Abstract
Familial juvenile nephronophthisis is an autosomal recessive, genetically heterogeneous kidney disorder representing the most frequent inherited cause of chronic renal failure in children. A gene, NPHP1, responsible for ∼85% of the purely renal form of nephronophthisis, has been mapped to 2q13 and characterized. The major NPHP1 gene defect is a large homozygous deletion found in ∼80% of the patients. In this study, by large-scale genomic sequencing and pulsed-field gel electrophoresis analysis, we characterized the complex organization of the NPHP1 locus and determined the mutational mechanism that results in the large deletion observed in most patients. We showed that the deletion is 290 kb in size and that NPHP1 is flanked by two large inverted repeats of ∼330 kb. In addition, a second sequence of 45 kb located adjacent to the proximal 330-kb repeat was shown to be directly repeated 250 kb away within the distal 330-kb repeat deleting the sequence tag site (STS) 804H10R present in the proximal copy. The patients' deletion breakpoints appear to be located within the 45-kb repeat, suggesting an unequal recombination between the two homologous copies of this smaller repeat. Moreover, we demonstrated a nonpathologic rearrangement involving the two 330-kb inverted repeats found in 11 patients and, in the homozygous state, in 2 (1.3%) control individuals. This could be explained by interchromosomal mispairing of the 330-kb inverted repeat, followed by double recombination or by a prior intrachromosomal mispairing of these repeats, leading to an inversion of the NPHP1 region, followed by an interchromosomal unequal crossover event. This complex rearrangement, as well as the common deletion found in most patients, illustrates the high level of rearrangements occurring in the centromeric region of chromosome 2.
Introduction
Familial juvenile nephronophthisis (MIM 256100) is a progressive tubulointerstitial kidney disorder with autosomal recessive inheritance. The disease is characterized by polyuria, growth retardation, and deterioration of renal function during childhood or adolescence. It accounts for 6%–10% of end-stage renal disease in children (Antignac et al. 1998). The most prominent histological changes that occur are characterized by tubular basement membrane thickening, tubular atrophy, interstitial fibrosis, and medullary cyst formation (Waldherr at al. 1982). Various extrarenal manifestations have been associated with nephronophthisis, especially Leber amaurosis (described as Senior-Løken syndrome [SLS]; Senior et al. 1961). Further abnormalities, such as cerebellar dysfunction or liver involvement, are, however, rarely observed (reviewed in Antignac et al. 1998). A gene (NPHP1) responsible for the vast majority (∼85%) of the purely renal form of nephronophthisis was mapped to chromosome 2q13 (Antignac et al. 1993; Medhioub et al. 1994), and it was subsequently identified by a positional cloning strategy (Hildebrandt et al. 1997; Saunier et al. 1997) by means of the knowledge of large homozygous deletions found in the majority of patients (Konrad et al. 1996). At the time, we had shown that these deletions, each covering ∼250 kb, were located between two large inverted repeats of ∼100 kb and that the telomeric repeat was at least partially deleted. The use of several STSs within the common deletion interval allowed the establishment of an accurate and easy diagnostic PCR-based assay (Konrad et al. 1996). Since then, different smaller deletions have been characterized, leading to the reduction of the NPHP1 interval and the cloning of the gene (Hildebrandt et al. 1997; Saunier et al. 1997). This gene encodes a new protein, nephrocystin, which contains an Src homology 3 (SH3) domain and whose function remains to be elucidated.
In this study, by large-scale genomic sequencing of a large part of the region and by pulsed-field gel electrophoresis (PFGE), we have extensively characterized a genomic interval of ∼900 kb covering the NPHP1 deletion region. We have shown that this region has a very complex organization containing large inverted repeats of ∼330 kb and direct repeats of 45 kb. All the deletions we have detected to date arise from the same mechanism of homologous recombination between the two 45-kb direct repeats, contrary to the suggestions of the initial data. Furthermore, we have shown that the large inverted repeats mediate a nonpathogenic rearrangement likely to arise through interchromosomal mispairing and homologous recombination.
Patients and Methods
Patients
Thirty-one unrelated patients with familial juvenile nephronophthisis, without extrarenal symptoms, and with evidence (detected by PCR) of a homozygous deletion of the NPHP1 region were selected for PFGE analysis. Although these patients are phenotypically identical, they could be classified into three groups with regard to the STS content of their deletions, as follows.
Group A
Group A comprised 27 unrelated patients, 7 of whom belonged to consanguineous families, with deletions encompassing markers 765F2L and 804/6 (Konrad et al. 1996). These two markers are located within the most common NPHP1 deletion.
Group B
Group B was made up of two unrelated patients (one from a consanguineous family) with deletions of 804H10R, which is located outside the previously described deletions (Konrad et al. 1996; Hildebrandt et al. 1997; Saunier et al. 1997).
Group C
Group C consisted of two unrelated patients, both from consanguineous families, with deletions encompassing 765F2L, 804/6, and 804H10R.
