Abstract
Crouzon syndrome and Pfeiffer syndrome are both autosomal dominant craniosynostotic disorders that can be caused by mutations in the fibroblast growth factor receptor 2 (FGFR2) gene. To determine the parental origin of these FGFR2 mutations, the amplification refractory mutation system (ARMS) was used. ARMS PCR primers were developed to recognize polymorphisms that could distinguish maternal and paternal alleles. A total of 4,374 bases between introns IIIa and 11 of the FGFR2 gene were sequenced and were assayed by heteroduplex analysis, to identify polymorphisms. Two polymorphisms (1333TA/TATA and 2710 C/T) were found and were used with two previously described polymorphisms, to screen a total of 41 families. Twenty-two of these families were shown to be informative (11 for Crouzon syndrome and 11 for Pfeiffer syndrome). Eleven different mutations in the 22 families were detected by either restriction digest or allele-specific oligonucleotide hybridization of ARMS PCR products. We molecularly proved the origin of these different mutations to be paternal for all informative cases analyzed (P=2.4×10-7; 95% confidence limits 87%–100%). Advanced paternal age was noted for the fathers of patients with Crouzon syndrome or Pfeiffer syndrome, compared with the fathers of control individuals (34.50±7.65 years vs. 30.45±1.28 years, P<.01). Our data on advanced paternal age corroborates and extends previous clinical evidence based on statistical analyses as well as additional reports of advanced paternal age associated with paternal origin of three sporadic mutations causing Apert syndrome (FGFR2) and achondroplasia (FGFR3). Our results suggest that older men either have accumulated or are more susceptible to a variety of germline mutations.
Introduction
Crouzon syndrome (MIM 123500) and Pfeiffer syndrome (MIM 101600) are autosomal dominantly inherited craniosynostosis conditions that are both genetically heterogeneous (Passos-Bueno et al. 1999). The majority of patients with Crouzon syndrome or Pfeiffer syndrome have mutations in the extracellular immunoglobulin III domain of the fibroblast growth-factor receptor 2 (FGFR2) gene. Crouzon syndrome affects cranial development and is characterized by craniosynostosis, exophthalmos, and midface hypoplasia. The birth prevalence of Crouzon syndrome is estimated to be 15–16/1 million births (Cohen and Kreiborg 1992), and it is also estimated that ∼30%–60% of cases are sporadic (al-Qattan and Phillips 1997). Pfeiffer syndrome involves craniofacial abnormalities as well as abnormalities of the hands and feet. Clinical phenotypes in Pfeiffer syndrome span a wide spectrum. The more severe types of Pfeiffer syndrome are due to de novo mutations only, whereas the less-severe types are due to inherited mutations as well as to de novo mutations (Cohen 1993; Jones 1997). The birth prevalence of Pfeiffer syndrome is assumed to be less than that of Crouzon syndrome.
Sporadic cases of Crouzon syndrome and Pfeiffer syndrome—as well as those of other dominant skeletal dysplasias, such as achondroplasia and Apert syndrome—have previously been associated with advanced paternal age (Jones et al. 1975). Such sporadic cases of achondroplasia and Apert syndrome have been shown to be caused exclusively by mutations arising in the paternal germline (Moloney et al. 1996; Wilkin et al. 1998). Unlike both Apert syndrome and achondroplasia, in which mutational heterogeneity is exceedingly limited, at least 32 different mutations have been found in patients with Crouzon syndrome and at least 26 mutations have been found in patients with Pfeiffer syndrome (Passos-Bueno et al. 1999). In the present study, we show that 11 different mutations that are responsible for sporadic cases of either Crouzon syndrome or Pfeiffer syndrome also arise in the paternal germline. Furthermore, we confirm advanced paternal age in the fathers of affected individuals.
Subjects and Methods
Subjects
A total of 41 families consisting of one or both unaffected parents and their affected child were studied. This sample consisted of 18 American families, 18 British families, 1 Israeli family, 1 French family, 1 Irish family, 1 Norwegian family, and 1 Polish family. Informed consent for phenotypic information collection and genetic analysis was obtained from the patient and/or the parent(s), after approval was given by the appropriate institutional review board. Blood for DNA isolation and analysis was obtained from the patient and parent(s). Of the 41 families, 22 were found to be informative (11 for Crouzon syndrome and 11 for Pfeiffer syndrome), where the proband was heterozygous for at least one polymorphism and at least one parent was homozygous for a given polymorphism. Of the 22 informative families and the 19 noninformative families, blood was obtained from both the parents and the patient in 20 and 17 families, respectively. In the remaining two families in each category, blood was available from only the mother and the patient. In these four cases, the father was clinically unaffected. Parental ages were obtained for all of the mothers and for 39/41 fathers, at the time of their child's birth. The ages of the parents of affected children were compared with the ages of the parents in a control group, for all of the British mothers and for 16/18 British fathers (Office of Population Censuses and Surveys 1960, 1974–97) and for 14/18 American mothers and fathers, where matched parental age data could be calculated from raw data available for the year of the child's birth (U.S. Department of Health and Human Services 1967, pp. I-12, I-122; 1974, pp. I-14, I-56; 1988a, pp. 14, 95; 1988b, pp. 14, 95; 1989, pp. 15, 99; 1990, pp. 15, 99; 1993, pp. 104, 109; 1994, pp. 108, 113; 1995a, pp. 108, 113; 1995b, pp. 108, 113; 1999, pp. 108, 113; Riccardi 1988).
