Abstract
Analysis of a genome screen of 504 brothers with prostate cancer (CaP) who were from 230 multiplex sibships identified five regions with nominally positive linkage signals, on chromosomes 2q, 12p, 15q, 16p, and 16q. The strongest signal in these data is found on chromosome 16q, between markers D16S515 and D16S3040, a region suspected to contain a tumor-suppressor gene. On the basis of findings from previous genome screens of families with CaP, three preplanned subanalyses were carried out, in the hope of increasing the subgroup homogeneity. Subgroups were formed by dividing the sibships into a group with a positive family history (FH+) that met criteria for “hereditary” CaP (n=111) versus those which did not meet the criteria (n=119) and by dividing the families into those with a mean onset age below the median (n=115) versus those with a mean onset age above the median (n=115). A separate subanalysis was carried out for families with a history of breast cancer (CaB+ [n=53]). Analyses of these subgroups revealed a number of potentially important differences in regions that were nonsignificant when all the families were analyzed together. In particular, the subgroup without a positive family history (FH−) had a signal in a region that is proximal to the putative site of the HPC1 locus on chromosome 1, whereas the late-age-at-onset group had a signal on 4q. The CaB+ subgroup revealed a strong linkage signal at 1p35.1.
Introduction
This year, prostate cancer (CaP) will be the most commonly diagnosed visceral cancer and the second leading cause of cancer mortality among men in the United States (Landis et al. 1999). The prevalence of CaP varies 20- to 30-fold worldwide. The highest frequency is found in African Americans, and the lowest frequency is found in Asian populations (Parkin et al. 1993; Whittemore 1994). Although immigrant (Staszewski and Haenszel 1965; Dunn 1975) and lifestyle and dietary studies (Whittemore et al. 1995a) point to the importance of environmental factors, twin (Grönberg et al. 1994; Ahlbom et al. 1997), “kinship” (Cannon et al. 1982; Holloway and Sofaer 1992a, 1992b), and family studies (Morganti et al. 1956; Woolf 1960; Steele et al. 1971; Krain 1974; Meikle et al. 1985; Steinberg et al. 1990; Ghadirian et al. 1991; Spitz et al. 1991; Keetch et al. 1995; Whittemore et al. 1995b) point to the importance of genetic factors. The two strongest predictors of increased risk for CaP, apart from age, are the presence of several affected first-degree relatives and an affected brother who had an unusually early age at onset (Keetch et al. 1995).
Segregation analysis has suggested that some cases of CaP are due to an autosomal susceptibility locus with an allele or alleles that collectively behave in a dominant and age-dependent fashion (Carter et al. 1992; Grönberg et al. 1997a; Schaid et al. 1998). Other investigators have argued either for a recessive mode of inheritance or, on the basis of an excess risk of CaP in men with affected brothers compared with men with affected fathers, for an X-linked mode of transmission (Monroe et al. 1995). Unlike breast cancer (Miki et al. 1994; Tavtigian et al. 1996) or colorectal cancer (Fearon et al. 1990; Groden et al. 1991), however, no susceptibility loci with alleles sufficient to cause CaP have yet been identified.
We report here the results of a genome screen of 230 multiplex sibships with CaP.
Families and Methods
Families
Since 1991, we have been collecting information about multiplex sibships with CaP. No ascertainment criteria, other than the presence of two or more brothers with documented CaP and a willingness to participate, have been used to recruit the members of the sample population. Approximately half of these subjects were patients of Washington University School of Medicine (WUSM) staff urologists, were referred by other urologists or CaP support groups, or responded to our publications soliciting participation. The remainder were referred by family members enrolled in our studies. The study protocol was approved by the Human Studies Committee of Washington University. Informed consent was obtained from all subjects. All probands and many of their affected brothers completed a family-history questionnaire that was used to partition the sample for various preplanned subanalyses.
A total of 513 subjects were genotyped. Subsequent analyses (see below) reduced this sample to 504 men with CaP. The subjects’ mean age at the time of diagnosis was 65.5 years (median, 65.4 years; range, 42–91 years).
The diagnosis of CaP was confirmed directly by WUSM pathologists or by examination of the medical records in 502 (99.6%) of the subjects. Pathologic documentation was missing from two subjects (0.4%); however, in these subjects, the diagnosis of CaP was affirmed by treatment records.
Seventy-six percent of the patients were treated primarily with radical prostatectomy, 10.3% were treated with radiation therapy, 2.6% were treated with primary hormonal therapy, 2.6% were managed with watchful waiting, and 8.5% received miscellaneous other treatments. Of the patients treated primarily with radical prostatectomy, 15.1% have also been treated with hormonal therapy and 7.8% have been treated with radiation therapy.
Genotyping
All samples were genotyped at the Center for Medical Genetics, Marshfield Medical Research Foundation, by use of Weber screening set 9 (Yuan et al. 1997), which consists of simple tandem-repeat polymorphisms, including 366 autosomal, 16 X-linked, and 4 Y-linked markers. Average marker heterozygosity was 77%, and average spacing on sex-equal maps was 9 cM (Broman et al. 1998).
A multipoint linkage analysis (see below) was used to rank the markers according to the estimated mean allele sharing among affected brothers. Regions around the highest-ranking nine markers were selected for further genotyping. An additional 38 microsatellite markers (∼4.2/signal) were genotyped at the Center for Medical Genetics, Marshfield Medical Research Foundation. The average spacing between adjacent markers for the 38 new intervals created by this second wave of genotyping was 2.2 cM.
Statistical Methods
Before conducting the linkage analysis, we assessed the marker genotypes to verify the status of each alleged sib pair, using two approaches: the RELATIVE program (Göring and Ott 1995) and a modified version of the RELPAIR program (Boehnke and Cox 1997; Broman and Weber 1998). The results were similar with both programs and revealed the presence of one half-sib (from an affected trio) and four sets of twins (one pair of which was from an affected trio) who, by genotyping, were shown to be MZ. An additional subject was dropped from the affected sample after the genome screen had been completed because a record review indicated that he did not have CaP. After these nine individuals had been deleted from the sample, 504 full sibs from 230 nuclear families remained for linkage analysis. The familial distribution of genotyped brothers used in this analysis was as follows: 188 affected pairs, 40 affected trios, and 2 affected quartets.
To determine if alleles at the microsatellite markers were in Hardy-Weinberg proportions in this sample, we carried out likelihood-ratio tests with the ASSOC program (Ott 1985), choosing, from each family at random, one genotyped sib per marker.