Large-Scale Genomic Sequencing
Four BAC clones (183K24, 187E16, 96G18, and part of 49G15) from a human BAC genomic library (Research Genetics) were sequenced as described elsewhere (Saunier et al. 1997).
PCR Analysis
Sequences derived from BAC sequencing were used to generate new STSs listed as in table 1. Primer sequences and PCR conditions were selected with the OLIGO 5.0 program (NBI) after identification and masking of repetitive sequences with the RepeatMasker program (Smit 1996). We performed PCR in a 25-μl volume containing 50 ng of genomic DNA or one YAC colony.
Table 1.
STSs Mapping to the NPHP1 Region
| Name | Primer Sequence | Amplified Product Size (bp) | Annealing Temperature (°C) |
| 47A | AAA TAC ACC CAA ACC CAC AG | 689 | 50 |
| CCC ATC AGA GCA AAC AAG T | |||
| 53A | GCC ACA AGG AGG AGG AA | 615 | 50 |
| CGC CAA ATA CCA TAC AAA GT | |||
| 57A | GGG CGG AGA CCA AGA ATA GA | 731 | 52 |
| CGC AGG CTG ACT CAA ACC A | |||
| 49.81B | TTT CTC TAT CTT TGG CGT AT | 586 | 53 |
| TCT TTC ACC TGG CAT CTA | |||
| C40 | CGA TGG ACC CAG AGC AGT TT | 598 | 55 |
| TGG CCA TAG AGG AGA CA | |||
| R30A | CAC ACC ACC TGA GCC AAT G | 510 | 52 |
| GTC AAG TGA GAA CGC CAC AA | |||
| C30124 | GTC CAC AAC AAT CTC TAG GG | 124 | 51 |
| GTC TCT CCA TAC TCC CTT CT | |||
| R30E | CTG GAT GGG ACG CTG TT | 177 | 50 |
| ATT GCC ACA TGG ATT CTC TG | |||
| N123 | AGA GGT GGC AGG GAA GTG AA | 149 | 55 |
| CCT GCA ACA TCC TGG CTA AAC |
Pulsed-Field Gel Electrophoresis
Agarose-embedded DNA from the patients and from YACs (879D3, 804H10, 765F2, 769G7, and 921H4) was digested with SfiI and NotI for the patients' DNA and with SfiI, SalI, MluI, ClaI, SacII, and NotI for the YAC DNA. Fragments were separated through 1% agarose gels in 0.5× Tris-borate EDTA buffer with a CHEF DRII system 5 (Biorad). Runs were performed for 24 h at 14°C and 200 V with 2–15 s of switch time for fragments of 25–250 kb and 60–120 s for fragments of 200 kb to 1.6 Mb. Fragments of 5–100 kb were separated by field inversion gel electrophoresis (FIGE Mapper; Biorad) under the following conditions: 180 V forward, 120 V reverse, with 0.1–2-s switch time for 16 h at 20°C. Gel blotting and hybridization procedures were performed as described elsewhere (Heidet et al. 1995) with probes 804H10R, 183K24BD, and 96G18BD (Konrad et al. 1996; Saunier et al. 1997), and 47A, 53A, 49.81B, C40, R30A, R30E, and N123 (table 1).
NPHP1 Mutation Screening
NPHP1 exons were amplified by PCR with flanking intronic primers. Primers of exons 2–10, 12, 14, 15, 18, and 19 have been previously described (Saunier et al. 1997). The other primers were as follows: exon 1: 5′-GACCACCGCAAGAGAACATT-3′ and 5′-GGCAAGCTCCCAGGATTAGG-3′; exon 11: 5′-AACAGAGTATTGAAACTTT-3′ and 5′-AGTACTGTTTAACCTGTATC-3′; exon 13: 5′-ATTCAAACCACACCAAACCC-3′ and 5′-TTCCTTGTCAATAGACACAT-3′; exon 16: 5′-GCACTACTGGGTGGTATATT-3′ and 5′-TGATCCCAAATTCACTGGAC-3′; exon 17: 5′-TTATAAGTTGGATTGTAGGG-3′ and 5′-CAGAGTATGAAGCATTACTG-3′; and exon 20A: 5′-CGATACCTGCCAGGCACTAA-3′ and 5′-GACAGTGATTTTTGGGTTCC-3′. After initial denaturation at 95°C for 3 min, PCR was performed with 30 cycles of denaturation for 1 min at 94°C, 1 min at annealing temperatures of 61°C for exon 1, 49°C for exon 17, and 50°C for exons 11, 13, 16, and 20, extension for 1 min at 72°C, and a final extension at 72°C for 10 min. PCR products were purified with the Wizard PCR Preps DNA purification System (Promega) and directly sequenced with an Applied Biosystems DNA sequencer (model 373A) and the BigDye terminator cycle sequencing kit (PE Biosystems) according to the manufacturer's protocol. According to the recommended nomenclature system for human gene mutations (Antonarakis and the Nomenclature Working Group 1998), the nucleotide numbering of the NPHP1 cDNA starts at the first methionine codon.