Paternal Identity and Sex Identification
Informative families for which there was DNA from the father were checked for paternity by analysis of either three polymorphic markers (D18S51, D19S253, and D21S11) (Urquart et al. 1995) or HLA class I markers. Typing for HLA class I (A, B, and C) allele groups comparable to serologically defined antigens was performed by use of the microsequence-specific primer (SSP) method. The tests were performed by use of the Pel-Freez SSP Unitray, according to the manufacturer's instructions. Ethnic background, when known, was matched to the corresponding allele frequencies. In cases where ethnic background was not available, the allele frequencies for white, African Caribbean, and Asian (from the Indian subcontinent) individuals (Urquart et al. 1995) were used to calculate the likelihood that the alleged father is the biological father. Statistical analysis based on Bayes theorem provided a ⩾95% probability that we had studied the biological fathers (Zachary et al. 1997). In the two informative case patients for which no blood samples were available from the fathers, the maternal contribution was homozygous for the allele that did not contain the mutation; thus, the paternal determination was made by exclusion.
Confirmation of the sex of maternal and paternal samples was made with the use of X-Y homologous primers (AMXY-1F and AMXY-2R) flanking the amelogenin gene on the X chromosome and the amelogenin-like sequence on the Y chromosome (Nakahori et al. 1991). The X-specific sequence has an additional 177-bp insert, which makes the size difference between the X and Y PCR products distinctive on gel electrophoresis.
Identification of Polymorphic Variants
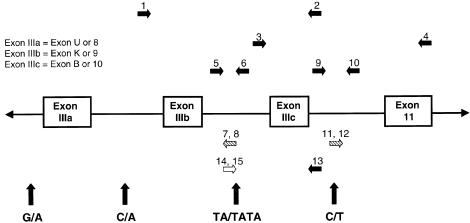
The upstream intron (intron IIIb) of exon IIIc (also referred to as “exon 10” or “exon B”) was PCR amplified by use of primers 1 and 2 (annealing at 54°C for 1 min and extension at 72°C for 4 min per cycle [35 cycles]) (table 1; fig. 1). The 1,663-bp PCR product was cloned by use of the TA Cloning Kit (Invitrogen). Plasmid DNA was isolated by use of the Qiagen Plasmid Midi-Kit. Clones were sequenced by the Johns Hopkins Genetic Research Core Facility, by means of both a 373A automated DNA sequencer (Applied Biosystems) and the fluorescent dideoxy terminator method (Smith et al. 1986), with use of these primers and other primers generated from the sequence. The downstream intron (intron IIIc) of exon IIIc was PCR amplified with the use of primers 3 and 4 (annealing at 50°C for 1 min and extension at 72°C for 4 min). The resulting 2,385-bp PCR product was cloned and was sequenced as described above. For confirmation, plasmid artificial chromosome (PAC) 304H11 and 71B17 clones located in this region were sequenced in the opposite direction. Both PAC clones were obtained by screening of a human PAC library (PAC-6541; Genome Systems) with a probe that was generated by PCR amplifying across FGFR2 exon 4 with intronic primers 5′-ACTTTGCTATGGAGAAGG-3′ and 5′-AAGAAGACAGGTGACAGG-3′. Sequence information was deposited in GenBank (accession number AF169399).
Table 1.
FGFR2 Oligonucleotides
| Positiona at |
||||
| Oligonucleotides | Sequence (5′→3′) | 5′ End | 3′ End | Description |
| 1 | CCCTTTAATGCCGCTGTTTAG | 532 | 552 | Intron IIIb amplification |
| 2 | AACCCAGAGAGAAAGAACAGTA | 2194 | 2173 | Intron IIIb amplification |
| 3 | ATCATTCCTGTGTCGTCTAGC | 1974 | 1994 | Intron IIIc amplification |
| 4 | TCCGCAGGGGGATACGTTTG | 4358 | 4338 | Intron IIIc amplification |
| 5 | TCCATGCCTGTTTAATACG | 1134 | 1152 | 1333TA/TATA polymorphism amplification |
| 6 | GACACCTCACCCATCCTC | 1582 | 1565 | 1333TA/TATA polymorphism amplification |
| 7 | CGTCCAGTAGTACATTCAT | 1340 | 1322 | TA ASO |
| 8 | CGTCCAGTATAGTACATTCAT | 1342 | 1322 | TATA ASO |
| 9 | TGCTTTCATCCCACTTTG | 2551 | 2568 | 2710C/T polymorphism amplification |
| 10 | AGCAAACCACAGTCTCTG | 2940 | 2923 | 2710C/T polymorphism amplification |
| 11 | TAGAATGGACTTTTGGTT | 2697 | 2714 | T ASO |
| 12 | TAGAATGGACTTTCGGTT | 2697 | 2714 | C ASO |
| 13 | AAAAAACCCAGAGAGAAAGAACAGTATA | 2198 | 2171 | ARMS amplification |
| 14 | GAGCTCATCTTAATGAATGTACTACT | 1310 | 1335 | TA ARMS primer |
| 15 | GAGCTCATCTTAATGAATGTACTATA | 1310 | 1335 | TATA ARMS primer |
Figure 1.
Partial map of the human FGFR2 gene. Unblackened arrow indicates the ARMS primer; diagonally hatched arrows, ASOs; blackened arrows, all other primers; vertical arrows, locations of the polymorphisms flanking exons IIIa and IIIc.
To search for polymorphic variants in the sequence, PCR products of 202–310 bp were amplified with the use of several overlapping primer sets generated from the sequence of the two flanking introns (IIIb and IIIc) and the genomic DNA from 20 control individuals. Heteroduplex analysis was done on 5 μl of the PCR product, by use of mutation-detection-enhancement (MDE; FMC Corporation) gel electrophoresis, according to the manufacturer's protocol. PCR products with variations in mobility were cloned and were sequenced.