Despite the evidence from three separate segregation analyses—all of which argue that a sizable proportion of CaP cases (particularly early-onset cases) are due to a highly penetrant dominant gene—a susceptibility locus has not yet been identified. Therefore, rather than compute linkage statistics under what may eventually prove to be a grossly inaccurate model, we preferred to compute allele-sharing statistics that do not require specification of the mode of transmission. Since the original implementation of the nonparametric-linkage (NPL) scoring algorithm has been shown to be overly conservative when data on parental genotypes are lacking (Davis and Weeks 1997; Badner et al. 1998), we decided to compute the Kong-and-Cox (KAC) statistic (Zlr score) as implemented in GENEHUNTER-PLUS (Kruglyak et al. 1996; Kong and Cox 1997), using the exponential model option, the “pairs” scoring function, and equal weights for each family. A related program, MAPMAKER/SIBS (version 2.0; Kruglyak and Lander 1995), was used to estimate the mean proportion of alleles shared identical by descent. Allele frequencies were estimated from the data, and for all subanalyses, allele frequencies were re-estimated for each data partition examined.
Under the null hypothesis, Zlr scores have a standard normal distribution. When the ith Zlr score from one data partition is compared with its complementary partition, the statistic
 |
is asymptotically N(0,1). “C” and “1-C” denote the two data partitions. Since all subanalyses partition the data by family units, and since the families are independent, the covariance term in the above expression is 0. With one notable exception (a predicted increase in the Zlr score of the early-age-at-onset partition for some chromosome 1 regions), we have no prior hypotheses about the direction of any differences that could result from partitioning of the data. Accordingly, for the sake of conservative consistency, all tests of the significance of D are two tailed.
Results
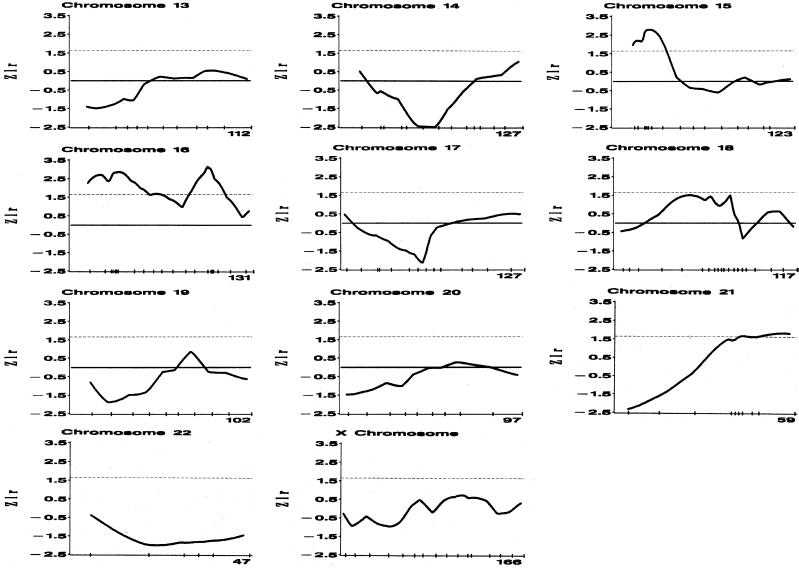
Multipoint Zlr scores for all chromosomes (except the Y chromosome) are displayed in figure 1. Five chromosomal regions gave nominal evidence for linkage (i.e., a Zlr score >1.645) at two or more adjacent markers: (1) a very broad region on 2q, extending ∼66 cM, from D2S1391 to D2S2968; (2) a narrow region on 12p, extending ∼3.03 cM, from D12S1615 to D12S1685; (3) a moderately sized region on 15q, extending ∼19.2 cM, from D15S822 to the dinucleotide repeat in the actin alpha cardiac-muscle gene; (4) a broad region on 16p, extending ∼39.1 cM, from ATA41E04 to the centromere; and (5) a moderate region on 16q, extending ∼16.8 cM, from D16S2624 to D16S3040. Table 1 reports the marker at which the maximum Zlr score occurs, for each of the aforementioned five chromosomal regions as well as the estimate of the mean proportion of alleles shared identical by descent. Only three markers gave nominal (i.e., P<.05) evidence of departure from Hardy-Weinberg equilibrium (GATA91H06, P=.006; ATA78D02, P=.047; and D18S970, P=.049), and none are located in any of the above five regions.
Figure 1.
Zlr scores for 420 markers genotyped on 504 brothers with CaP who were from 230 multiplex sibships. The check marks show the positions of the markers. The total length of each chromosome (in cM) is shown in the lower-right corner.
Table 1.
Allele Sharing in ASPs, for Five Chromosomal Regions That Yield Nominal Evidence for Linkage
| Chromosome | Marker | Posiiton(cM) | Zlr | Mean ASP Allele Sharing |
| 2q | D2S2228 | 224.33 | 2.78 | .557 |
| 12p | D12S1685 | 7.67 | 2.00 | .533 |
| 15q | D15S1010 | 23.89 | 2.77 | .544 |
| 16p | D16S3103 | 32.07 | 2.81 | .546 |
| 16q | D16S3096 | 99.44 | 3.15 | .563 |
We preplanned three subanalyses that involved dichotomizing the total sample into subsets that previous research has suggested may increase homogeneity in the subgroups. χ2 Analyses indicate that the various family partitions are not significantly pairwise-dependent in these data (table 2). As noted above, allele frequencies were re-estimated whenever a new partition of the families was constructed. Since chromosome 16 yielded moderate-to-strong signals for all family partitions, the Zlr scores for this chromosome are presented separately.
Table 2.
Distribution of Families Cross-Classified According to Criteria Used to Partition the Data for Preplanned Subanalyses
| Early Age at Onset |
Late Age at Onset |
|||||||
| FH+ |
FH− |
FH+ |
FH− |
|||||
| CaB+ | CaB− | CaB+ | CaB− | CaB+ | CaB− | CaB+ | CaB− | |
| No. of families | 16 | 41 | 14 | 44 | 10 | 44 | 13 | 48 |
| No. of ASPs | 31 | 67 | 14 | 44 | 23 | 78 | 13 | 48 |
It is unclear how best to report the results of a whole-genome screen when various subgroups are analyzed separately. The fact that these subgroup analyses were preplanned does not mean that a price need not be paid for performance of multiple tests. On the other hand, for a complex and heterogeneous phenotype, such as CaP, complete genetic characterization—including precise specification of all gene-gene and gene-environment interactions—may require sample sizes that are an order of magnitude larger than currently available. We have chosen to report all adjacent markers (i.e., two or more) for which the Zlr score is nominally significant at P < .05. Since these P values are uncorrected for multiple tests, most will prove to be false positives.