Results
Genomic Map of the Region
Three BAC clones (96G18, 187E16, and 183K24) covering about 325 kb of the NPHP1 locus had been previously partially sequenced and characterized (Saunier et al. 1997; fig. 1A). We further extended the sequence analysis upstream to the 49G15 BAC clone and increased the percentage coverage of the aforementioned three BACs by primer walking. A total of 365 kb from STS 804H10R (i.e., the proximal end of YAC 804H10) to the distal end of BAC 183K24 (183K24BD) were almost entirely sequenced (96% coverage), and ∼190 kb upstream of 804H10R (part of BAC 49G15) were partially sequenced (83% coverage). As previously shown by PFGE, within this 365-kb sequence, the 100 kb between STSs 769G7R and 183K24BD were present in two inverted copies that exhibit >97% homology (Konrad et al. 1996; Saunier et al. 1997). To further study regions of interest around the markers 804H10R, 769G7R, and 183K24BD and to define patients' deletion breakpoints, we generated new STSs (see Patients and Methods) and checked, by PCR of YAC DNA, whether these were single copies or were also duplicated distal to the NPHP1 region. To build a long-range restriction map of the region, some of these STSs were used as probes to hybridize YAC DNA digested with different rare-cutting restriction enzymes (fig. 1B). By combining these different approaches, we were able to show that the NPHP1 gene is included in a 175-kb genomic region framed by two large inverted repeats of 300–330 kb (fig. 1C). We named the proximal and the distal copies 330RI and 330RII, respectively. One STS (804H10R) present in the centromeric repeat was not found in 330RII, as shown by its failure to hybridize YACs covering the distal region (fig. 2). Furthermore, we showed by PCR that four STSs—C30124, N123, R30E, and R30A—located 20–60 kb upstream of the first exon of NPHP1, were also duplicated downstream of the gene. By using these markers as well as STSs 57A and 183K24BD as hybridization probes (fig. 3), we were able to show that a ∼45-kb sequence, located 20 kb upstream of NPHP1 exon 1, was directly repeated 250 kb away, downstream of the gene, and within 330RII, deleting the STS 804H10R (fig. 1C) and removing a nearby SfiI site (shown in red in fig. 1B). We named the proximal and the distal copies “45RI” and “45RII,” respectively.
Figure 1.
Characterization of the genomic region surrounding the NPHP1 gene. A, Diagram of the sequenced region. The thick line represents the entirely sequenced interval, whereas the dashed thick line represents the partially sequenced region. BAC clones are represented as solid lines, and the BAC ends mapped to the region are shown as open rectangles. B, Restriction map of the NPHP1 region and YAC contig spanning the region. STS positions and restriction sites (F = SfiI; C = ClaI; S = SalI; M = MluI; Sc = SacII; N = NotI) are indicated by vertical lines. NotI sites used to define the extent of the deletion are shown in red. The markers in green and blue are localized within the 45-kb direct and the 330-kb inverted repeats, respectively, and the markers in red are those initially found to be deleted by PCR analysis of individuals with nephronophthisis. C, Schematic representation of the genomic region surrounding the NPHP1 gene. The shaded blue and green arrows represent, respectively, the inverted 330-kb repeats (named “330RI” and “330RII”) and the direct 45-kb repeats (named “45RI” and “45RII”). The NPHP1 gene is represented by an open box, and the direction of transcription is indicated by the small arrow above it. The red rectangle indicates the region around 804H10R in 330RI, deleted by the distal copy of the direct 45-kb repeat in 330RII.
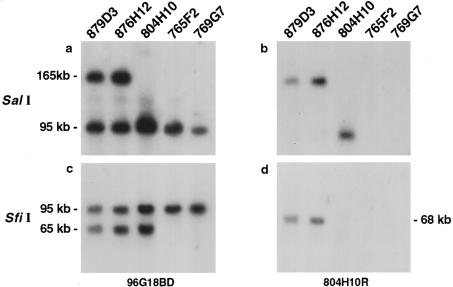
Figure 2.
Southern blot hybridization of SalI (a, b) and SfiI (c, d) digested DNA from five YACs spanning the NPHP1 region with 96G18BD (a, c) and 804H10R (b, d). Only a 165-kb SalI fragment and a 68-kb SfiI fragment were detected with the 804H10R probe in the two YACs covering the whole region (879D3 and 876H12), and no fragment was detected in the YACs covering only the distal copy of the inverted repeats (765F2 and 769G7), demonstrating that 804H10R escapes duplication.
Figure 3.
