Abstract
Rec8 syndrome (also known as “recombinant 8 syndrome” and “San Luis Valley syndrome”) is a chromosomal disorder found in individuals of Hispanic descent with ancestry from the San Luis Valley of southern Colorado and northern New Mexico. Affected individuals typically have mental retardation, congenital heart defects, seizures, a characteristic facial appearance, and other manifestations. The recombinant chromosome is rec(8)dup(8q)inv(8)(p23.1q22.1), and is derived from a parental pericentric inversion, inv(8)(p23.1q22.1). Here we report on the cloning, sequencing, and characterization of the 8p23.1 and 8q22 breakpoints from the inversion 8 chromosome associated with Rec8 syndrome. Analysis of the breakpoint regions indicates that they are highly repetitive. Of 6 kb surrounding the 8p23.1 breakpoint, 75% consists of repetitive gene family members—including Alu, LINE, and LTR elements—and the inversion took place in a small single-copy region flanked by repetitive elements. Analysis of 3.7 kb surrounding the 8q22 breakpoint region reveals that it is 99% repetitive and contains multiple LTR elements, and that the 8q inversion site is within one of the LTR elements.
Rec8 syndrome (also known as “recombinant 8 syndrome” and “San Luis Valley syndrome” [MIM 179613]) was first reported (Fujimoto et al. 1975) in a Hispanic female infant who died from heart failure at age 6 wk. Since that time, multiple other cases have been identified, almost all of known Hispanic ancestry with ancestral origin in the San Luis Valley of southern Colorado and northern New Mexico (Fujimoto et al. 1975; Sujansky et al. 1993). Rec8 syndrome is associated with significant morbidity and mortality, and affected individuals typically have congenital heart defects, mental retardation, seizures, a characteristic facial appearance, and other manifestations. Cardiac defects are found in 93% of individuals with Rec8 syndrome and are a major contributor to the mortality of children with Rec8 syndrome, who have an average age at death of ∼6 years (Sujansky et al. 1993).
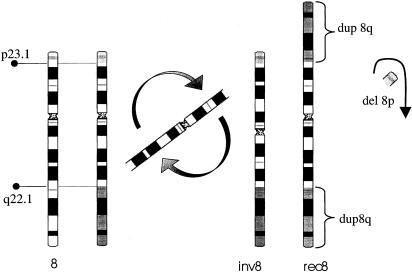
The recombinant chromosome is rec(8)dup(8q)inv(8)(p23.1q22.1) and is deleted for 8p23.1→pter and duplicated for 8q22→qter (fig. 1). In all cases, at least one of the parents of a child with Rec8 syndrome carries a pericentric inversion, inv(8)(p23.1q22.1). Carriers of the Inv8 chromosome are apparently phenotypically normal, and it has been estimated that an Inv8 carrier has a 6.2% chance in each pregnancy of having a child with Rec8 syndrome (Smith et al. 1987).
Figure 1.
Chromosome 8 rearrangements. The normal chromosome 8 is shown, with 8p23.1 and 8q22 breakpoints indicated. The Inv8 chromosome results from a pericentric inversion. The Rec8 chromosome contains a duplication of 8q22→8qter, whereas the 8p23.1→8pter fragment is lost.
It is apparent that not all regions of the genome are equally likely to participate in chromosomal rearrangements, and there are several indications that 8p23.1 may be a relatively unstable region of the human genome. Multiple cases of chromosomal rearrangement involving 8p23.1 have been described in the literature, including duplications, deletions, and inverted duplications. Several cases of isolated del(8p23.1) have been described (Fryns et al. 1989; Fagan et al. 1988; Blennow and Brondum-Nielsen 1990; Pecile et al. 1990; Hutchinson et al. 1992; Pettenati et al. 1992; Wu et al. 1996). In general, clinical findings in these patients are variable and may include mental retardation, behavior problems, congenital heart defects, seizures, and genitourinary abnormalities. Multiple cases of “inverted duplication” of 8p have been described, producing a chromosome with a deletion of material distal to the 8p23.1 breakpoint and an inverted duplication of a variable amount of material proximal to the 8p23.1 breakpoint, with clinical manifestations ranging from insignificant to significant developmental delay (Dill et al. 1987; Nevin et al. 1990; Henderson et al. 1992; Feldman et al. 1993; Minelli et al. 1993; Barber et al. 1994; Engelen et al. 1994; Mitchell et al. 1994; Guo et al. 1995; Floridia et al. 1996; Barber et al. 1998).