The first subanalysis involved partitioning the families according to whether they met the “Hopkins” criteria for “hereditary” CaP due to features of their family history (FH). To be classified as belonging to the FH-positive (FH+) group, a family must contain either (1) two or more brothers with a diagnosis of CaP at age ⩽55 years, (2) at least three first-degree relatives with a diagnosis of CaP, or (3) three consecutive generations with CaP (Carter et al. 1992). Since our study design required, at a minimum, the presence of at least one affected sib pair (ASP), any family meeting criterion 3 would necessarily also meet criterion 2. Only six of the families under study met criterion 1, and five of them also had an affected father. Consequently, virtually all of our FH+ families were so classified because they met criterion 2. One hundred eleven families containing a total of 199 ASPs met the criteria for FH+. Table 3 reports the distribution of nominally significant Zlr scores achieved in either partition. A number of interesting contrasts are apparent.
Table 3.
Nominally Significant KAC Zlr Scores and P Values for Families with CaP, Subdivided According to Whether They Are FH+ or FH−
|
KAC Zlr Score (P) inb |
|||
| Markera | Position(cM) | FH+ Families | FH− Families |
| D1S534 | 151.88 | 1.65 (.050) | |
| D1S1653 | 164.09 | …* | 1.86 (.031) |
| D1S1679 | 170.84 | …** | 2.28 (.011) |
| D1S1677 | 175.62 | …** | 2.72 (.003) |
| D1S2141 | 233.38 | 2.10 (.018) | …* |
| D1S549 | 239.66 | 1.84 (.034) | …* |
| D2S1384 | 200.43 | 2.02 (.022) | |
| D2S2944 | 210.43 | 2.23 (.012) | |
| D2S434 | 215.78 | 2.15 (.016) | |
| D2S2228 | 224.33 | 2.22 (.013) | |
| D2S2390 | 225.67 | 1.76 (.039) | |
| D2S1363 | 227.00 | 1.85 (.032) | |
| D2S159 | 228.61 | 1.94 (.026) | |
| D2S427 | 236.70 | 2.25 (.012) | |
| D2S2968 | 251.94 | 2.25 (.012) | |
| D2S125 | 260.63 | 1.72 (.042) | |
| D3S4529 | 112.42 | 1.78 (.038) | |
| D3S2459 | 119.09 | 2.15 (.016) | |
| D3S1591 | 121.67 | 2.01 (.022) | |
| D3S3045 | 124.16 | 1.94 (.026) | |
| D3S1616 | 124.16 | 1.94 (.026) | |
| D3S3695 | 124.83 | 1.73 (.042) | |
| D8S1119 | 101.01 | 1.67 (.047) | |
| GAAT1A4 | 110.20 | 2.36 (.009) | …*** |
| D15S822 | 12.30 | 1.80 (.036) | |
| D15S1002 | 14.58 | 1.92 (.027) | |
| D15S1048 | 19.12 | 1.89 (.029) | |
| D15S165 | 20.24 | 1.65 (.050) | |
| D15S184 | 21.58 | 1.83 (.033) | 2.01 (.022) |
| D15S1010 | 23.89 | 2.34 (.009) | |
| ACTC | 31.46 | 2.74 (.003) | |
| GATA173A03 | 54.40 | 1.87 (.031) | |
| D18S535 | 64.48 | 1.70 (.045) | |
| GATA81H03 | 66.66 | 1.80 (.036) | |
| D18S970 | 68.30 | 1.94 (.026) | |
| D18S363 | 71.32 | 2.09 (.018) | |
| D18S851 | 74.93 | 1.95 (.025) | |
| D18S539 | 74.93 | 1.95 (.025) | |
| ATA82B02 | 106.81 | 1.75 (.040) | |
Chromosome 16 markers are not included.
Asterisks denote level of significance between the respective Zlr scores of each subgroup: * = .05>P>.01, ** = .01>P>.001, and *** = P<.001.
Although no nominally significant evidence of linkage of chromosomes 1, 3, 8, or 18 was obtained when all of the families were analyzed together, the FH+/FH− partitioning reveals such evidence. With respect to the signals on chromosome 1, the two regions are separated by ∼57.8 cM, so, despite their synteny, they are unlinked. Accordingly, it is noteworthy that, for the two blocks of adjacent chromosome 1 markers, when one group or the other attained a nominally significant Zlr score, the partitioning resulted in a significant difference between the two subgroups. Of the 33 non–chromosome 1 markers listed in table 3, only one, on chromosome 8 (GAAT1A4), reveals a significant between-subgroup difference.
A consistent finding in cancer genetics (Giardiello 1997) is that families with an early age at onset appear to have higher “genetic loading” and, in some cases, a single major locus with alleles sufficient to cause the cancer. Therefore, we subdivided our sample according to each family’s mean age at onset. The means were ranked, and a median split resulted in 115 families in each subgroup. Nominally significant Zlr scores for this partition are reported in table 4. Interestingly, with the exception of two contiguous markers on chromosome 12 and two contiguous markers on chromosome 15, the highest Zlr scores are observed in the families with the latest mean age at onset. Of the 35 markers listed in table 4, the median split resulted in 13 (37%) significant between-group differences in their Zlr scores. And in all but 2 of these 13 significant differences, the linkage signal occurs in the late-age-at-onset group.
Table 4.
Nominally Significant KAC Zlr Scores and P Values Families with CaP, Subdivided by Median Age at Onset[Note]
|
KAC Zlr Score (P) in |
|||
| Marker | Position(cM) | Families with Mean Age at Onset in Lower 50th Percentile | Families with Mean Age at Onset in Upper 50th Percentile |
| D1S547 | 267.51 | …* | 2.06 (.020) |
| D1S1609 | 274.53 | …* | 2.01 (.022) |
| D2S2944 | 210.43 | 1.99 (.024) | |
| D2S434 | 215.78 | 2.16 (.016) | |
| D2S2228 | 224.33 | 2.92 (.002) | |
| D2S2390 | 225.67 | 2.57 (.005) | |
| D2S1363 | 227.00 | 2.35 (.009) | |
| D2S159 | 228.61 | 2.36 (.009) | |
| D2S427 | 236.70 | 2.16 (.015) | |
| D2S2968 | 251.94 | 1.67 (.048) | |
| D4S2367 | 78.43 | …* | 2.28 (.011) |
| D4S3243 | 88.35 | …* | 1.85 (.032) |
| D4S1647 | 104.94 | …** | 2.72 (.003) |
| D4S2623 | 114.04 | …** | 2.85 (.002) |
| D4S2394 | 129.92 | …** | 1.92 (.027) |
| ATA34E08 | 33.02 | …*** | 1.87 (.031) |
| D11S1392 | 43.16 | …** | 2.17 (.015) |
| D12S1685 | 7.67 | 1.85 (.032) | |
| GATA49D12 | 17.72 | 1.80 (.036) | |
| D12S2070 | 125.31 | …** | 1.65 (.050) |
| D12S395 | 136.82 | …** | 1.98 (.024) |
| D15S1002 | 14.58 | 1.65 (.049) | |
| D15S1048 | 19.12 | 2.10 (.018) | |
| D15S165 | 20.24 | 2.37 (.009) | |
| D15S184 | 21.58 | 2.50 (.006) | |
| D15S1010 | 23.89 | 3.01 (.001) | |
| ACTC | 31.46 | 3.00 (.001) | |
| D15S657 | 104.86 | 1.71 (.043) | …* |
| D15S642 | 122.14 | 1.68 (.047) | …* |
| D21S1440 | 36.77 | 2.22 (.013) | |
| D21S270 | 38.08 | 2.05 (.020) | |
| D21S1255 | 39.22 | 2.06 (.020) | |
| D21S2055 | 40.49 | 2.08 (.019) | |
| D21S1893 | 43.67 | 2.15 (.016) | |
| D21S266 | 45.87 | 2.12 (.017) | |
Note.— See footnotes to table 3.