SfiI-digested DNA of four YACs spanning the NPHP1 region successively hybridized with probes 57A (a), 183K24BD (b), R30A (c), and N123 (d). All markers are duplicated; two SfiI fragments are present in YACs covering the whole NPHP1 region (879D3 and 876H12), whereas only one fragment is detected in YACs covering only the telomeric part of the region (765F2 and 769G7), allowing us to distinguish both copies. Note that R30A and 183K24BD detect the same 80-kb telomeric fragment, and that 57A and N123 detect the same 85-kb telomeric fragment.
Mechanism of Deletions In Nephronophthisis: Homologous Recombination between Direct Repeats
Twenty patients from group A, with homozygous deletions of the whole NPHP1 gene as identified by PCR of 765F2L and 804/6 (fig. 4), were shown to have the same hybridization pattern on PFGE: hybridization of the NotI-digested patient DNA with 183K24BD, located within the 330-kb repeats, detected a ∼510-kb fragment (fig. 5, lanes 2–10) instead of the ∼800-kb fragment observed in controls (fig. 5, lane 1). Hybridization of SfiI-digested DNA revealed only the proximal fragments (95 kb and 45 kb) with probes 96G18BD and 183K24BD, as the 80- and 65-kb telomeric fragments present in the control (fig. 6, lane 1) were absent (fig. 6, lanes 2–10). Additionally, hybridization with R30A and R30E probes located within the 45-kb repeats failed to detect the 80-kb telomeric SfiI fragment. However, hybridization with N123 also located in the 45-kb repeats revealed a normal 85-kb telomeric fragment, but no centromeric fragment (data not shown). It thus appeared that the breakpoints in these 20 patients all fall within the 45-kb repeats. In an attempt to further refine the localization of the breakpoints, we performed Southern-blot analyses with more-frequently-cutting enzymes (EcoRI, BamHI, HindIII, and PstI), with probes located in the 45-kb repeats. Unfortunately, no abnormal fragment could be found (data not shown). These results are consistent with deletions arising through homologous recombination between the two 45-kb direct repeats. Additionally, the more detailed characterization of the NPHP1 region allowed us to determine the exact size of the deletion (previously estimated as ∼250 kb) to be 290 kb.
Figure 4.
A, Schematic representation of the common NPHP1 deletion involving an homologous recombination between the 45-kb direct repeats and leading to a deletion of 290 kb; top, normal chromosome; bottom, deleted chromosome. B, Model of unequal recombination by chromosome misalignment followed by formation of a loop structure. C, Model of unequal recombination by unequal crossing-over between two different chromosomes, leading in theory to either a deletion (middle) or duplication (bottom) of the NPHP1 gene.
Figure 5.
PFGE of NotI-digested DNA of 12 patients and a control individual hybridized with 183K24BD. Lane 1, control individual showing a normal ∼800-kb NotI fragment. Lanes 2–10, patients from group A with homozygous deletion of makers 765F2L and 804/6, as detected by PCR. An abnormal ∼510 kb NotI fragment is detected. Lane 11, patient from group C only deleted for 804H10R by PCR, with an apparently normal 800-kb NotI fragment. Lanes 12 and 13, patients from group A deleted for 765F2L and 804/6 with two distinct NotI fragments (∼510 kb and ∼550 kb).
Figure 6.
PFGE of SfiI-digested DNA of 11 patients and a control individual successively hybridized with 96G18BD and 183K24BD. Hybridization with probes 96G18BD (a) and 183K24BD (b) reveals two SfiI fragments in the control individual (lane 1), 95-kb and 45-kb proximal fragments, and 80- and 65-kb telomeric fragments, respectively. Lanes 2–10, patients from group A with deletion of markers 765F2L and 804/6 by PCR. The normal proximal fragments detected by both probes are missing. Lanes 11 and 12, patients from group A with two distinct abnormal NotI fragments (fig. 5). Hybridization with 183K24BD reveals the normal 80-kb telomeric SfiI fragment.
Characterization of a Nonpathogenic Rearrangement Mediated by the 330-kb Inverted Repeats
The two patients from group B, one of whom was the product of a consanguineous marriage, were found by PCR to have a homozygous deletion of 804H10R, an STS that is located outside of the common NPHP1 deletion interval. No fragment was detected when EcoRI-digested DNA was hybridized with 804H10R, confirming the presence of a deletion encompassing the whole marker (data not shown). Furthermore, by using probes (57A and 183K24BD) that cover ∼80kb around 804H10R and map within the 330-kb repeats, the 68- and 45-kb proximal SfiI fragments detected by these probes, respectively, were absent (data not shown). Surprisingly, an apparently normal-sized ∼800-kb fragment was detected when NotI-digested DNA was hybridized with 183K24BD (fig. 5, lane 11), excluding the presence of a large deletion. An hypothesis to reconcile these apparently discordant results is the replacement of 804H10R by a third copy of the 45-kb repeat in the opposite orientation. The genomic map of these two patients obtained by hybridizing their SfiI-digested DNA with probes 57A, 183K24BD, 96G18BD, R30A, and N123 is in agreement with this hypothesis. The absent 68-kb (probes 57A and N123) and 45-kb (183K24BD and R30A) proximal SfiI fragments are replaced by the duplication of the 85- and 80-kb distal fragments respectively. A summary of these data is schematically represented in figure 7A.
