenko 1997), which suggests that these correlations might be attributable to both genetic and environmental common variance. Whether the variable nature of linkages to specific phenotypes that was observed in different samples is explainable by the underlying commonality of cognitive processes involved in poor reading, which is reflected through different phenotypic measures with differential success, or by some other factor (e.g., specific patterns of brain activity) that is manifested in different families through different phenotypes, or whether there is a different explanation for the observed pattern of the results is to be determined in future studies. Another characteristic of our sample possibly deserves attention. Specifically, despite the fact that identical population-based cutoff points were used in dichotomization of the four process-based phenotypes, the number of affected individuals varied for each phenotype. Specifically (with some affected individuals having two or more phenotypes), 38% of the sample had phonemic-awareness deficiency; 37.3% had phonological-decoding problems; 35.7% had rapid-naming difficulties; and 40% had a lifelong diagnosis of dyslexia; but only 20% had single-word-reading deficit; only 18% had vocabulary deficit; and only 20% had spelling difficulties. Given that we used allele-sharing techniques, the high frequency of other phenotypes in our sample might have influenced the obtained results. In other words, this study poses two questions. The first question is whether differential population-based cutoff points should be used for different phenotypes. The second question is whether large numbers of affected individuals jeopardize the power of IBD-based model-free methods to detect linkage. To conclude, our study, in concordance with two of the three recent publications on the region of interest, suggests that a locus on 6p21.3 influences dyslexia-related processes.
Acknowledgments
We thank Leslie Hart for collecting blood samples and Lina Golovyan and Jed Pauls for technical assistance with genotyping. This study was supported in part by grants HD21887 (to F.B.W., principal investigator) and MH00508 (a Research Scientist Award, to D.L.P.). We are very grateful to all the families who contributed to the study. Without their cooperation, this research would not have been possible.
Electronic-Database Information
The URL for data in this article is as follows:
- Genome Database, http://www.gdb.org/ [Google Scholar]
References
- Blachman B (ed) (1997) Foundations of reading acquisition. Lawrence Erlbaum, Mahwah, NJ [Google Scholar]
- Cardon LE, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed]
- ——— (1995) Quantitative trait locus for reading disability: correction. Science 268:1553 [DOI] [PubMed]
- Davis S, Schroeder M, Goldin LR, Weeks DE (1996) Nonparametric simulation-based statistics for detecting linkage in general pedigrees. Am J Hum Genet 58:867–880 [PMC free article] [PubMed]
- Denckla MA, Rudel RG (1976) Naming of object drawing by dyslexic and other learning disabled children. Brain Lang 3:1–16 [DOI] [PubMed]
- Dunn LM, Dunn LM (1981) The Peabody picture vocabulary tests, reviewed. American Guidance Service, Circle Pines, MN [Google Scholar]
- Elston RC (1997) Algorithms and inferences: the challenge of multifactorial diseases. Am J Hum Genet 60:255–262 [PMC free article] [PubMed]
- ——— (1998) Methods of linkage analysis—and the assumptions underlying them. Am J Hum Genet 63:931–934 [DOI] [PMC free article] [PubMed]
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, et al (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13:399–408 [DOI] [PubMed]
- Felton RH, Naylor CE, Wood FB (1990) Neuropsychological profile of adult dyslexics. Brain Lang 39:485–497 [DOI] [PubMed]
- Field LL, Kaplan BJ (1998) Absence of linkage of phonological coding dyslexia to chromosome 6p23-p21.3 in a large family data set. Am J Hum Genet 63:1448–1456 (erratum: 64:334) [DOI] [PMC free article] [PubMed]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, et al (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed]
- Gayán J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, et al (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed]
- Grigorenko EL (1997) Linkage analyses on chromosome 1, 6, and 15. Paper presented at the 4th World Congress on Dyslexia, September 24–26, Halkidiki, Macedonia [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL (1997) Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet 60:27–39 [PMC free article] [PubMed]
- Jastak J, Wilkinson GS (1984) Wide range achievement test: revised edition. Jastak, Wilmington, DE [Google Scholar]
- Lalouel JM, Rao DC, Morton NE, Elston RC (1983) A unified model for complex segregation analysis. Am J Hum Genet 35:816–826 [PMC free article] [PubMed]
- Lathrop M, Lalouel J, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed]
- Lindamood CH, Lindamood PC (1971) Lindamood auditory conceptualization test. Teaching Resources, Boston [Google Scholar]
- Lyon GR (1995) Toward a definition of dyslexia. Annals of Dyslexia 35:3–27 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed]
- Richardson E, DiBenedetto B (1985) Decoding skills. Western Psychological Services, Los Angeles [Google Scholar]
- Rosner J (1979) Test of auditory analysis skills. Academic Therapy, Navato, CA [Google Scholar]
- Siegel LS, Himel N (1998) Socioeconomic status, age, and the classification of dyslexics and poor readers: the danger of using IQ scores in the definition of reading disability. Dyslexia 4:90–103 [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed]
- Weeks DE, Lange K (1988) The affected-pedigree-member method of linkage analysis. Am J Hum Genet 42:315–326 [PMC free article] [PubMed]
- Wechsler D (1974) Manual for the Wechsler intelligence scale for children, rev. The Psychological Corporation, New York [Google Scholar]
- ——— (1981) Wechsler adult intelligence scale: review. Harvard University Press, Cambridge [Google Scholar]
- Woodcock RW, Johnson MB (1977) Woodcock-Johnson psychoeducational battery. Teaching Resources, Hingham, MA [Google Scholar]