On the basis of recent observations by Cerhan et al. (1999), we partitioned the families according to whether they were positive for breast cancer (CaB+). To qualify for classification within the CaB+ group, the proband had to report the presence of breast cancer in a sister, mother, biological aunt, or grandmother. These self-reports were not verified. Fifty-three of the 230 multiplex families contained one or more cases of breast cancer (no cases of male breast cancer were reported). These 53 families contained a total of 81 ASPs. Five adjacent markers on 1p and seven adjacent markers on 21q in the CaB+ partition yielded nominally significant Zlr scores (table 5).
Table 5.
Nominally Significant Scores for Families with CaP That Are CaB+[Note]
| Marker | Position(cM) | Zlr (P) |
| D1S552 | 45.33 | 1.89 (.029) |
| D1S1622 | 56.74 | 3.78 (<.001) |
| D1S3721 | 72.59 | 2.20 (.014) |
| D1S2134 | 75.66 | 1.87 (.030) |
| D1S3728 | 89.49 | 2.37 (.009) |
| D21S1440 | 36.77 | 2.68 (.003) |
| D21S270 | 38.08 | 2.86 (.002) |
| D21S1255 | 39.22 | 2.83 (.002) |
| D21S2055 | 40.49 | 2.90 (.002) |
| D21S1893 | 43.67 | 2.36 (.009) |
| D21S266 | 45.87 | 2.21 (.013) |
| D21S1446 | 57.77 | 2.03 (.021) |
Note.— See footnote “a” to table 3.
The most consistent finding from our genome screen is the suggestion of susceptibility loci on chromosome 16. Table 6 reports the Zlr scores for all 22 markers typed on chromosome 16, both for the entire sample and for the various partitions. It is perhaps noteworthy that, whereas the various subgroups reported in tables 3–5 suggest a degree of chromosomal specificity, little is evident for chromosome 16. Only for families in the late-age-at-onset partition (and only for 16q markers) is there an absence of even a nominally significant signal. With the exception of markers D16S539 and D16S2621 in the early- versus late-age-at-onset comparison, none of the data partitions resulted in a significant difference between any of the subgroups.
Table 6.
Chromosome 16 Multipoint KAC Zlr Scores for the Total Sample and for the Three Data Partitions
| KAC Zlr Score in |
|||||||
| Marker | Position( cM) | All Families | FH+ Families | FH− Families | Families with Early Age at Onset | Families with Late Age at Onset | CaB+ Families |
| ATA41E04 | 11.46 | 2.17 | 2.07 | 1.08 | 1.40 | 1.71 | 2.48 |
| D16S748 | 22.65 | 2.71 | 1.93 | 1.94 | 1.64 | 2.20 | 2.14 |
| D16S3062 | 27.05 | 2.34 | 1.65 | 1.71 | 1.16 | 2.16 | 1.99 |
| D16S405 | 28.30 | 2.38 | 1.60 | 1.78 | 1.31 | 2.08 | 1.87 |
| D16S764 | 29.97 | 2.61 | 1.54 | 2.18 | 1.66 | 2.05 | 1.92 |
| ATA63G01 | 30.81 | 2.80 | 1.66 | 2.34 | 1.89 | 2.12 | 1.93 |
| D16S3103 | 32.07 | 2.81 | 1.74 | 2.28 | 1.96 | 2.07 | 1.69 |
| D16S403 | 43.89 | 2.37 | 1.43 | 1.86 | 2.08 | 1.20 | 1.10 |
| D16S769 | 50.60 | 1.99 | 1.88 | .90 | 1.70 | 1.05 | .62 |
| Centromere | |||||||
| D16S753 | 57.79 | 1.61 | 2.14 | .16 | 1.16 | 1.04 | −.37 |
| D16S3396 | 63.78 | 1.67 | 2.46 | −.04 | 1.28 | 1.12 | −.46 |
| D16S3253 | 71.77 | 1.40 | 2.43 | −.38 | 1.26 | .75 | −.12 |
| GATA67G11 | 81.15 | .95 | 1.92 | −.64 | 1.25 | −.01 | .16 |
| D16S2624 | 87.62 | 1.81 | 2.30 | .12 | 2.11 | .36 | .93 |
| D16S3049 | 97.03 | 2.80 | 2.00 | 1.84 | 2.27 | 1.57 | 1.60 |
| D16S3096 | 99.44 | 3.15 | 2.06 | 2.30 | 2.82 | 1.56 | 2.08 |
| D16S516 | 100.39 | 3.07 | 2.12 | 2.14 | 2.83 | 1.46 | 1.95 |
| D16S504 | 101.23 | 3.08 | 2.13 | 2.12 | 2.86 | 1.42 | 2.02 |
| D16S3040 | 104.45 | 2.48 | 2.23 | 1.19 | 2.62 | .77 | 1.62 |
| D16S402 | 113.52 | 1.48 | 1.44 | .49 | 2.25 | −.34 | 1.35 |
| D16S539 | 124.73 | .41 | .90 | −.48 | 1.97 | −1.53 | .53 |
| D16S2621 | 130.41 | .82 | .80 | .18 | 2.08 | −1.04 | .84 |
Discussion
Susceptibility loci that predispose to diseases with a late mean age at onset are notoriously difficult to map. The proband’s parents are usually deceased, and, even if DNA were available on all members of the sibship, the sibship may be too small to allow unambiguous reconstruction of the parental genotypes. Depending on the particulars of the disease, the proband’s offspring are unlikely to be old enough to be informative. These difficulties certainly apply to CaP, in which the sex-limited nature of the disease further reduces the available information. These factors help to explain why no undisputed susceptibility locus has yet been identified for CaP and why it is proving so difficult to achieve unambiguous replication in linkage studies.