In addition to its tendency to participate in chromosomal rearrangements, 8p23.1 has been noted by Ohashi et al. (1994) to contain a sequence that can function as a neocentromere. 8p23.1 also represents a region of the genome that is evolutionarily unstable, as evidenced by comparison of human and macaque banding patterns and FISH analysis (Small et al. 1985; Weinberg et al. 1992).
In spite of this evidence that 8p23.1 is an unstable segment of the genome, the exact molecular basis of these rearrangements has not been defined. Here we report on the cloning and characterization of the 8p23.1 and 8q22 breakpoints from the Inv8 chromosome associated with Rec8 syndrome. Analysis of the sequences surrounding these breakpoints indicates that they contain multiple repetitive elements that may have mediated the original inversion event.
Material from two Rec8 syndrome patients and three Inv8 carriers was used to construct lymphoblastoid cell lines (Neitzel et al. 1986) and hamster-human hybrid cell lines (Moore et al. 1977), segregating the abnormal chromosome 8 from other chromosome 8 material. Rec8-1 is a female Rec8 syndrome patient with a 46,XX,rec(8)dup(8q)inv(8)(p23.1q22.1)pat karyotype. During the neonatal period, the patient was noted to be cyanotic and to have a heart murmur, brachycephaly, and lowset ears. Cardiac evaluation at age 2 d revealed a ventricular septal defect, patent ductus arteriosus, and pulmonary atresia. On karyotypic analysis, the father (Inv8-1) and brother of Rec8-1 were found to be Inv8 carriers, with 46,XY,inv(8)(p23q22). The mother's karyotype was normal. Rec8-2 is a Rec8 syndrome female with a karyotype of 46,XX,rec(8)dup(8q)inv(8)(p23.1q22.1). Rec8-2 had profound mental retardation, moderate conductive hearing loss, atrial septal defect, patent ductus arteriosus, ventricular septal defect, and pulmonary stenosis. Inv8-2 is a female Inv8 carrier with a karyotype of 46,XX,inv(8)(p23.1q22) and the mother of a female child with Rec8 syndrome diagnosed at age 2 years. Inv8-3 is a female inversion 8 carrier, with a karyotype of 46,XX,inv(8)(p23.1q22). An “Inv8 mapping panel” was created to facilitate mapping, and, in addition to the Inv8 and Rec 8 LCLs and hybrids mentioned earlier, includes HeLa; CHO Gly-B (Jones et al. 1981); Cl17, a hamster-human hybrid line containing normal chromosome 8 as its only human material (Jones et al. 1981); and 21q+ (Drabkin et al. 1985), a hamster-human hybrid containing 8q22–8qter translocated to human chromosome 21.