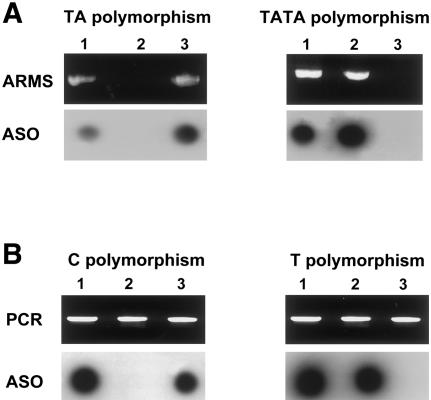
We found one polymorphism in intron IIIb and another polymorphism in intron IIIc. Intron IIIb, which has a TA/TATA polymorphism at nucleotide position 1333 (nucleotide numbers are derived from GenBank [accession number AF169399]), was PCR amplified for allele-specific oligonucleotide (ASO) hybridization or for amplification refractory mutation system (ARMS) analysis (see below). For detection by use of ASO, part of intron IIIb was PCR amplified with primers 5 and 6 (58°C annealing, 449-bp product; see table 1 and fig. 1). A total of 10–20 μl product was added to 80 μl 0.4 M NaOH and 25 mM Na2 EDTA solution, and samples were dotted onto a Zetabind filter. ASOs (ASO 7 and ASO 8) were developed for each of the polymorphic alleles. After hybridization, the filter was washed at 52°C (for ASO 7) or 54°C (for ASO 8), was autoradiographed, and was developed for analysis (fig. 2).
Figure 2.
Polymorphism detection in three unrelated control individuals. A, 1333TA/TATA polymorphism. Top panels show gel electrophoresis of ARMS PCR products derived from primers 13 and 14 (top left) or from primers 13 and 15 (top right). Bottom panels show PCR products that were amplified with primers 5 and 6, were dotted onto filters, and were hybridized with either ASO 7 (bottom left) or ASO 8 (bottom right). Lane 1 represents the heterozygous individual; lane 2, the individual that is homozygous for the TATA polymorphism; lane 3, the individual that is homozygous for the TA polymorphism. B, 2710C/T polymorphism. Top panels show gel electrophoresis of PCR products amplified with the use of primers 9 and 10. Bottom panels show the same PCR products dotted onto filters and hybridized with either ASO 12 (bottom left) or ASO 11 (bottom right). No ARMS PCR products are shown. Lane 1 represents the heterozygous individual; lane 2, the individual that is homozygous for the T polymorphism; lane 3, the individual that is homozygous for the C polymorphism.
Intron IIIc, which contains a C/T polymorphism at nucleotide position 2710, was amplified with the use of primers 9 and 10 (54°C annealing, 390-bp product). After ASO hybridization, the filter was washed at 52°C (for ASO 11) or 53°C (for ASO 12), was autoradiographed, and was developed for analysis (fig. 2). To determine the frequency of these polymorphisms, genomic DNA from at least 50 unrelated CEPH control individuals was amplified with the use of either ARMS primers (see data below) or primers 5 and 6, for ASO-hybridization analysis for the 1333TA/TATA polymorphism, and with the use of primers 9 and 10, for ASO-hybridization analysis for the 2710C/T polymorphism.
Design of ARMS Analysis
Mutations in our patients with Crouzon syndrome or Pfeiffer syndrome lie either in the 191-bp exon IIIa (also referred to as “exon 8” or “exon U”), in the 145-bp exon IIIc, or in the splice-acceptor site for exon IIIc of the FGFR2 gene (fig. 1). For patients with mutations in exon IIIa, we used the ARMS developed by Moloney et al. (1996), to amplify individual alleles by use of primers specific for single-nucleotide polymorphisms, a G/A variant in the upstream intron, and a C/A variant in the downstream intron. On the basis of the identification of a polymorphism upstream of exon IIIc, we developed ARMS primers with which to distinguish the TA and TATA variants. A segment of genomic DNA was amplified by use of either primers 13 and 14 (annealing at 64°C) or primers 13 and 15 (annealing at 63°C; see table 1 and fig. 1). PCR amplification yielded products of 889 bp and 891 bp, respectively. For each affected child and the parent(s), both PCR reactions were done to assess whether the family was informative. If the family was informative, we proceeded to analyze for a specific mutation by means of either a previously published restriction digest or ASO-hybridization detection. Specific mutations, types of mutations, and methods of mutation detection are summarized in table 2. By using the polymorphism data, we ascertained the phase of the mutation, and therefore the origin of the mutation was known.
Table 2.