We report here the results of a linkage study of 230 multiplex sibships with CaP, using a total of 420 highly polymorphic markers. Although five different chromosomal regions gave nominal evidence of a possible susceptibility locus, none of the signals in the total sample is sufficiently strong to meet the Lander and Kruglyak's (1995) threshold for “suggestive” linkage (i.e., a P=.00074 or Zlr≈3.18). And, although the usefulness of this criterion has been questioned (Curtis 1996; Witte et al. 1996), it is clear that oligogenic phenotypes—even those with 100% heritability—may result in increased sib-pair allele sharing that is only a few percentage points over the null value of 50% (Suarez et al. 1994).
Linkage studies have been successful in the identification of disease-susceptibility loci, including many that predispose to cancer (Fearon et al. 1990; Hall et al. 1990; Groden et al. 1991; Miki et al. 1994; Tavtigian et al. 1996). However, there are surprisingly few linkage studies of CaP. The first and only complete genome screen published to date was reported by Smith et al. (1996) and presented evidence of a susceptibility locus (HPC1 [MIM 601518]) on the long arm of chromosome 1 at 1q24-q25. A subsequent reanalysis of an expanded collection of the multiplex pedigrees in their study suggested that families with an early age at onset were primarily responsible for the linkage signal at HPC1 (Grönberg et al. 1997b, 1999). This claim remains controversial. Two other studies have produced modest support for the existence of HPC1. Cooney et al. (1997) reported an analysis of 59 multiplex families and obtained an NPL Z-score of 1.58 (P=.057) at D1S466. An analysis of the 20 families that met criteria for “hereditary” CaP produced an NPL Z-score of 1.72 (P=.045) at D1S466. Hsieh et al. (1997) obtained equivocal results in a sample of 92 multiplex families. When these families were subdivided according to the family’s mean age at onset, a nominally significant signal in the younger group was detected at D1S452 (two-point Z=2.04, P=.023), and another modest signal was detected at D1S2883 (two-point Z=1.91, P=.030), in the late-age-at-onset partition. Since these two markers are only about 5.5 cM apart, these results suggest that there could be two different CaP-susceptibility loci on 1q.
Three of the markers we typed map within the putative HPC1 region, and none approach nominal significance in the total sample. However, nominally significant linkage is obtained for a block of four proximal markers in the FH− partition and for two distal markers in the FH+ subgroup. Since our FH+ signal occurs approximately 20 cM from the closest HPC1 marker, this should not be interpreted as a replication. Three other studies have been unable to confirm the existence of HPC1 (McIndoe et al. 1997; Berthon et al. 1998; Eeles et al. 1998).
Two other regions of chromosome 1 have also been reported to harbor CaP-susceptibility loci. Berthon et al. (1998) reported an NPL Z score of 3.1 (P<.001) in the vicinity of 1q42.2-q43 in 47 French and German families. The NPL Z score increased to 3.32 in a subset of nine families in which the mean age at onset was <60 years. Homogeneity analysis led Berthon et al. (1998) to estimate that this putative susceptibility locus (PCAP; MIM 602759) accounts for <50% of the “hereditary” CaP cases in their data. Only two markers from the Weber 9 set map within this region, and, in our total sample, the Zlr score is <1.0, for both markers. In our subgroup analyses, we obtained nominally significant evidence of linkage for these two markers—but, in the families that we studied, the signal comes from the late-age-at-onset partition. Recently, Gibbs et al. (1999a) reported negative LOD scores for four markers from this distal region of chromosome 1q, and Whittemore et al. (1999) reported negative NPL Z-scores for the same four markers.
In a separate report, Gibbs et al. (1999b) presented evidence for a rare susceptibility locus, at 1p36, that appears to be important only in families that also have primary brain cancer. Although we did not preplan to analyze our families according to the presence of brain cancer, we conducted such an analysis of just chromosome 1p, once the report by Gibbs et al. (1999b) appeared. Only 13 families in our sample have a history of brain cancer, so we have little power to confirm the linkage. Three of the markers that we genotyped are located in the vicinity of the signal reported by Gibbs et al. (1999b), and, for all three markers, nonsignificant positive Zlr scores were obtained (Zlr=0.98, 1.15, and 1.49 at D1S1597, D1S3669, and D1S552, respectively).
Recently, Xu et al. (1998) presented evidence of an X-linked susceptibility locus (HPCX [MIM 300147]), at Xq27-q28, that, they estimate, accounts for ∼16% of “hereditary” cases of CaP. In this region, the only X-linked marker genotyped in the sibships that we studied was GATA31E08, at Xq27.1. For the entire sample, we obtained a multipoint Zlr score of −0.163, and none of the various data partitions produced a Zlr score >0.81.
The strongest linkage signal in our genome screen of the entire sample occurred on the long arm of chromosome 16, at 16q23.2. Analysis of the various subsamples indicated that no family partition disproportionately accounts for these signals. A maximum Zlr score of 3.15 is obtained at D16S3096.
Loss of heterozygosity (LOH) studies in CaP tumors have consistently found an increased loss on chromosome 16q (as well as 8p and 10q [Carter et al. 1990; Bergerheim et al. 1991; Cher et al. 1995; Elo et al. 1997; Osman et al. 1997]). Indeed, the pattern and distribution of LOH on 16q has led to speculation that up to three distinct susceptibility loci important for tumorigenesis, metastasis, or both may be present (Suzuki et al. 1996; Latil et al. 1997). One of these regions is located in the vicinity of our strongest signal. All previous studies of LOH in CaP tumors have been carried out in unrelated individuals. If the moderate signal that we have observed in these data is not a type I error, then it raises the possibility that a proportion of the families in our sample may be segregating an allele at a tumor-suppressor gene in this region; and, according to the Knudson (1971) model, all that is required to initiate tumorigenesis is a second somatic mutation in a single prostate cell.
Although we were able to verify the diagnosis of CaP by histological means or medical-record review in all but two of our subjects (and those two received treatment consistent with the diagnosis), the information regarding a family history positive for breast cancer was obtained from the probands, and no attempt to verify it was made. Two genomic regions—a broad region containing five markers and covering ∼45 cM on chromosome 1p and a 21-cM region on chromosome 21q—yielded nominally significant Zlr scores. The Zlr score at D1S1622 (3.78) corresponds to a LOD score >3 and meets criteria for suggestive linkage.
The short arm of chromosome 1 frequently shows allelic loss in breast cancer tumors (Schwab et al. 1996; Bieche et al. 1999; Perri et al. 1999). The c-myc promoter–binding protein, MPB1, which suppresses tumorigenicity in breast cancer cells, has been mapped to the p35-pter region of chromosome 1 (White et al. 1997). In a recent study, Millikan et al. (1999) report frequent LOH at two 1p36 markers (D1S243 and D1S160), but no evidence of linkage was obtained from an analysis of families with a history of early-onset bilateral breast cancer. Our results in the CaB+ partition raise the possibility that one or more tumor-suppressor genes capable of inhibiting tumorigenesis in both breast and prostate cells may be located on the short arm of chromosome 1.