Figure 7.
A, Genomic SfiI map of the NPHP1 region when 804H10R is missing and replaced by an additional copy of 45RII. B, Reciprocal rearrangement leading to the duplication of 804H10R. C, Models for nonpathogenic DNA rearrangements mediated by mispairing of the inverted repeats and double homologous recombination (C) or by a crossing-over between the inverted repeats leading to an inversion of the whole NPHP1 region (C′), followed by a crossing-over, localized telomeric of the 804H10R marker (C″). Centromeric and telomeric copies of 57A and 183K24BD are indicated in orange and blue, respectively.
This rearrangement does not actually involve the NPHP1 gene and thus is unlikely to explain the phenotype in patients from group B. The absence of the 804H10R PCR product upon amplification of DNA from 2 of 152 control individuals (1.3%) supported the hypothesis that this rearrangement was nonpathogenic. This result was further confirmed by performing Southern blotting analysis by using the probe C40, which is 5 kb from 804H10R and located within the 330-kb repeats (fig. 1B). Hybridization with this probe, which normally detects the same 8.7-kb PstI fragment as 804H10R as well as a telomeric 7-kb PstI fragment located within 330RII, resulted in the detection of only the 7-kb fragment for the DNA found to be deleted for 804H10R by PCR (fig. 8) of the two patients and the two control individuals. Careful analysis of Southern blots containing DNA from 152 controls allowed us to detect individuals heterozygous for this nonpathogenic rearrangement: individuals heterozygous for the deletion of 804H10R have a more intense 7-kb PstI fragment in comparison with the 8.7-kb fragment (21%), and individuals heterozygous for the reciprocal event leading to a duplication of 804H10R (fig. 7B) present with a stronger 8.7-kb fragment (13%; fig. 8). To determine the cause of nephronophthisis in the two group B patients, we searched for point mutations in the NPHP1 gene by PCR and direct sequencing of all the 20 exons of the gene. In one patient, we detected an homozygous single-base deletion at position 639 of exon 7, causing a frame shift and generating a stop codon leading to a putative truncated polypeptide. In the other patient, two heterozygous missense mutations in exon 6 (a T→C substitution at position 539 leading to the substitution of a leucine by a proline at position 180 in the SH3 domain) and exon 8 (an A→G substitution at position 859, changing an arginine to a serine at position 287). Neither of these two missense mutations was detected in a panel of 50 unrelated control individuals who were also screened.
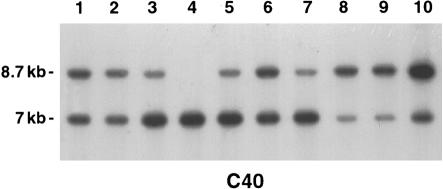
Figure 8.
Southern blot analysis of PstI-digested DNA of control individuals hybridized with probe C40. Lanes 3, 5, and 7 show that the 7-kb fragment is more intense than the 8.7-kb fragment, suggesting that each individual has only one copy of 804H10R and three copies of 45RII. Lanes 8–10 show that the 8.7-kb fragment is more intense than the 7-kb fragment, suggesting the occurrence of the reciprocal event (i.e., three copies of 804H10R and one copy of 45RII) in these individuals. Lane 4 shows a control individual carrying a homozygous deletion of 804H10R as detected by PCR. Lanes 1, 2, and 6 depict control individuals showing two bands of the same intensity; therefore, they are not carrying a nonpathogenic rearrangement.
Two additional unrelated patients from group C who originated from consanguineous marriages had no PCR products with markers 765F2L, 804/6 (located in the common deletion), or 804H10R involved in the nonpathogenic rearrangement. The 183K24BD probe, hybridized to NotI-digested DNA of these two patients, detected a ∼550-kb fragment (data not shown), a fragment slightly larger than the fragment detected in patients without PCR products for only 765F2L and 804/6 (fig. 5, lanes 2–10). In these two patients, SfiI-digested DNA hybridized to 183K24BD demonstrated the presence of the distal fragment, but not the proximal one (data not shown). The pattern observed in these two patients is consistent with the homozygous pathogenic deletions occurring on chromosomes that also carry the nonpathogenic rearrangement.