Informative Patients, Their Diagnoses and Mutations, and the Methods Used to Detect the Mutations
| Patient ID Number | Diagnosisa | Age of Fatherb (years) | Age of Motherb (years) | Exon | Mutation | Type of Mutation | Method Used to DetectMutationc | Reference |
| C 6 | CZ | 35 | 35 | IIIa | S267P (T→C) | Missense | BslI+ | Oldridge et al. (1995) |
| C 4 | CZ | 60 | 31 | IIIa | C278F (G→T) | Missense | BbsI+ | Oldridge et al. (1995) |
| C 11 | CZ | 44 | 33 | IIIa | Q289P (A→C) | Missense | BsaJI+ | Oldridge et al. (1995) |
| FCAB 1452 | CZ | 41 | 35 | IIIc | C342S (G→C) | Missense | BsaAI or StyI+ | Present study |
| 1580 | CZ | NAd | 22 | IIIc | C342Y (G→A) | Missense | RsaI+ | Present study |
| C 12 | CZ | 28 | 28 | IIIc | C342Y (G→A) | Missense | RsaI+ | Oldridge et al. (1995) |
| FCJO OM12 | CZ | 35 | 28 | IIIc | C342Y (G→A) | Missense | RsaI+ | Present study |
| 1836 | CZ | 33 | 31 | IIIc | C342W (C→G) | Missense | ASO | Present study |
| FCKR OM6 | CZ | 34 | 38 | IIIc | A344A (G→A) | Splice | AciI− | Li et al. (1995) |
| FCCD 1946 | CZ | 31 | 27 | IIIc | A344A (G→A) | Splice | AciI− | Present study |
| 1839 | CZ | 39 | 36 | IIIc | S347C (C→G) | Missense | ASO | Present study |
| 1583 | P | 35 | 34 | IIIa | C278F (G→T) | Missense | BbsI+ | Present study |
| FPPB 28 | P | 39 | 32 | IIIa | C278F (G→T) | Missense | BbsI+ | Meyers et al. (1996) |
| FPKH 1552 | P | 26 | 27 | IIIa | C278F (G→T) | Missense | BbsI+ | Present study |
| FPAM 57 | P | 25 | 23 | IIIa | W290C (G→C) | Missense | AlwI− | Przylepa et al. (1998) |
| 1647 | P | 33 | 32 | IIIc | C342S (G→C) | Missense | StyI+ | Present study |
| 1963 (case 2) | P | 33 | 36 | IIIc | C342R (T→C) | Missense | CfoI+ | Rutland et al. (1995) |
| 1956 (case 4) | P | 38 | 28 | IIIc | C342R (T→C) | Missense | CfoI+ | Rutland et al. (1995) |
| FPRW 39 | P | 38 | 39 | IIIc | C342R (T→C) | Missense | CfoI+ | Meyers et al. (1996) |
| FPEW OM32 | P | 32 | 27 | IIIc | C342R (T→C) | Missense | CfoI+ | Meyers et al. (1996) |
| FPRA OM20 | P | 22 | 22 | IIIc | C342Y (G→A) | Missense | RsaI+ | Present study |
| 1833 (patient 4) | P | 30 | 32 | IIIc | 940-2A→T | Splice | EaeI+ | Oldridge et al. (1999) |
CZ = Crouzon syndrome; P = Pfeiffer syndrome.
Age of parent at time of patient's birth.
+ = mutation created a restriction site; − = mutation destroyed a restriction site.
NA = not available.
Results
Parental Ages
The mean paternal and maternal ages were determined for the families in the present study (table 3). The mean parental ages for all of the families in both the informative (mean paternal age 34.81±7.92 years; mean maternal age 30.68±4.89 years) and noninformative (mean paternal age 32.94±6.18 years; mean maternal age 28.32±4.31 years) groups are similar. Statistical analysis with use of the t-test was not able to reject the null hypothesis that the mean parental ages in each group were the same, thus arguing against an age bias in families that are informative. The apparent difference between the mean paternal ages of the fathers of the families that are informative (for Crouzon syndrome 38.00±9.06 years; for Pfeiffer syndrome 31.91±5.66 years) versus the fathers of the families that are noninformative (for Crouzon syndrome 31.00±4.69 years; for Pfeiffer syndrome 36.83±7.36 years) is most likely a result of the small sample sizes—in particular, the six families that are noninformative for Pfeiffer syndrome. Since, in this study, Crouzon syndrome and Pfeiffer syndrome are both the result of FGFR2 mutations and since identical FGFR2 mutations can be found in either syndrome, patients can be viewed on a phenotypic continuum where patients with Crouzon syndrome have a craniosynostotic phenotype and where patients with Pfeiffer syndrome have this phenotype as well as abnormalities of the digits. With this in mind and given the fact that there is no significant difference between the mean parental ages of the informative and noninformative groups, we chose to combine these parental ages and to compare our data against the average parental ages in the general population to look for evidence of a parental age effect.
Table 3.
Parental Ages in Families with Crouzon Syndrome and Pfeiffer Syndrome
| Families | No. of Individuals | Mean ± SD Age (Range) (years) | No. of Individuals | Mean ± SD Age (Range) (years) | Δage(years) |
| Fathers |
Mothers |
||||
| Informative: | |||||
| Crouzon syndrome | 10 | 38.00 ± 9.06 (28–60) | 11 | 31.18 ± 4.67 (22–38) | 6.82 |
| Pfeiffer syndrome | 11 | 31.91 ± 5.66 (22–39) | 11 | 30.18 ± 5.29 (22–39) | 1.73 |
| Total | 21 | 34.81 ± 7.92a (22–60) | 22 | 30.68 ± 4.89b (22–39) | 4.13c |
| Noninformative: | |||||
| Crouzon syndrome | 12 | 31.00 ± 4.69 (25–40) | 13 | 28.54 ± 4.03 (24–35) | 2.46 |
| Pfeiffer syndrome | 6 | 36.83 ± 7.36 (28–48) | 6 | 27.83 ± 5.23 (22–35) | 9.00 |
| Total | 18 | 32.94 ± 6.18 (25–48) | 19 | 28.32 ± 4.31 (22–35) | 4.63c |
| Informative and Noninformative: | |||||
| Crouzon syndrome | 22 | 34.18 ± 7.71 (25–60) | 24 | 29.75 ± 4.45 (22–38) | 4.43 |
| Pfeiffer syndrome | 17 | 33.65 ± 6.55 (22–48) | 17 | 29.35 ± 5.23 (22–39) | 4.30 |
| Total | 39 | 33.95 ± 7.14 (22–60) | 41 | 29.59 ± 4.73 (22–39) | 4.39d |
| Informative: | |||||
| Matched affected | 17e | 35.35 ± 8.46 (22–60) | 18f | 30.44 ± 4.82 (22–38) | 4.91c |
| Matched control | 17e | 30.66 ± 1.57 (27.39–32.95) | 18f | 27.47 ± 1.81 (24.10–30.30) | |
| Difference | 4.69g | 2.97h | |||
| Informative and Noninformative: | |||||
| Matched affected | 30i | 34.50 ± 7.65 (22–60) | 32j | 29.88 ± 4.66 (22–38) | 4.62d |
| Matched control | 30i | 30.45 ± 1.28 (27.39–32.95) | 32j | 27.18 ± 1.53 (24.10–30.30) | |
| Difference | 4.05h | 2.70h | |||
P=.42 (compared with average paternal age for all noninformative families).