As is the case with most complex diseases, polymorphisms in a number of candidate genes have been proposed as increasing the risk for CaP. Alleles at these loci are not believed to be necessary or sufficient to cause CaP, any more than the Apo ε4 allele is sufficient to cause Alzheimer disease; rather, they are risk factors in the epidemiological sense. Among these loci are the steroid 5-alpha-reductase 2 gene (Reichardt et al. 1995), on 2p23; the vitamin D–receptor gene (Taylor et al. 1996; Ingles et al. 1998), on 12q12-q14; the homeobox 3A gene (Abbaszadegan et al. 1998), on 8p21; and the X-linked androgen-receptor (AR) gene, on Xq11-q12, which contains in its first exon two polymorphic trinucleotide repeats—a 5′ CAG repeat and a 3′ GGC repeat. Given the central role played by androgens in the development and maintenance of normal prostate, and given that the length of the CAG repeat is inversely correlated with transcriptional activity, it is not surprising that these AR polymorphisms have received a great deal of attention. Hardy et al. (1996) found a significant correlation between the CAG-repeat number and an early age at onset of CaP, whereas Giovannucci et al. (1997) found that men with shorter repeats were at particularly high risk for distant metastatic and fatal CaP. A recent case-control study in a French and German sample, however, found no association between these polymorphisms and risk for CaP (Correa-Cerro et al. 1999). Although we did not type any of these candidate genes, our genome screen revealed no signals in the regions where these candidates map.
Prior to conducting any of the linkage analyses, we preplanned to partition our sample according to variables that reasonably might produce greater homogeneity in the subgroups. Two of these partitions were based on FH: families that met the Hopkins criteria for hereditary CaP were compared with families that may be sporadically multiplex. The second subanalysis focused on sibships from families that were CaB+. The third partitioning used age at onset to divide the families into two equal groups according to whether the sibship’s mean age at onset was below or above the sample’s median. The use of a median split in the present study is entirely arbitrary, since age at onset in our sample does not deviate from normality (Shapiro-and-Wilk [1965] test; W=.988, P=.82). For a number of well-known diseases, including various cancers, either strong FH+ (usually with a dominant-type transmission pattern) or an unusually early age at onset suggests a single segregating susceptibility gene with high penetrance. And, indeed, this association has been exploited successfully to map, clone, and characterize a number of large-effect susceptibility loci (e.g., BRCA1 and BRCA2) in stringently ascertained pedigrees; however, it is unlikely that genes such as BRCA1 or BRCA2 would be identified in a simple sib-pair study, just as it is unlikely that any of the highly penetrant genes that give rise to Alzheimer disease (i.e., amyloid beta A4–precursor protein, presenilin 1, or presenilin 2) would be identified in a random sample of affected sibs. These major genes are simply too rare. On the other hand, a genome screen of a random sample of sib pairs concordant for Alzheimer disease can detect the linkage signal in the vicinity of the Apo E locus on chromosome 19q, as recently demonstrated by Kehoe et al. (1999).
In the two analyses that compared linkage signals from complementary data partitions (tables 3 and 4), additional nominally significant signals were detected in the partitions—namely, the FH− and the late-age-at-onset partitions—that, on a priori grounds, might be expected to yield a larger proportion of sporadic cases. This excess could be a measure of the increased type I–error rate occasioned by the smaller sample sizes that result from subdivision. Alternatively, some of these signals may reflect the presence of true susceptibility loci that exert an effect, for instance, later in life. Further work will be required to eliminate the false-positive signals.
Acknowledgments
We thank Dr. Nancy J. Cox for her assistance with GENEHUNTER-PLUS. This work was supported in part by awards from the Urological Research Foundation (to B.K.S., J.K.B., J.S.W., and W.J.C.), the CaP CURE Foundation (support to B.K.S. and W.J.C.), U.S. Public Health Service grants MH31302 (to B.K.S.), HG00198 (to K.W.B), GM28356 and RR03655 (both to R.C.E.), and HV48141 (to J.L.W.); and U.S. Army grant DAMD17–98–18589 (to J.S.W.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://www.marshfield.org/genetics (for markers and their positions [in cM])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih/gov.Omim (for HPCX [MIM 300147], HPC1 [MIM 601518], and PCAP [MIM 602759] )
References
- Abbaszadegan MR, Brown KM, Voeller JH, Sciavolino P, Abate-Shen C, Gelmann EP (1998) Polymorphic allele of NKX3.1, a prostate-specific homeobox gene is a possible risk factor for prostate cancer. Proc Am Assoc Cancer Res 39:365 [Google Scholar]
- Ahlbom A, Lichtenstein P, Malmstrom H, Feychting M, Hemminki K, Pedersen N (1997) Cancer in twins: genetic and nongenetic familial risk factors. J Natl Cancer Inst 89:287–293 [DOI] [PubMed]
- Badner JA, Gershon ES, Goldin LR (1998) Optimal ascertainment strategies to detect linkage to common disease alleles. Am J Hum Genet 63:880–888 [DOI] [PMC free article] [PubMed]
- Bergerheim USR, Kunimi K, Collins VP, Ekman P (1991) Deletion mapping of chromosomes 8, 10, and 16 in human prostatic carcinoma. Genes Chromosomes Cancer 3:215–220 [DOI] [PubMed]
- Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, Wohr G, Latil A (1998) Predisposing gene for early onset prostate cancer, localized on chromosome 1q42.2-43. Am J Hum Genet 62:1416–1424 [DOI] [PMC free article] [PubMed]
- Bieche I, Khodja A, Lidereau R (1999) Deletion mapping of chromosomal region 1p32-pter in primary breast cancer. Genes Chromosomes Cancer 24:255–263 [DOI] [PubMed]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed]
- Broman KW, Weber JL (1998) Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet 63:1563–1564 [DOI] [PMC free article] [PubMed]