Finally, seven patients from group A—all from nonconsanguineous marriages and homozygously deleted for 765F2L and 804/6 but not for 804H10R—have two distinct NotI fragments, ∼510 and ∼550 kb, detected by 183K24BD (fig. 5, lanes 12 and 13). The 183K24BD probe hybridized to SfiI-digested DNA detected in all these patients the normal 45-kb proximal and 80-kb distal SfiI fragments (fig. 6b, lanes 11 and 12), which initially suggested that they might have a smaller deletion (Saunier et al. 1997). However, these additional data indicated that these patients are heterozygous for the nonpathogenic rearrangement involving 804H10R and the NPHP1 deletions, mediated by the same mechanism of homologous recombination, had occurred on two different chromosomes 2, explaining the two distinct NotI fragments. This was confirmed by the pattern obtained with hybridization of NotI-digested DNA from the patients' parents with 804H10R, since the ∼550kb NotI fragment corresponding to the deleted allele was completely absent in one of each parent (fig. 9).
Figure 9.
PFGE of NotI-digested DNA of a patient and her parents successively hybridized with 183K24BD (a) and 804H10R (b). Lane 3 depicts an affected individual. Lanes 1 and 2 depict the father and the mother of the patient, respectively. Each parent is heterozygous for the NPHP1 deletion, as shown by hybridization with 183K24BD, but the father lacks the 804H10R marker on the deleted allele. Furthermore, the abnormal NotI fragment detected by 183K24BD in the father (∼550 kb) is slightly larger than the abnormal fragment detected in the mother (∼510 kb), in agreement with the occurrence of a 290-kb deletion on a chromosome carrying the nonpathogenic rearrangement and, thus, two copies of the 45-kb repeat. This proves that the child bears the nonpathogenic rearrangement on her paternal allele in addition to the 290-kb deletion on both alleles, as was suggested by the SfiI pattern.
Discussion
The most frequent mutation observed in familial juvenile nephronophthisis is a large homozygous deletion encompassing the whole NPHP1 gene in the centromeric region of chromosome 2 (Konrad et al. 1996; Hildebrandt et al. 1997; Saunier et al. 1997). In this report, the high level of genomic detail obtained by sequencing and PFGE analysis allowed us to characterize the very complex organization of the NPHP1 region; it also allowed us to demonstrate the mechanism leading to this common NPHP1 gene deletion mutation. We showed that the NPHP1 gene is flanked by two large inverted repeats of ∼330 kb, the distal one interrupted by a ∼45-kb sequence, which is in turn directly repeated upstream of the NPHP1 gene. The patients' deletion breakpoints appear to be located within the 45-kb repeat. This would suggest an unequal recombination event between two homologous but nonallelic copies of 45-kb repeats, which leads to the 290-kb deletion. However, the extremely high sequence homology that exists between the two repeats, as suggested by the conservation of all the restriction sites in both copies, precluded easy identification of the exact position of the deletion breakpoints. More refined studies, such as those performed by Reiter et al. (1997), might allow us to define the breakpoints. Low-copy number repeats have been implicated in mediating rearrangements of other human disorders, as reviewed in Lupski (1998). Large rearrangements have been described between repeats on the X chromosome, leading to steroid sulfatase gene deletions (Li et al. 1992), and reciprocal duplication and deletion products of unequal crossing-over events on chromosomal band 17p11.2 are responsible for Charcot-Marie-Tooth disease type 1A (Pentao et al. 1992) and hereditary neuropathy with liability to pressure palsies (Chance et al. 1994), respectively. In these cases, rearrangements arise from unequal crossing-over and homologous recombination between 24-kb repeats that are 1.5 Mb apart (Reiter et al. 1997). Similar mechanisms have also been reported for deletion formation in Smith-Magenis syndrome (Chen et al. 1997) and velocardiofacial syndrome (Edelmann et al. 1999). In the case of NPHP1 deletion, the unequal recombination between two homologous but nonallelic 45-kb repeats may occur either by chromosome misalignment, followed by an unequal crossing-over, or by the formation of a loop structure on a single chromosome, as illustrated in figure 4B and 4C. We did not detect any de novo deletions, although haplotype analyses in NPHP1 families strongly suggest that the deletions are not due to a founder effect (Konrad et al. 1996). We found no evidence of the expected reciprocal event of the deletion: a duplication of NPHP1 region (the ∼1,000 kb NotI fragment as illustrated in fig. 4C) in patients or control individuals. However, such a duplication could lead to a distinct disease phenotype as shown in Charcot-Marie-Tooth disease type 1A (Pentao et al. 1992) and may therefore not be present in normal controls. Thus, our results did not allow us to determine whether the NPHP1 deletion are mediated by inter- or intrachromosomic recombination.
We also detected a more common nonpathogenic DNA rearrangement involving the two 330-kb inverted repeats flanking the NPHP1 gene. This rearrangement can be explained by various mechanisms. One such mechanism is an interchromosomal mispairing of the two copies of the inverted repeats followed by a double recombination, leading to the replacement of a short genomic sequence containing the 804H10R STS by an additional copy of the 45-kb repeat (fig. 7C). Another such mechanism is a prior intrachromosomal mispairing of the inverted repeats and an unequal crossover leading to the inversion of the whole NPHP1 region (fig. 7C′), followed by recombination between two different chromosomes (fig. 7C″), leading to the same rearrangement as in the first mechanism described.