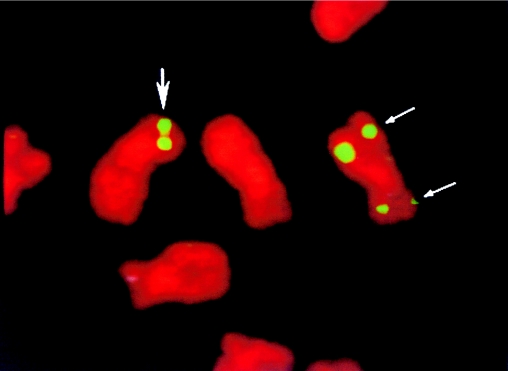
Previous studies by Shechter et al. (1994) reported the isolation of YAC B20C12, encoding the squalene synthase (SS) gene on chromosome 8. Additional SS YACs 737E5 and 779E9 were identified through screening the CEPH megaYAC library (Green and Olson 1990; Chumakov 1992), and SS P1 clones P36G5 and P72A2 by screening the Dupont/Merck P1 library (Sternberg 1990; Pierce and Sternberg 1992). FISH analyses (Lichter et al. 1990; Patterson et al. 1993) of SS YACs B20C12, 737E5, and 779E9 and SS P1 clones P36G5 and P72A2, against metaphase chromosomes from an Inv8 LCL, revealed that these clones span the 8p23.1 breakpoint (fig. 2). To further limit the breakpoint candidate region, breakpoint-spanning SS P1 clone P36G5 was digested with BamHI, and fragments were subcloned into λZAP or pUC19 and were analyzed. The ends of individual subclones were sequenced, and the sequences were used to design primer pairs for detection of each end (for primer pairs, see table 1 in online edition)(table 1). PCR analysis of the Inv8 mapping panel reveals that the 6-kb subclone P36-21 spans the breakpoint (data not shown). P36-21 was completely sequenced (partial sequence found in fig. 3; complete sequence GenBank accession number AF181099), and additional PCR analyses of the Inv8 mapping panel limited the breakpoint to a 25-bp region (data not shown). Sequences flanking this region were used to design probes just proximal and distal to the breakpoint.
Figure 2.
FISH with P1 clone P36G5 against Inv8 lymphoblastoid cell line metaphase chromosomes. Thick arrow, hybridization to the normal chromosome 8; thin arrows, double signal on the Inv8 chromosome.
Table1.
Primers for Analysis of P36G5 Subclones
| Subclone | Size (kb) | Primer Sequence (5′→3′) |
| P36-1 | 6 | M3F/left: GTACTCCCTTCTCCGAGCC |
| M3F/right: CCTGGCACATAACGGGTAC | ||
| M3R/left: ACTCCCTTCCTCGCATTCT | ||
| M3R/right: AGAATCAATTGCTCCCCCT | ||
| P36-21 | 6 | M3F/left: TTTTGCTTTCTCTGGCCCTA |
| M3F/right: ACTGGGATTAGCAGTGGTGG | ||
| M3R/left: CTGGGTTTTGCTTTACAACG | ||
| M3R/right: GTGCAGACCGAAACAGACAAG | ||
| ZB20 | 5 | T3/left: TCTGTCACAAGGCCACAAAG |
| T3/right: CGACAGAAAGAAACTCCATC | ||
| T7/left: ACTTTCATGAGGCCCAGCTC | ||
| T7/right: CAGAGAGGAACTGCAGAGGG | ||
| P36-15 | 5 | M3R/left: TGTTCCCTGAAATAAAGCGG |
| M3R/right: GATTCAAGTGCAGGCAGTCA | ||
| M3F/left: CTGAGTATTGGAGAGAGAC | ||
| M3F/right: TCTGGAGAAATCGTTCACC | ||
| ZB16 | 4 | T3/left: GACACTCCTCAGCTCTTGGG |
| T3/right: ACAGGCCTTACTGCCTCTCG | ||
| T7/left: TGGCGAAATAGAAATCCAGG | ||
| T7/right: GGTTTTGTTTGTATGACTCGGA | ||
| ZB6 | 2 | T7/left: ATGTCCTGAAATACAGCCCG |
| T7/right: GCAAGCAGTGAGCTACCCTC | ||
| T3/l eft: TGCTTTAGAAAGCCCCTTCA | ||
| T3/right: TCAAACTTTTGAGCAGCCCT | ||
| ZB10 | 2 | T3/left: TGGAGGAAAATGACTCACCC |
| T3/right: AGGTGCTGGGATTACAGGTG | ||
| T7/left: TTCCAGTTGTCCCTTTTTGG | ||
| T7/right: CTTGGTTCCAAATTCTCCCA |
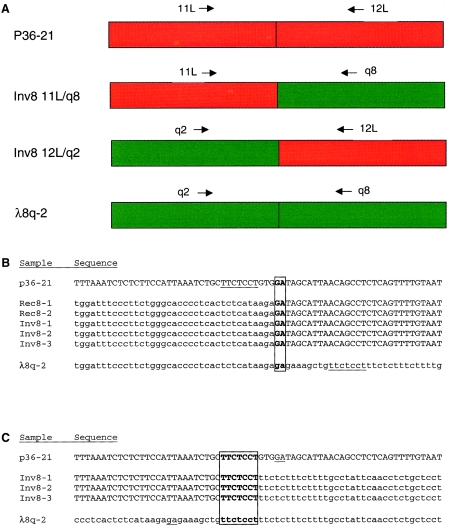
Figure 3.