- Cannon L, Bishop DT, Skolnick M, Hunt S, Lyon JL, Smart CR (1982) Genetic epidemiology of prostate cancer in the Utah Mormon genealogy. Cancer Surv 1:47–69 [Google Scholar]
- Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC (1992) Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci USA 89:3367–3371 [DOI] [PMC free article] [PubMed]
- Carter BS, Ewing CM, Ward WS, Treiger BF, Aalders TW, Schalken JA, Epstein JI, et al (1990) Allelic loss of chromosomes 16q and 10q in human prostate cancer. Proc Natl Acad Sci USA 87:8751–8755 [DOI] [PMC free article] [PubMed]
- Cerhan JR, Parker AS, Putnam SD, Chiu BC-H, Lynch CF, Cohen MB, Torner JC, et al (1999) Family history and prostate cancer risk in a population-based cohort of Iowa men. Cancer Epidemiol Biomarkers Prev 8:53–60 [PubMed]
- Cher ML, Ito T, Weidner N, Carroll PR, Jensen RH (1995) Mapping of regions of physical deletion on chromosome 16q in prostate cancer cells by fluorescence in situ hybridization (FISH). J Urol 153:249–254 [DOI] [PubMed]
- Cooney KA, McCarthy JD, Lange E, Huang L, Miesfeldt S, Montie JE, Oesterling JE (1997) Prostate cancer susceptibility locus on chromosome 1q: a confirmatory study. J Natl Cancer Inst 89:955–959 [DOI] [PubMed]
- Correa-Cerro L, Wohr G, Haussler J, Berthon P, Drelon E, Mangin P, Fournier G, et al (1999) (CAG)nCAA and GGN repeats in the human androgen receptor gene are not associated with prostate cancer in a French-German population. Eur J Hum Genet 7:357–362 [DOI] [PubMed]
- Curtis D (1996) Genetic dissection of complex traits. Nat Genet 12:356–358 [DOI] [PubMed]
- Davis S, Weeks DE (1997) Comparison of nonparametric statistics for detection of linkage in nuclear families: single-marker evaluation. Am J Hum Genet 61:1431–1444 [DOI] [PMC free article] [PubMed]
- Dunn JE (1975) Cancer epidemiology in populations of the United States—with emphasis of Hawaii and California—and Japan. Cancer Res 35:3240–3245 [PubMed]
- Eeles RA, Durocher F, Edwards S, Teare D, Badzioch M, Hamoudi R, Gill S, et al (1998) Linkage analysis of chromosome 1q markers in 136 prostate cancer families. Am J Hum Genet 62:653–658 [DOI] [PMC free article] [PubMed]
- Elo JP, Harkonen P, Kyllonen AP, Lukkarinen O, Poutanen M, Vihko R, Vihko P (1997) Loss of heterozygosity at 16q24.1-q24.2 is significantly associated with metastatic and aggressive behavior of prostate cancer. Cancer Res 57:3356–3359 [PubMed]
- Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, et al (1990) Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 247:49–56 [DOI] [PubMed]
- Ghadirian P, Cadotte M, Lacroix A, Perret C (1991) Family aggregation of cancer of the prostate in Quebec: the tip of the iceberg. Prostate 19:43–52 [DOI] [PubMed]
- Giardiello FM (1997) Genetic testing in hereditary colorectal cancer. JAMA 278:1278–1281 [PubMed]
- Gibbs M, Chakrabarti L, Stanford JL, Goode EL, Kolb S, Schuster EF, Buckley VA, et al (1999a) Analysis of chromosome 1q42.2-43 in 152 families with high risk of prostate cancer. Am J Hum Genet 64:1087–1095 [DOI] [PMC free article] [PubMed]
- Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, et al (1999b) Evidence for a rare prostate cancer-susceptibility locus at chromosome 1p36. Am J Hum Genet 64:776–787 [DOI] [PMC free article] [PubMed]
- Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, et al (1997) The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci USA 94:3320–3323 [DOI] [PMC free article] [PubMed]
- Göring HHH, Ott J (1995) Verification of sib relationship without knowledge of parental genotypes. Am J Hum Genet Suppl 57:A192 [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, et al (1991) Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66:589–600 [DOI] [PubMed]
- Grönberg H, Damber L, Damber JE (1994) Studies of genetic factors in prostate cancer in a twin population. J Urol 152:1484–1489 [DOI] [PubMed]
- Grönberg H, Damber L, Damber JE, Iselius L (1997a) Segregation analysis of prostate cancer in Sweden: support for dominant inheritance. Am J Epidemiol 146:552–557 [DOI] [PubMed]
- Grönberg H, Smith J, Emanuelsson M, Jonsson BA, Bergh A, Carpten J, Isaacs W, et al (1999) In Swedish families with hereditary prostate cancer, linkage to the HPC1 locus on chromosome 1q24–25 is restricted to families with early-onset prostate cancer. Am J Hum Genet 65:134–140 [DOI] [PMC free article] [PubMed]
- Grönberg H, Xu J, Smith JR, Carpten JD, Isaacs SD, Freije D, Bova GS, et al (1997b) Early age at diagnosis in families providing evidence of linkage to the hereditary prostate cancer locus (HPC1) on chromosome 1. Cancer Res 57:4707–4709 [PubMed]
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250:1684–1689 [DOI] [PubMed]
- Hardy DO, Scher HI, Bogenreider T, Sabbatini P, Zhang ZF, Nanus DM, Catterall JF (1996) Androgen receptor CAG repeat lengths in prostate cancer: correlation with age of onset. J Clin Endocrinol Metab 81:4400–4405 [DOI] [PubMed]
- Holloway SM, Sofaer JA (1992a) Coefficients of relationship by isonymy among registrations for five common cancers in Scottish males. J Epidemiol Commun Health 46:368–372 [DOI] [PMC free article] [PubMed]
- Holloway SM, Sofaer JA (1992b) Coefficients of relationship by isonymy among Scottish males with early and late onset cancers. Dis Markers 10:109–114 [PubMed]
- Hsieh CL, Oakley-Girvan I, Gallagher RP, Wu AH, Kolonel LN, Teh CZ, Halpern J, et al (1997) Re: prostate cancer susceptibility locus on chromosome 1q: a confirmatory study. J Natl Cancer Inst 89:1893–1894 [DOI] [PubMed]
- Ingles SA, Coetzee GA, Ross RK, Henderson BE, Kolonel LN, Crocitto L, Wang W, et al (1998) Association of prostate cancer with vitamin D receptor haplotypes in African-Americans. Cancer Res 58:1620–1623 [PubMed]
- Keetch DW, Rice JP, Suarez BK, Catalona WJ (1995) Familial aspects of prostate cancer: a case control study. J Urol 154:2100–2102 [PubMed]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, et al (1999) A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet 8:237–245 [DOI] [PubMed]
- Knudson GG (1971) Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 68:820–823 [DOI] [PMC free article] [PubMed]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed]