P=.11 (compared with average maternal age for all noninformative families).
.01<P<.05 by the t-test.
P<.01 by the t-test.
Eleven British and 6 American fathers, for whom matched control data for paternal ages at the year of the affected child's birth were available, were used in this calculation. This subset of families included families informative for either Crouzon syndrome or Pfeiffer syndrome.
Twelve British and 6 American mothers, for whom matched control data for maternal ages at the year of the affected child's birth were available, were used in this calculation. This subset of families included families informative for either Crouzon syndrome or Pfeiffer syndrome.
P=.022 by the one-tailed paired t-test.
P<.01 by the one-tailed paired t-test.
Sixteen British and 14 American fathers, for whom matched control data for paternal ages at the year of the affected child's birth were available, were used in this calculation. This subset of families included families that are either informative or noninformative for Crouzon syndrome or Pfeiffer syndrome.
Eighteen British and 14 American mothers, in whom matched control data for maternal ages at the year of the affected child's birth were available, were used in this calculation. This subset of families included families that are either informative or noninformative for Crouzon syndrome and Pfeiffer syndrome.
An exact analysis of a parental age effect can be done by comparing the ages of parents of affected children with the ages of a control population for which parental age data also exist, specific for the exact year of the affected child's birth (referred to here as “matched data”). Because there was no statistical difference between the parental ages in the informative versus the noninformative groups, we were able to use those informative and noninformative families for which matched data were available. This was possible for all of the British mothers, for 16/18 British fathers, and for 14/18 American families. The difference between the paternal age at the time of the child's birth and the mean paternal age in the general population for that year of birth was calculated. For the British families, paternal age data differed, depending on whether the child was born in or out of wedlock; this variable was taken into consideration when the calculations were made. Similarly, the difference between the maternal age at the time of the child's birth and the mean maternal age in the general population for that year was also calculated. Significant increases in both paternal age (+4.05 years, P<.01) and maternal age (+2.70 years, P<.01) were noted, thus suggesting an advanced parental age effect on sporadic mutations causing both Crouzon syndrome and Pfeiffer syndrome. A significant increase in paternal age (+4.69 years, P=.022) was noted for the cases proved molecularly to be paternal in origin (informative cases). Of most significance, the differences between mean paternal age and mean maternal age (referred to as “Δage” in table 3) were observed when data were generated from the informative, noninformative, or combined groups, irrespective of whether these groups were matched for the year of the child’s birth. Taken together, these results are consistent in demonstrating an increased paternal age in sporadic cases of Crouzon syndrome and Pfeiffer syndrome.
Polymorphism Frequency
The frequency of the TA variant of the 1333TA/TATA polymorphism in intron IIIb was determined to be 68.75%, whereas the frequency of the other variant, TATA, was found to be 31.25% (n=112). The frequency of the C polymorphism in intron IIIc was determined to be 73.94%, whereas the other variant, T, was only found in 26.06% of the alleles (n=142). Figure 2 depicts the identification of these polymorphic variants in unrelated control individuals by means of the two previously described methods (ARMS and ASO).
Thirty-eight control individuals were screened for both polymorphisms. In this subsample, the TATA variant was only present when the T variant was present as well. We analyzed the haplotypes present in the 15 informative patients with mutations in exon IIIc, to determine whether a certain haplotype was predisposed to the mutation. In 11/15 patients, the mutation occurred on the TATA-T background, and, in the remaining four cases, the mutation occurred in the TA-C background (P=.042). With this sample size there is no clear predisposition to mutations occurring on one particular haplotype.
Identification of Origin of Mutation
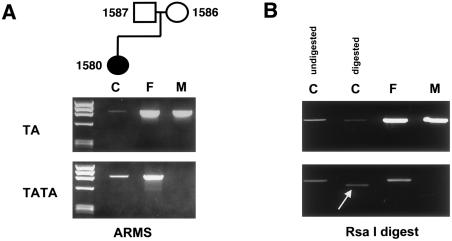
The parental origin of mutations in sporadic cases of Crouzon and Pfeiffer syndromes was determined by the use of ARMS allele-specific PCR followed by either restriction digest or ASO hybridization specific for mutations. For 22 informative cases, all were of paternal origin for 11 different mutations. For example, figure 3 shows an informative family in which the C342Y mutation in exon IIIc of the affected patient (patient 1580) was detected by restriction-enzyme digestion. ARMS PCR amplification of the TA polymorphism (with the use of primers 13 and 14) resulted in a product for the affected child and both parents. ARMS PCR amplification of the TATA polymorphism (with the use of primers 13 and 15) yielded a product from only the affected child and the normal father. Therefore, both the child and the father are heterozygous, whereas the mother is homozygous for the TA polymorphism. Digestion of the child's allele with the TATA polymorphism, the allele inherited from the father, with the use of RsaI results in a smaller DNA fragment, since the mutation created a new restriction site. The father’s TATA allele did not digest. Digestion of the other allele with the TA polymorphism from the child and the father, as well as digestion of both of the mother’s alleles, with RsaI, did not result in smaller DNA fragments, because these alleles do not contain the mutation. Therefore, the origin of the mutation is determined to be paternal.
Figure 3.