A, Schematic illustrating structure of P36-21 (red) and λ8q-2 (green) and novel Inv8 junction fragments detected with primer pairs 11L/q8 and 12L/q2. B, Sequence analysis of 12L/q2 PCR products. Sequence of P36-21 (normal 8p23.1) is indicated in upper case, and of λ8q-2 (normal 8q22) in lower case. Sequence from Rec8 patients and Inv8 carriers is indicated with material corresponding to P36-21 in upper case and to λ8q-2 in lower case. The boxed and bolded “GA” indicates the site of recombination. Underlined “TTCTCCT” denotes site of recombination found in 11L/q8 PCR products. C, Sequence analysis of 11L/q8 PCR products. Sequence of P36-21 (normal 8p23.1) is indicated in uppercase, and of λ8q-2 (normal 8q22) in lowercase. Sequence from Inv8 carriers is indicated with material corresponding to P36-21 in upper case and to λ8q-2 in lower case. The boxed and bolded “TTCTCCT” indicates the site or recombination. Underlined “GA” denotes site of recombination found in 12L/q2 products.
Approximately 5 × 105 plaques from an Inv8 hybrid λDASH library were screened with breakpoint-flanking probes, and a single clone (Inv8/8p-prox) was isolated. Clone Inv8/8p-prox was partially sequenced and was compared to P36-21 to identify the breakpoint. Novel sequence of Inv8/8p-prox (representing presumptive 8q material) was used to probe a HeLa λDASH library, and two clones were identified (λ8q-2 and λ8q-8). In addition, this probe was used to screen the CEPH megaYAC library (Chumakov 1992) by PCR and two YACs were identified: 811C1 and 900B3. Both of these YACs have been positioned in the Whitehead Institute database and are part of chromosome 8q contig WC-100, localized to 8q22.1–22.3. Comparison of λ8q-2 sequence (partial sequence in fig. 3; complete sequence GenBank accession number AF181100) with Inv8 junction fragment and P36-21 sequences verified the molecular location of the rearrangement (fig. 3).
To characterize the rearrangement and determine whether Inv8 and Rec8 chromosomes all contained the same breakpoints, we used sequences from P36-21 (normal 8p23.1) and λ8q-2 (normal 8q22) to design PCR assays specifically for the rearranged chromosomes. Primer combination 12L and q2 is specific for the novel junction fragment proximal-8p/distal-8q and detects both the Inv8 and Rec8 chromosomes, whereas combination 11L and q8 is specific for the distal-8p/proximal-8q novel junction fragment and detects only the Inv8 chromosome (fig. 3). PCRs were done with 12L/q2 and 11L/q8 on Inv8 LCLs Inv8-3 and Inv8-2, Inv8 hybrid Inv8-1, Rec8 hybrid Rec8-2, and Rec8 LCL Rec8-1, and products were purified and sequenced. All Inv8 and Rec8 samples produced a product of identical size and sequence with primers 12L/q2 (fig. 3b), verifying that these breakpoints are identical and supporting the hypothesis of a founder effect leading to dispersion of a single Inv8 chromosome throughout this population. As expected, primers 11L/q8 produce a product only with the Inv8 LCLs and hybrids and fail to produce a product with Rec8 LCL Rec8-1 and Rec8 hybrid Rec8-2. The Inv8 products were all of identical size and sequence (fig. 3c). Comparison of the rearranged sequences found in 12L/q2 with that of normal 8p and 8q reveals that there is only a 2-bp region of identity at the breakpoint, and that formation of this product did not result in the loss of any material. Comparison of the rearranged sequences found in p11L/q8 with that of normal 8p and 8q reveals that the best alignment is obtained by crossing over in a 7-bp region of identity, and that formation of this product did not result in the loss of any material. The 2-bp region of identity found in 12L/q2 is not contained within the 7-bp region of identity found in 11L/q8, but is 4 bp away on P36-21 and 9 bp away on λ8q-2, indicating that the rearrangements took place at slightly offset locations.