- Krain LS (1974) Some epidemiologic variables in prostatic carcinoma in California. Prev Med 3:154–159 [DOI] [PubMed]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed]
- Landis SH, Murray T, Bolden S, Wingo PA (1999) Cancer statistics, 1999. CA Cancer J Clin 49:8–31 [DOI] [PubMed]
- Latil A, Cussenof O, Fournier G, Driouch K, Lidereau R (1997) Loss of heterozygosity at chromosome 16q in prostate adenocarcinoma: identification of three independent regions. Cancer Res 57:1058–1062 [PubMed]
- McIndoe RA, Stanford JL, Gibbs M, Jarvik GP, Brandzel S, Neal CL, Li S, et al (1997) Linkage analysis of 49 high-risk families does not support a common familial prostate cancer-susceptibility gene at 1q24–25. Am J Hum Genet 61:347–353 [DOI] [PMC free article] [PubMed]
- Meikle AW, Smith JA, West DW (1985) Familial factors affecting prostatic cancer risk and plasma sex-steroid levels. Prostate 6:121–128 [DOI] [PubMed]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71 [DOI] [PubMed]
- Millikan RC, Ingles SA, Diep AT, Xue S, Zhou N, Florentine BD, Sparkes RS, et al (1999) Linkage analysis and loss of heterozygosity for chromosome arm 1p in familial breast cancer. Genes Chromosomes Cancer 25:354–361 [PubMed]
- Monroe KR, Yu MC, Kolonel LN, Coetzee GA, Wilkens LR, Ross RK, Henderson BE (1995) Evidence of an X-linked or recessive genetic component to prostate cancer risk. Nat Med 1:827–829 [DOI] [PubMed]
- Morganti G, Gianferrari L, Cresseri A (1956) Recherches clinico-statistiques et génétiques sur les néoplasies de la prostate. Acta Genet Stat Med 6:304–305 [PubMed] [Google Scholar]
- Osman I, Scher H, Dalbagni G, Reuter V, Zhang ZF, Cordon-Cardo C (1997) Chromosome 16 in primary prostate cancer: a microsatellite analysis. Int J Cancer 71:580–584 [DOI] [PubMed]
- Ott J (1985) A chi-square test to distinguish allelic association from other causes of phenotypic association between two loci. Genet Epidemiol 2:79–84 [DOI] [PubMed]
- Parkin DM, Pisani P, Ferlay J (1993) Estimates of the worldwide incidence of 18 major cancers in 1985. Int J Cancer 54:594–606 [DOI] [PubMed]
- Perri P, Praml C, Savelyeva L, Pillmann A, Schwab M (1999) Fine mapping of distal 1p loci reveals TP73 at D1S468. Cytogenet Cell Genet 84:111–114 [DOI] [PubMed]
- Reichardt JKV, Makridakis N, Henderson BE, Yu MC, Pike MC, Ross RK (1995) Genetic variability of the human SRD5A2 gene: implications for prostate cancer risk. Cancer Res 55: 3973–3975 [PubMed]
- Schaid DJ, McDonnell SK, Blute ML, Thibodeau SN (1998) Evidence for autosomal dominant inheritance of prostate cancer. Am J Hum Genet 62:1425–1438 [DOI] [PMC free article] [PubMed]
- Schwab M, Praml C, Amler LC (1996). Genomic instability in 1p and human malignancies. Genes Chromosomes Cancer 16:211–229 [DOI] [PubMed]
- Shapiro SS, Wilk MB (1965) An analysis of variance test for normality (complete samples). Biometrika 52:591–611 [Google Scholar]
- Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, Isaacs SD, Brownstein MJ, et al (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 [DOI] [PubMed]
- Spitz MR, Currier RD, Fueger JJ, Babaian RJ, Newel GR (1991) Familial patterns of prostate cancer: a case-control analysis. J Urol 146:1305–1307 [DOI] [PubMed]
- Staszewski J, Haenszel W (1965) Cancer mortality among the Polish-born in the United States. J Natl Cancer Inst 35:291–297 [PubMed]
- Steele R, Lees RE, Kraus AS, Rao C (1971) Sexual factors in the epidemiology of cancer of the prostate. J Chronic Dis 24:29–37 [DOI] [PubMed]
- Steinberg GD, Carter BS, Beaty TH, Child B, Walsh PC (1990) Family history and the risk of prostate cancer. Prostate 17:337–347 [DOI] [PubMed]
- Suarez BK, Hampe CL, Van Eerdewegh P (1994) Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR (eds) Genetic approaches to mental disorders. American Psychiatric Press, Washington, DC, pp 23–46 [Google Scholar]
- Suzuki H, Komiya A, Emi M, Kuramochi H, Shiraishi T, Yatani R, Shimazaki J (1996) Three distinct commonly deleted regions of chromosome arm 16q in human primary and metastatic prostate cancers. Genes Chromosomes Cancer 17:225–233 [DOI] [PubMed]
- Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, et al (1996) The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 12:333–337 [DOI] [PubMed]
- Taylor JA, Hirvonen A, Watson M, Pittman G, Mohler JL, Bell DA (1996) Association of prostate cancer with vitamin D receptor gene polymorphism. Cancer Res 56:4108–4110 [PubMed]
- White RA, Adkison LR, Dowler LL, Ray RB (1997). Chromosomal localization of the human gene encoding c-myc promoter-binding protein (MPB1) to chromosome 1p35-pter. Genomics 39:406–408 [DOI] [PubMed]
- Whittemore AS (1994) Prostate cancer. Cancer Surv 19-20:309–322 [PubMed]
- Whittemore AS, Kolonel LN, Wu AH, John EM, Gallagher RP, Howe GR, Burch JD, et al (1995a) Prostate cancer in relation to diet, physical activity, and body size in blacks, whites, and Asians in the United States and Canada. J Natl Cancer Inst 87:652–661 [DOI] [PubMed]
- Whittemore AS, Lin IG, Oakley-Girvan I, Gallagher RP, Halpern J, Kolonel LN, Wu AH, et al (1999) No evidence of linkage for chromosome 1q42.2-43 in prostate cancer. Am J Hum Genet 65:254–256 [DOI] [PMC free article] [PubMed]
- Whittemore AS, Wu AH, Kolonel LN, John EM, Gallagher RP, Howe GR, West DW, et al (1995b) Family history and prostate cancer risk in black, white, and Asian men in the United States and Canada. Am J Epidemiol 141:732–740 [DOI] [PubMed]
- Witte JS, Elston RC, Schork NJ (1996) Genetic dissection of complex traits. Nat Genet 12:355–356 [DOI] [PubMed]
- Woolf CM (1960) An investigation of the familial aspects of carcinoma of the prostate. Cancer 13:739–744 [DOI] [PubMed] [Google Scholar]
- Xu J, Meyers D, Freije D, Isaacs S, Wiley K, Nusskern D, Ewing C, et al (1998) Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet 20:175–179 [DOI] [PubMed]
- Yuan B, Vaske D, Weber JL, Beck J, Sheffield VC (1997) Improved set of short-tandem-repeat polymorphisms for screening the human genome. Am J Hum Genet 60:459–460 [PMC free article] [PubMed]