We did not obtain evidence that led us to favor one hypothesis over the other. However, inversion events involving two inverted repeats on the same chromosome have been described in several disorders. For example, inversions in the factor VIII gene causing severe hemophilia A involve a recombination event between two inverted repeats located in intron 22 and located upstream of the gene (Lakich at al. 1993). Additionally, nonpathogenic rearrangements involving mispairing of inverted repeats leading to the maintenance of repeat sequence homogeneity and inversion of the region have been previously reported (Small et al. 1997) around the emerin locus at Xq28 and were found in 33% of women in the heterozygous state. In this study, the replacement of 804H10R by the 45-kb repeat was found in 1.3% and in 21% of controls in the homozygous and heterozygous states, respectively. We were also able to detect the reciprocal event in the heterozygous state. This represents a key element for the validation of our hypothesis concerning the mechanism of the origin of this complex rearrangement. This could also explain the results of Nothwang et al. (1998b), who found a duplication of 804H10R but no duplication of two markers (57N19T and 278P6T) located in 45 RI upstream of the NPHP1 gene when they built a PAC contig of the NPHP1 region. It can be supposed that the human DNA used to establish this PAC library (Genome System) bears the predicted duplication of 804H10R and the deletion of the 45RII copy.
These repetitive structures appear to predispose the human genome to frequent deletion and duplication. Their persistence through evolution may thus make particular regions of the genome more susceptible to rearrangement, consequently leading to genetic disorders (Lupski 1998). The human chromosome 2 has arisen from the fusion of two ancestral ape chromosomes (Yunis and Prakash 1982), the telomere-telomere fusion point being located at band 2q13 (Ijdo et al. 1991), telomeric to the NPHP1 locus (Hildebrandt et al. 1996). Telomeric regions are known to contain several repeat sequences (Allshire et al. 1988). The close proximity of these identical telomeres at the centromere of chromosome 2 would be expected to make this region prone to rearrangements. Indeed, this has been illustrated by centromeric inversions found in 0.1% of controls (Vejerslev and Friedrich 1984; MacDonald and Cox 1985; Djalali et al. 1986). Additionally, several genes have been found to be duplicated within the 2q13 region, BENE (Saunier et al. 1997) and RANBP2L1 (Nothwang et al. 1998a), with some on either side of the centromere such as IGO (Huber et al. 1990) and LIS2 (Reiner et al. 1995).
The elucidation of the complex structure of the NPHP1 region allowed us to show that all the patients with nephronophthisis without extrarenal symptoms that we have studied to date do indeed have the same deletion, although this deletion occurs on different ancestral chromosomes. The detailed genomic organization of the NPHP1 region will also help us to characterize the rearrangements involved in patients with nephronophthisis associated with extrarenal symptoms who also have deletions in the same region (Caridi et al. 1998).
Acknowledgments
We thank E. Boye for helpful discussion and critical review of the manuscript. Sequence analysis was performed in Généthon, CNRS-URA 1922, 91000 Evry, France. This study was supported by the Association Française contre les Myopathies (AFM), the MENESR (ACC-SV95), the Fondation pour la Recherche Médicale, and the Association pour l'Utilisation du Rein Artificiel.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for familial juvenile nephronophthisis [MIM 256100])
- RepeatMasker program, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
References
- Allshire RC, Gosden JR, Cross SH, Cranston G, Rout D, Sugawara N, Szostak JW, et al (1988) Telomeric repeat from T. thermophila cross hybridizes with human telomeres. Nature 14:656–659 [DOI] [PubMed]
- Antignac C, Arduy CH, Beckmann JS, Benessy F, Gros F, Medhioub M, Hildebrandt F, et al (1993) A gene for familial juvenile nephronophthisis (recessive medullary cystic kidney disease) maps to chromosome 2p. Nat Genet 3:342–345 [DOI] [PubMed]
- Antignac C, Kleinknecht C, Habib R (1998) Nephronophthisis. In: Cameron D, Davison AM, Cameron JS, Grünfeld J-P, Kerr DNS, Ritz E, Winearls G (eds) Clinical nephrology. Oxford University Press, Oxford and New York, pp 2417–2426 [Google Scholar]
- Antonarakis SE and the Nomenclature Working Group (1998) Recommendations for a nomenclature system for human gene mutations. Hum Mutat 11:1–3 [DOI] [PubMed]