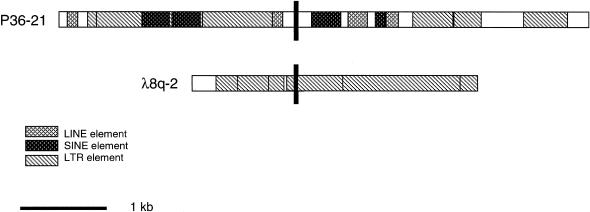
Approximately 6 kb of P36-21 (8p23.1) sequence and 4 kb of λ8q-2 (8q22) sequence were analyzed by the BLAST algorithm (Altschul et al. 1990), RepeatMasker (A. F. A. Smit and P. Green), and MAR-Finder. BLAST analysis did not produce hits of any significant homology outside of repetitive elements (data not shown). RepeatMasker analysis revealed that both P36-21 and λ8q-2 contain multiple repetitive elements (fig. 4). P36-21 is 6,062 bp in length and is 75% repetitive, containing four Alu elements, two LINE2 elements, and five LTR elements. Formation of the Inv8 chromosome takes place at two sites that are slightly offset, one at bp 2,618–2,624 (11L/q8) and one at bp 2,628–2,629 (12L/q2), within a small 370-bp unique sequence region flanked by a LINE/L2 element and an Alu element. A single potential matrix attachment region (MAR) was identified in P36-21, between 800 bp and 1400 bp, by use of the MAR-Finder program. This same region was also shown by RepeatMasker to contain an LTR10C element and an AluSq element. The sequence of λ8q-2 is 3724 bp in length and contains multiple LTR elements, totaling 3,688 bp, or 99.03% of the sequence. Recombination at the 8q22 break occurs at two places that are slightly offset, one at bp 2,560–2,566 (11L/q8) and one at bp 2,550–2,551 (12L/q2) within an LTR element.
Figure 4.
RepeatMasker analysis of P36-21 and λ8q-2. Repetitive elements are as indicated. Breakpoint location is indicated by a solid, vertical bar.
The junction sequences of several constitutional chromosomal rearrangements have been characterized, leading to speculation on the molecular mechanisms behind such rearrangements. In some cases, such as deletion of an α-globin gene leading to α-thalassemia (Ottolenghi et al. 1974; Taylor et al. 1974), recombination is between homologous segments of two members of a gene family. In other cases, such as in the LDL-receptor (Hobbs et al. 1986), α-globin (Nicholls et al. 1987), β-globin (Vanin et al. 1983; Henthorn et al. 1986), and apolipoprotein B genes (Huang et al. 1989), rearrangement has been shown to be mediated by Alu repetitive elements. It has been proposed that Alu elements function as recombinatorial hotspots (Calabretta et al. 1982; Barsh et al. 1983), and Rudiger et al. (1995) have speculated that a highly conserved 26-bp core sequence enhances recombination. Other repetitive elements, such as LINE-1 (L1), have also been shown to participate in chromosomal rearrangements (Toriello et al. 1996). In still other cases (e.g., Budarf et al. 1995), there is minimal or no obvious sequence homology to mediate the rearrangement, and the cause is speculative. In at least one case of a constitutional translocation t(8;17), resulting in isolated lissencephaly (Kurahashi et al. 1998), there was no sequence homology between the parental chromosomes, but the region immediately surrounding the 17p13.3 breakpoint had five Alu repetitive elements and the region surrounding the 8p11.2 breakpoint contained three L1 sequences, leading to speculation that the repetitive elements themselves contribute to instability and recombination.