Determination of the origin of mutation by means of ARMS PCR followed by restriction digest. A, top panel, primers 13 and 14 were used to amplify 889 bp, starting from the TA variant and extending 3′ to exon IIIc (in which the mutation is located). Unmarked lane indicates size markers φX174 DNA digested with HaeIII. Templates for ARMS PCR are as follows: affected child (subject 1580) (lane C), unaffected father (subject 1587) (lane F), and unaffected mother (subject 1586) (lane M). A, bottom panel, primers 13 and 15 were used to amplify 891 bp from the TATA variant to 3′ of the exon IIIc. Templates for ARMS PCR are the same as those for the top panel. B, The G→A nucleotide change for the C342Y mutation creates an RsaI restriction site in the patient. Top panel indicates digest of ARMS PCR products amplified with the use of primers 13 and 14. Bottom panel indicates digest of ARMS PCR products amplified with the use of primers 13 and 15. Samples are as follows: affected child, undigested (left lane C); affected child, digested (right lane C); unaffected father, digested (lane F); and unaffected mother, digested (lane M). The arrow indicates the smaller digested product from the child’s allele containing the TA polymorphism.
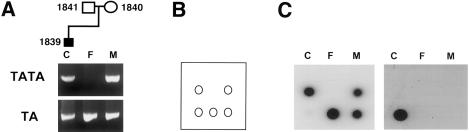
Figure 4 shows another informative family in which the S347C mutation in exon IIIc of the affected child was detected by use of ASO hybridization. As in the previous case, ARMS primers were used to amplify individual alleles, each of which contained one of the polymorphic variants. In this family, the affected child and the normal mother are heterozygous, whereas the father is homozygous for the TA polymorphism. ASOs 5′-GGGTAATTCTATTGGGAT-3′ and 5′-GGGTAATTGTATTGGGAT-3′ were used to hybridize to the normal and mutant alleles, respectively. The former ASO hybridized to the normal allele in the affected child as well as to both alleles in both unaffected parents. The latter ASO, which recognized the mutation, hybridized only to the child's allele containing the TA polymorphism that was inherited from the father. The origin of the mutation is again determined to be paternal.
Figure 4.
Determination of the origin of mutation by means of ARMS PCR followed by ASO hybridization. A, ARMS PCR products spanning the region containing the TA/TATA polymorphism and exon IIIc, which contains the mutation. Templates for PCR: affected child (subject 1839) (lane C), unaffected father (subject 1841) (lane F), and unaffected mother (subject 1840) (lane M). Top lanes were PCR amplified with the use of primers 13 and 15, which recognize the TATA polymorphism. Bottom lanes were PCR amplified with the use of primers 13 and 14, which recognize the TA polymorphism. B, diagram of the filter dotted with ARMS PCR products in the same orientation that is seen in A. C, Left filter is hybridized with ASO (GGGTAATTCTATTGGGAT), which recognizes the normal allele. Right filter is hybridized with ASO (GGGTAATTGTATTGGGAT), which recognizes the C→G nucleotide change in the S347C mutation in exon IIIc.
For those patients that had mutations in exon IIIa, we used ARMS primers developed by Moloney et al. (1996) to recognize polymorphisms flanking exon IIIa. Detection of those mutations required restriction digests.
Discussion
We have molecularly proved a paternal origin of mutation in all 22 informative cases of sporadic Crouzon syndrome and Pfeiffer syndrome. Furthermore, in our analysis of parental age, we have shown that there is an advanced parental age effect for these syndromes. These data provide strong evidence that the parental age effect is primarily paternal. Although the association between older paternal age and sporadic mutations—hypothesized to be paternal in origin—has been noted since the beginning of this century, Jones et al. described the first association of older paternal age with Crouzon syndrome and Pfeiffer syndrome in 1975. A comparison of the original data from 1975 with data from the present study is shown in table 4. Both sets of data are similar, with the added observation that we have now molecularly proved the origin of mutation to be paternal.
Table 4.
Comparison of Two Parental Age Studies
| Study and Syndrome | No. of Individuals | Mean ± SD Age(years) | No. of Individuals | Mean ± SD Age(years) | Δage(years) |
| Fathers |
Mothers |
||||
| Present study: | |||||
| Crouzon syndromea | 22 | 34.18 ± 7.71 | 24 | 29.75 ± 4.45 | 4.43 |
| Pfeiffer syndromea | 17 | 33.65 ± 6.55 | 17 | 29.35 ± 5.23 | 4.30 |
| Jones et al. (1975): | |||||
| Crouzon syndrome | 38 | 33.90 ± 8.30 | 38 | 28.60 ± 5.90 | 5.30 |
| Pfeiffer syndrome | 10 | 29.20 ± 4.00 | 10 | 27.00 ± 5.30 | 2.20 |
Informative and noninformative families.
More than 10 genetic disorders are associated with advanced paternal age; half of them (Apert syndrome, achondroplasia, multiple endocrine neoplasia type 2B [Carlson et al. 1994], Crouzon syndrome, and Pfeiffer syndrome) have definitively been shown to be the result of mutations that are paternal in origin. The long observed paternal-age effect was explained by Penrose (1955) in his “copy error hypothesis,” which was based on the different mechanisms of gametogenesis in males and females. In males, 36 divisions give rise to the spermatogonia that are present before puberty begins. From puberty onward, the spermatogonia stem cells divide every 16 days, which is equivalent to 23 divisions per year. Thus, in a 30-year-old male, for example, 430 divisions of the stem cells have occurred (Heller and Clermont 1963; Vogel and Rathenberg 1975; Crow 1997). If one compares the processes of spermatogeneis and oogenesis, a major difference is immediately apparent: ∼24 divisions occur in the female, to produce all of the oocytes already present at birth (Chandley 1991).