- Caridi G, Murer L, Bellantuono R, Sorino P, Caringella DA, Gusmano R, Ghiggeri GM (1998) Renal-retinal syndromes: association of retinal anomalies and recessive nephronophthisis in patients with homozygous deletion of the NPH1 locus. Am J Kidney Dis 32:1059–1062 [DOI] [PubMed]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 [DOI] [PubMed]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, et al (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed]
- Djalali M, Steinbach P, Bullerdiek J, Holmes-Siedle M, Verschraegen-Spae MR, Smith A (1986) The significance of pericentric inversions of chromosome 2. Hum Genet 72:32–36 [DOI] [PubMed]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, et al (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 [DOI] [PubMed]
- Heidet L, Dahan K, Zhou J, Xu Z, Cochat P, Gould JD, Leppig KA, et al (1995) Deletions of both alpha 5(IV) and alpha 6(IV) collagen genes in Alport syndrome and in Alport syndrome associated with smooth muscle tumours. Hum Mol Genet 4:99–108 [DOI] [PubMed]
- Hildebrandt F, Cybulla M, Strahm B, Nothwang HG, Singh-Sawhney I, Berz K, Nicklin M, et al (1996) Physical mapping of the gene for juvenile nephronophthisis (NPH1) by construction of a complete YAC contig of 7 Mb on chromosome 2q13. Cytogenet Cell Genet 73:235–239 [DOI] [PubMed]
- Hildebrandt F, Otto E, Rensing C, Nothwang HG, Vollmer M, Adolphs J, Hanusch H, et al (1997) A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet 17:149–153 [DOI] [PubMed]
- Huber C, Thiebe R, Hameister H, Smola H, Lotscher E, Zachau HG (1990) A human immunoglobulin kappa orphon without sequence defects may be the product of a pericentric inversion. Nucleic Acids Res 18:3475–3478 [DOI] [PMC free article] [PubMed]
- Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA (1991) Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA 88:9051–9055 [DOI] [PMC free article] [PubMed]
- Konrad M, Saunier S, Heidet L, Silbermann F, Benessy F, Calado J, Le Paslier D, et al (1996) Large homozygous deletions of the 2q13 region are a major cause of juvenile nephronophthisis. Hum Mol Genet 5:367–371 [DOI] [PubMed]
- Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J (1993) Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 5:236–241 [DOI] [PubMed]
- Li XM, Yen PH, Shapiro LJ (1992) Characterization of a low copy repetitive element S232 involved in the generation of frequent deletions of the distal short arm of the human X chromosome. Nucleic Acids Res 20:1117–1122 [DOI] [PMC free article] [PubMed]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed]
- MacDonald IM, Cox DM (1985) Inversion of chromosome 2 (p11p13): frequency and implications for genetic counselling. Hum Genet 69:281–283 [DOI] [PubMed]
- Medhioub M, Cherif D, Benessy F, Silbermann F, Gubler MC, Le Paslier D, Cohen D, et al (1994) Refined mapping of a gene (NPH1) causing familial juvenile nephronophthisis and evidence for genetic heterogeneity. Genomics 22:296–301 [DOI] [PubMed]
- Nothwang HG, Rensing C, Kubler M, Denich D, Brandl B, Stubanus M, Haaf T, et al (1998a) Identification of a novel Ran binding protein 2 related gene (RANBP2L1) and detection of a gene cluster on human chromosome 2q11-q12. Genomics 47:383–392 [DOI] [PubMed]
- Nothwang HG, Stubanus M, Adolphs J, Hanusch H, Vossmerbaumer U, Denich D, Kubler M, et al (1998b) Construction of a gene map of the nephronophthisis type 1 (NPHP1) region on human chromosome 2q12-q13. Genomics 47:276–285 [DOI] [PubMed]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300 [DOI] [PubMed]
- Reiner O, Bar-Am I, Sapir T, Shmueli O, Carrozzo R, Lindsay EA, Baldini A, et al (1995) LIS2, gene and pseudogene, homologous to LIS1 (lissencephaly 1), located on the short and long arms of chromosome 2. Genomics 30:251–256 [DOI] [PubMed]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR (1997) The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet 6:1595–1603 [DOI] [PubMed]
- Saunier S, Calado J, Heilig R, Silbermann F, Benessy F, Morin G, Konrad M, et al (1997) A novel gene that encodes a protein with a putative src homology 3 domain is a candidate gene for familial juvenile nephronophthisis. Hum Mol Genet 6:2317–2323 [DOI] [PubMed]
- Senior B, Friedmann AI, Braudo JL (1961) Juvenile familial nephropathy with tapetoretinal degeneration: a new oculorenal dystrophy. Am J Ophthalmol 52:625–633 [DOI] [PubMed] [Google Scholar]
- Small K, Iber J, Warren ST (1997) Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet 16:96–99 [DOI] [PubMed]
- Smit AF (1996) The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6:743–748 [DOI] [PubMed]
- Vejerslev LO, Friedrich U (1984) Experiences with unexpected structural chromosome aberrations in prenatal diagnosis in a Danish series. Prenat Diagn 4:181–186 [DOI] [PubMed]
- Waldherr R, Lennert T, Weber HP, Fodisch HJ, Scharer K (1982) The nephronophthisis complex: a clinicopathologic study in children. Virchows Arch 394:235–254 [DOI] [PubMed]
- Yunis JJ, Prakash O (1982) The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530 [DOI] [PubMed]