In the case of the Inv8 chromosome, we have shown that the inversion event takes place between two highly repetitive regions, and that the exact break occurred in a small single-copy region of 8p23.1 and in the midst of an expanse of LTR elements in 8q22, supporting the hypothesis that repetitive elements are themselves recombinogenic.
Acknowledgments
We thank Miles Brennan for technical assistance. Special thanks to Billie Carstens of the Colorado Genetics Laboratory for preparing figure 1. This work was supported by funding from The Dorothea Haus Ross Foundation and grant 9808369P from the American Heart Association, Desert/Mountain Affiliate.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, (for complete P36-21 sequence [accession number AF181099] and complete λ8q-2 sequence [accession number AF181100]), [Google Scholar]
- MAR-Finder, http://www.ncgr.org/MarFinder/intro.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for Rec8 syndrome [MIM 179613])
- RepeatMasker, http://ftp.genome.washington.edu/RM/RepeatMasker.html
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu/
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed]
- Barber JCK, James RS, Patch C, Temple IK (1994) Prototelomeric sequences are deleted in cases of short arm inverted duplication of chromosome 8. Am J Med Genet 50:296–299 [DOI] [PubMed]
- Barber JCK, Joyce CA, Collinson MN, Nicholson JC, Willatt LR, Dyson HM, Bateman MS, et al (1998) Duplication of 8p23.1: a cytogenetic anomaly with no established clinical significance. J Med Genet 35:491–496 [DOI] [PMC free article] [PubMed]
- Barsh GS, Seeburg PH, Gelinas RE (1983) The human growth hormone gene family: structure and evolution of the chromosome locus. Nucleic Acids Res 11:3939–3958 [DOI] [PMC free article] [PubMed]
- Blennow E, Brondum-Nielsen K (1990) Partial monosomy 8p with minimal dysmorphic signs. J Med Genet 27:327–329 [DOI] [PMC free article] [PubMed]
- Budarf ML, Collins J, Gong W, Roe B, Wang Z, Bailey LC, Sellinger B, et al (1995) Cloning a balanced translocation associated with DiGeorge syndrome and identification of a disrupted candidate gene. Nat Genet 10:269–278 [DOI] [PubMed]
- Calabretta B, Robberson DL, Barrera-Saldana HA, Lambrou TP, Saunders GF (1982) Genome instability in a region of human DNA enriched in Alu repeat sequences. Nature 296:219–225 [DOI] [PubMed]
- Chumakov I, Rigault P, Guillou S, Ougen P, Billaut A, Guasconi G, Gervy P, et al (1992) Continuum of overlapping clones spanning the entire human chromosome 21q. Nature 359:380–387 [DOI] [PubMed]
- Dill FJ, Schertzer M, Sandercock J, Tischler B, Wood S (1987) Inverted tandem duplication generates a duplication deficiency of chromosome 8p. Clin Genet 32:109–113 [DOI] [PubMed]
- Drabkin HA, Diaz M, Bradley CM, LeBeau MM, Rowley JD, Patterson D (1985) Isolation and analysis of the 21q+ chromosome in the acute myelogenous leukemia 8;21 translocation: evidence that c-mos is not translocated. Proc Natl Acad Sci USA 82:464–468 [DOI] [PMC free article] [PubMed]
- Engelen JJM, de Die-Smulders CEM, Fryns JP, Hoovers JMN, Albrechts JCM, Loots WJG, Jacobs ME, et al (1994) Partial trisomy and monosomy 8p due to inversion duplication. Clin Genet 45:203–207 [DOI] [PubMed]
- Fagan K, Wilkinson I, Allen M, Brownlea S (1988) The coagulation factor VII regulator is located on 8p23.1. Hum Genet 79:365–367 [DOI] [PubMed]
- Feldman GL, Weiss L, Phelan MC, Schroer RJ, VanDyke DL (1993) Inverted duplication of 8p: ten new patients and review of the literature. Am J Med Genet 47:482–486 [DOI] [PubMed]
- Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, Rossi E, Gimelli G, et al (1996) The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am J Hum Genet 58:785–796 [PMC free article] [PubMed]
- Fryns JP, Kleczkowska A, Vogels A, Van den Berghe H (1989) Normal phenotype and slight mental retardation in de novo distal 8p deletion (8pter-8p23.1:). Ann Genet 32:171–173 [PubMed]
- Fujimoto A, Wilson MG, Towner JW (1975) Familial inversion of chromosome no. 8: an affected child and a carrier fetus. Humangenetik 27:67–73 [PubMed]
- Green ED, Olson MV (1990) Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci USA 87:1213–1217 [DOI] [PMC free article] [PubMed]
- Guo W-J, Callif-Daley F, Zapata MC, Miller ME (1995) Clinical and cytogenetic findings in seven cases of inverted duplication of 8p with evidence of a telomeric deletion using fluorescence in situ hybridization. Am J Med Genet 58:230–236 [DOI] [PubMed]
- Henderson KG, Dill FJ, Wood S (1992) Characterization of an inversion duplication of the short arm of chromosome 8 by fluorescent in situ hybridization. Am J Med Genet 44:615–618 [DOI] [PubMed]
- Henthorn PS, Mager DL, Huisman TH, Smithies O (1986) A gene deletion ending within a complex array of repeated sequences 3′ to the human beta-globin gene cluster. Proc Natl Acad Sci USA 83:5194–5198 [DOI] [PMC free article] [PubMed]
- Hobbs HH, Brown MS, Goldstein JL, Russell DW (1986) Deletion of exon encoding cysteine-rich repeat of low density lipoprotein receptor alters its binding specificity in a subject with familial hypercholesterolemia. J Biol Chem 261:13114–13120 [PubMed]
- Huang LS, Ripps ME, Korman SH, Deckelbaum RJ, Breslow JL (1989) Hypobetalipoproteinemia due to an apolipoprotein B gene exon 21 deletion derived by Alu-Alu recombination. J Biol Chem 264:11394–11400 [PubMed]
- Hutchinson R, Wilson M, Voullaire L (1992) Distal 8p deletion (8p23.1-8pter): a common deletion? J Med Genet 29:407–411 [DOI] [PMC free article] [PubMed]
- Jones C, Patterson D, Kao F-T (1981) Assignment of the gene coding for phosphoribosylglycineamide formyltransferase to human chromosome 14. Somatic Cell Genet 7:399–409 [DOI] [PubMed]
- Kurahashi H, Sakamoto M, Ono J, Honda A, Okada S, Nakamura Y (1998) Molecular cloning of the chromosomal breakpoint in the LIS1 gene of a patient with isolated lissencephaly and balanced t(8;17). Hum Genet 103:189–192 [DOI] [PubMed]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC (1990) High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247:64–69 [DOI] [PubMed]
- Minelli A, Floridia G, Rossi E, Clementi M, Tenconi R, Camurri L, Bernardi F, et al (1993) D8S7 is consistently deleted in inverted duplications of the short arm of chromosome 8 (inv dup 8p). Hum Genet 92:391–396 [DOI] [PubMed]
- Mitchell JJ, Vekemans M, Luscombe S, Hayden M, Weber B, Richter A, Sparkes R, et al (1994) U-type exchange in a paracentric inversion as a possible mechanism of origin of an inverted tandem duplication of chromosome 8. Am J Med Genet 49:384–387 [DOI] [PubMed]
- Moore EE, Jones C, Kao F-T, Oates DC (1977) Synteny between glycinamide ribonucleotide synthetase and superoxide dismutase (soluble). Am J Hum Genet 29:389–396 [PMC free article] [PubMed]
- Neitzel H (1986) A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet 73:320–326 [DOI] [PubMed]
- Nevin NC, Morrison PJ, Jones J, Reid MM (1990) Inverted tandem duplication of 8p12-p23.1 in a child with increased activity of glutathione reductase. J Med Genet 27:135–136 [DOI] [PMC free article] [PubMed]