Examination of the processes of gametogenesis allows one to hypothesize about the types of mutations one would see in older men versus older women. Thus, in men, it is easy to see that, given a fixed error-of-replication rate, the more divisions that occur (i.e., the older the male), the more mutations that occur (i.e., the more children born with a specific disorder), for a supposedly linear rate of mutation that is dependent on the age of the male (i.e., the number of divisions). These mutations would most likely be point mutations (Vogel and Rathenberg 1975; Chandley 1991; Crow 1997). During examination of the data for the frequency of sporadic cases of achondroplasia, Apert syndrome, Crouzon syndrome, or Pfeiffer syndrome versus paternal age, a linear relationship is not evident. In fact, the curve steepens as the rate of increase in the mutation rate itself increases with age (Vogel and Rathenberg 1975; Risch et al. 1987; Crow 1997). One explanation for this could be that the efficiency of the proofreading of the replication machinery in spermatogenesis declines with age (Crow 1997). This suggests that there are other as-yet-unknown mechanisms that contribute to this advanced paternal-age effect.
It is interesting to note that, in our study, 11 different point mutations occurred in the informative families and that, for every one of these mutations, the origin of mutation was paternal. Furthermore, nine of these mutations were missense, and the other two were splice-site mutations. In the two previous reports of paternal origin of mutation for Apert syndrome and achondroplasia (Moloney et al. 1996; Wilkin et al. 1998), two point mutations caused each disorder. These mutations were all missense mutations, and two of the four mutations occurred within a CpG dinucleotide and account for >97% of all reported cases. The germline frequencies of these mutations were 5.0×10-6 and 2.7×10-6 for the Apert mutations (Moloney et al. 1996) and 5.5–28×10-6 for the achondroplasia mutations (Bellus et al. 1995). Spontaneous deamination of a methylated cytosine could be the mechanism for the more common of the Apert syndrome and achondroplasia mutations, each of which occurs within a CpG dinucleotide.
In this study, however, none of the 11 mutations occurred within the context of a CpG dinucleotide. Among the mutations that occurred in the informative patients (table 2), the C278F, C342R, and C342Y mutations occurred most frequently, which reflects the overall occurrence of each mutation in Crouzon syndrome and Pfeiffer syndrome (Passos-Bueno et al. 1999). The frequencies of these mutations would most certainly be less than those of the mutations causing Apert syndrome and achondroplasia. This is more evidence to suggest that perhaps another mechanism is responsible for the advanced paternal-age effect. It is intriguing that all of the mutations previously mentioned for achondroplasia, Apert syndrome, Crouzon syndrome, and Pfeiffer syndrome are point mutations, occur in FGFR genes, are paternal in origin, and have an advanced paternal-age effect. One such mechanism could be the effect of cis sequences on mutations. We examined the haplotype background on which the mutation occurred. No significant difference between backgrounds was found.
Oldridge et al. (1997) proposed that “the high apparent rates for several FGFR mutations could arise by a selective advantage conferred to the mutated male germ cell.” With any mutation, there is a bias in detecting those mutations that cause a phenotype. Inherent in this is the fact that the cell with the mutation must be able to survive, and, in this case, maybe those mutations that are occurring at an increasing rate in the sperm of older men somehow confer a selective advantage over other sperm without the mutations. We anticipate that future studies will clarify the underlying mechanism(s) responsible for the paternal origin of mutations as well as for the paternal-age effect.
Acknowledgments
We would like to thank all of the families who participated in this study, as well as A. T. Midro, P. R. Njolstad, and C. A. Stevens, for contributing samples, and Marilyn C. Jones, Donald Day, and Linda S. Cowan, for contributing information on previously reported cases. We are very grateful to Roxann Ashworth, for help with sequencing, and to Michael Posner, for helpful discussions. This study was supported by National Institute of Health grants T32 GM07814 (to R.L.G.), T32 GM7471 (to S.A.B.), and RR 00052 and RO1 DE1441 (to E.W.J.), and by Wellcome Trust awards (to D.J. and A.O.M.W). This work was presented at the 49th Annual Meeting of the American Society of Human Genetics, San Francisco, October 19–23, 1999.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CEPH, http://www.cephb.fr/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for oligonucleotide primers and nucleotide numbers [accession number AF169399])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Crouzon syndrome [MIM 123500] and Pfeiffer syndrome [MIM 101600])
References
- al-Qattan MM, Phillips JH (1997) Clinical features of Crouzon's syndrome patients with and without a positive family history of Crouzon's syndrome. J Craniofac Surg 8:11–13 [DOI] [PubMed]
- Bellus GA, Hefferon TW, Ortiz de Luna RI, Hecht JT, Horton WA, Machado M, Kaitila I, et al (1995) Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet 56:368–373 [PMC free article] [PubMed]
- Carlson KM, Bracamontes J, Jackson CE, Clark R, Lacroix A, Wells SA Jr, Goodfellow PJ (1994) Parent-of-origin effects in multiple endocrine neoplasia type 2B. Am J Hum Genet 55:1076–1082 [PMC free article] [PubMed]
- Chandley AC (1991) On the parental origin of de novo mutation in man. J Med Genet 28:217–223 [DOI] [PMC free article] [PubMed]
- Cohen MM Jr (1993) Pfeiffer syndrome update, clinical subtypes, and guidelines for differential diagnosis. Am J Med Genet 45:300–307 [DOI] [PubMed]