- Nicholls RD, Fischel-Ghodsian N, Higgs DR (1987) Recombination at the human alpha-globin gene cluster: sequence features and topological constraints. Cell 49:369–378 [DOI] [PubMed]
- Ohashi H, Wakui K, Ogawa K, Okano T, Niikawa N, Fukushima Y (1994) A stable acentric marker chromosome: possible existence of an intercalary ancient centromere at distal 8p. Am J Hum Genet 55:1202–1208 [PMC free article] [PubMed]
- Ottolenghi S, Lanyon WG, Paul J, Williamson R, Weatherall DJ, Clegg JB, Pritchard J, et al (1974) The severe form of alpha thalassaemia is caused by a haemoglobin gene deletion. Nature 251:389–391 [DOI] [PubMed]
- Patterson D, Hart I, Lai LW, Brahe C, Moscetti A, Tassone F, Raimondi E, et al (1993) Molecular cytogenetic characterization of a Chinese hamster/human hybrid cell line containing a der (21)t(Ypter→cenY::cen21→21qter) chromosome. Genomics 15:177–179 [DOI] [PubMed]
- Pecile V, Petroni MG, Fertz MC, Filippi G (1990) Deficiency of distal 8p—report of two cases and review of the literature. Clin Genet 37:271–278 [DOI] [PubMed]
- Pettenati MJ, Rao N, Johnson C, Hayworth R, Crandall K, Huff O, Thomas IT (1992) Molecular cytogenetic analysis of a familial 8p23.1 deletion associated with minimal dysmorphic features, seizures, and mild mental retardation. Hum Genet 89:602–606 [DOI] [PubMed]
- Pierce JC, Sternberg NL (1992) Using bacteriophage P1 system to clone high molecular weight genomic DNA. Methods Enzymol 216:549–574 [DOI] [PubMed]
- Rudiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23:256–260 [DOI] [PMC free article] [PubMed]
- Shechter I, Conrad DG, Hart I, Berger RC, McKenzie TL, Bleskan J, Patterson D (1994) Localization of the squalene synthase gene (FDFT1) to human chromosome 8p22-p23.1. Genomics 20:116–118 [DOI] [PubMed]
- Small MF, Stanyon R, Smith DG, Sineo L (1985) High-resolution chromosomes of Rhesus macaques (Macaca mulatta). Am J Primatol 9:63–67 [DOI] [PubMed] [Google Scholar]
- Smith ACM, Spuhler K, Williams TM, McConnell T, Sujansky E, Robinson A (1987) Genetic risk for recombinant 8 syndrome and the transmission rate of balanced inversion 8 in the Hispanic population of the southwestern United States. Am J Hum Genet 41:1083–1103 [PMC free article] [PubMed]
- Sternberg N (1990) Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc Natl Acad Sci USA 87:103–107 [DOI] [PMC free article] [PubMed]
- Sujansky E, Smith ACM, Prescott KE, Freehauf CL, Clericuzio C, Robinson A (1993) Natural history of the recombinant (8) syndrome. Am J Med Genet 47:512–525 [DOI] [PubMed]
- Taylor JM, Dozy A, Kan YW, Varmus HE, Lie-Injo LE, Ganesan J, Todd D (1974) Genetic lesion in homozygous alpha thalassaemia (hydrops fetalis). Nature 251:392–393 [DOI] [PubMed]
- Toriello HV, Glover TW, Takahara K, Byers PH, Miller DE, Higgins JV, Greenspan DS (1996) A translocation interrupts the COL5A1 gene in a patient with Ehlers-Danlos syndrome and hypomelanosis of Ito. Nat Genet 13:361–365 [DOI] [PubMed]
- Vanin EF, Henthorn PS, Kioussis D, Grosveld F, Smithies O (1983) Unexpected relationships between four large deletions in the human beta-globin gene cluster. Cell 35:701–709 [DOI] [PubMed]
- Weinberg J, Stanyon R, Jauch A, Cremer T (1992) Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma 101:265–270 [DOI] [PubMed]
- Wu B-L, Schneider GH, Sabatino DE, Bozovic JZ, Cao B, Korf BR (1996) Distal 8p deletion (8)(p23.1): an easily missed chromosomal abnormality that may be associated with congenital heart defect and mental retardation. Am J Med Genet 62:77–83 [DOI] [PubMed]