- Cohen MM Jr, Kreiborg S (1992) Birth prevalence studies of the Crouzon syndrome: comparison of direct and indirect methods. Clin Genet 41:12–15 [DOI] [PubMed]
- Crow JF (1997) The high spontaneous mutation rate: is it a health risk? Proc Natl Acad Sci USA 94:8380–8386 [DOI] [PMC free article] [PubMed]
- Heller CG, Clermont Y (1963) Spermatogenesis in man: an estimate of its duration. Science 140:184–185 [DOI] [PubMed] [Google Scholar]
- Jones KL (ed) (1997) Smith's recognizable patterns of human malformation, 5th ed. WB Saunders, Philadelphia [Google Scholar]
- Jones KL, Smith DW, Harvey MA, Hall BD, Quan L (1975) Older paternal age and fresh gene mutation: data on additional disorders. J Pediatr 86:84–88 [DOI] [PubMed]
- Li X, Park W-J, Pyeritz RE, Jabs EW (1995) Effect on splicing of a silent FGFR2 mutation in Crouzon syndrome. Nat Genet 9:232–233 [DOI] [PubMed]
- Meyers GA, Day D, Goldberg R, Daentl DL, Pryzlepa KA, Abrams LJ, Graham JM Jr, et al (1996) FGFR2 exon IIIa and IIIc mutations in Crouzon, Jackson-Weiss, and Pfeiffer syndromes: evidence for missense changes, insertions and a deletion due to alternative RNA splicing. Am J Hum Genet 58:491–498 [PMC free article] [PubMed]
- Moloney DM, Slaney SF, Oldridge M, Wall SA, Sahlin P, Stenman G, Wilkie AOM (1996) Exclusive paternal origin of new mutations in Apert syndrome. Nat Genet 13:48–53 [DOI] [PubMed]
- Nakahori Y, Hamano K, Iwaya M, Nakagome Y (1991) Sex identification by polymerase chain reaction using X-Y homologous primer. Am J Med Genet 39:472–473 [DOI] [PubMed]
- Office of Population Censuses and Surveys (1974–97) Birth statistics, series FM1. Her Majesty's Stationery Office, London [Google Scholar]
- ——— (1960) Registrar General’s statistical review of England and Wales. Part II. Tables, population. Her Majesty's Stationery Office, London [Google Scholar]
- Oldridge M, Lunt PW, Zackai EH, McDonald-McGinn DM, Muenke M, Moloney DM, Twigg SR, et al (1997) Genotype-phenotype correlation for nucleotide substitutions in the IgII–IgIII linker of FGFR2. Hum Mol Genet 6:137–143 [DOI] [PubMed]
- Oldridge M, Wilkie AOM, Slaney SF, Poole MD, Pulleyn LJ, Rutland P, Hockley AD, et al (1995) Mutations in the third immunoglobulin domain of the fibroblast growth factor receptor-2 gene in Crouzon syndrome. Hum Mol Genet 4:1077–1082 [DOI] [PubMed]
- Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SR, Johnson D, et al (1999) De novo Alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am J Hum Genet 64:446–461 [DOI] [PMC free article] [PubMed]
- Passos-Bueno MR, Wilcox WR, Jabs EW, Sertié AL, Alonso LG, Kitoh H (1999) Clinical spectrum of fibroblast growth factor receptor mutations. Hum Mutat 14:115–125 [DOI] [PubMed]
- Penrose LS (1955) Parental age and mutation. Lancet 2:312–313 [DOI] [PubMed] [Google Scholar]
- Pryzlepa KA, Moloney DM, Wall SA, Gagnon D, Hoganson G, Yin M, Wilkie AOM, et al (1998) A FGFR2 mutation causing type 2 Pfeiffer syndrome. J Craniofac Genet Dev Biol 18:6–7 [Google Scholar]
- Riccardi VM (1988) American paternal age data for selected years from 1876 to 1981. Neurofibromatosis 1:93–99 [PubMed]
- Risch R, Reich EW, Wishnick MM, McCarthy JG (1987) Spontaneous mutation and parental age in humans. Am J Hum Genet 41:218–248 [PMC free article] [PubMed]
- Rutland P, Pulleyn LJ, Reardon W, Baraitser M, Hayward R, Jones B, Malcolm S, et al (1995) Identical mutations in the FGFR2 gene cause both Pfeiffer and Crouzon syndrome phenotypes. Nat Genet 9:173–176 [DOI] [PubMed]
- Smith LM, Sander JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, et al (1986) Fluorescence detection in automated DNA sequence analysis. Nature 321:674–679 [DOI] [PubMed]
- Urquart A, Oldroyd NJ, Kimpton CP, Gill P (1995) Highly discriminating heptaplex short tandem repeat PCR system for forensic identification. Biotechniques 18:116–118, 120–121 [PubMed]
- U.S. Department of Health and Human Services, Public Health Service. (1967) Vital statistics of the United States 1965. Vol 1: Natality. Washington, DC, U.S. Department of Health and Human Services [Google Scholar]
- ——— (1974) Vital statistics of the United States 1969. Vol 1: Natality. Washington, DC, U.S. Department of Health and Human Services [Google Scholar]
- ——— (1988a) Vital statistics of the United States 1984. Vol 1: Natality. Washington, DC, U.S. Department of Health and Human Services [Google Scholar]
- ——— (1988b) Vital statistics of the United States 1986. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1989) Vital statistics of the United States 1987. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1990) Vital statistics of the United States 1988. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1993) Vital statistics of the United States 1989. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1994) Vital statistics of the United States 1990. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1995a) Vital statistics of the United States 1991. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1995b) Vital statistics of the United States 1992. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- ——— (1999) Vital statistics of the United States 1993. Vol 1: Natality. Washington, DC,U.S. Department of Health and Human Services [Google Scholar]
- Vogel F, Rathenberg R (1975) Spontaneous mutation in man. Adv Hum Genet 5:223–318 [DOI] [PubMed]
- Wilkin DJ, Szabo JK, Cameron R, Henderson S, Bellus GA, Mack ML, Kaitila I, et al (1998) Mutations in fibroblast growth-factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. Am J Hum Genet 63:711–716 [DOI] [PMC free article] [PubMed]
- Zachary AA, Bias WB, Word CJ (1997) Principles and applications of genetic identification. In: Rose NR, Conway de Macario E, Folds JD, Lane HC, Nakamura RM (eds) Manual of clinical laboratory immunology R: transplantation immunology and immunogenetics. AMS Press, Washington, DC, pp 1141–1151 [Google Scholar]