Abstract
mtDNA haplotypes of representatives of the cosmopolitan peoples of north-central Mexico were studied. Two hundred twenty-three samples from individuals residing in vicinities of two localities in north-central Mexico were analyzed. A combination of strategies was employed to identify the origin of each haplotype, including length variation analysis of the COII and tRNALYS intergenic region, nucleotide sequence analysis of control region hypervariable segment 1, and RFLP analysis of PCR products spanning diagnostic sites. Analysis of these data revealed that the majority of the mtDNA haplotypes were of Native American origin, belonging to one of four primary Native American haplogroups. Others were of European or African origin, and the frequency of African haplotypes was equivalent to that of haplotypes of European derivation. These results provide diagnostic, discrete character, molecular genetic evidence that, together with results of previous studies of classical genetic systems, is informative with regard to both the magnitude of African admixture and the relative maternal contribution of African, European, and Native American peoples to the genetic heritage of Mexico. Phylogenetic analysis revealed that African sequences formed a basal, paraphyletic group.
Introduction
According to widespread popular belief, the present day peoples of Mexico are, by and large, descendants of Native American and European (Spanish) ancestors. Historical accounts also document African slavery in Mexico during the 16th–18th centuries (Beltrán 1944). Although records from this period are incomplete, estimates of the number of African slaves brought to Mexico are in the range of 200,000–500,000 (Beltrán 1944; Curtin 1969; Muhammad 1995). The actual number may be higher, since many slaves were imported illegally, without documentation, and since African ancestry was often not reported for census data (Beltrán 1944; Tjarks 1978; Muhammad 1995). The contributions of Africans to the genes and culture of the peoples of Mexico have been largely denied and forgotten in popular culture. Consequently, these Africans have been culturally and genetically assimilated to a greater extent than has been the case in other regions of the Americas.
Various classical genetic systems (blood groups, blood enzymes, and blood proteins) have been used to estimate the genetic composition of the peoples inhabiting Mexico (Crawford et al. 1974, 1976; Schanfield et al. 1978; Tiburcio et al. 1978; Lisker et al. 1986, 1988, 1990, 1994, 1995). Such studies have found that Native Americans provide the largest ancestral contribution to the contemporary peoples of Mexico. Studies assuming a trihybrid model have estimated various frequencies of African and European admixture with Native American populations. In general, estimates of European gene frequencies were greater than those of African gene frequencies, when African gene frequencies were considered. Recently, Lisker et al. (1996) compiled admixture estimates, using data derived from classical genetic systems reported in previous studies in Mexico (Crawford et al. 1974, 1976; Tiburcio et al. 1978; Lisker and Babinsky 1986; Lisker et al. 1986, 1988, 1990, 1995; Cerda-Flores and Garza-Chapa 1989). African or European admixture was identified in all regions and groups analyzed (Lisker et al. 1996). Even relatively isolated Native American populations showed some degree of African or European admixture when classical genetic systems were used with the trihybrid model (Lisker et al. 1996).
Previous studies of mtDNA variation in Mexico have focused on geographically and culturally isolated Native American populations, to avoid admixture (Schurr et al. 1990; Torroni et al. 1994a). That approach is useful for recovery of the history of the peopling of the Americas, but it does not provide information regarding ancestry of the general, cosmopolitan (relatively recent admixture of indigenous peoples with peoples of diverse geographic origins) population. We assessed mtDNA haplotypes among the general cosmopolitan population, to provide information regarding both Mexican history and prehistory.
Most Native Americans share common mtDNA mutations that define four primary haplogroups (A, B, C, and D), reflecting descent from Asian colonization of the New World (Schurr et al. 1990; Ward et al. 1991; Horai et al. 1993; Torroni et al. 1993). The majority of mtDNA haplotypes of European origin are members of one of seven European haplogroups (H, I, J, K, T, V, and W) or two haplogroups (U and X) that Europeans share with Africans or a small number of Pacific Northwest Native Americans, respectively (Torroni 1994b, 1996, 1998; Forster et al. 1996; Richards et al. 1996). The mtDNA haplotypes of the majority of individuals of African ancestry are members of either macrohaplogroup L or haplogroup L3 (Chen et al. 1995; Watson et al. 1997). Various mtDNA screening methods may be used to identify these haplogroups, and thus the geographic region of origin of individual haplotypes may be determined in cosmopolitan populations. Here, we provide analysis of these diagnostic, discrete character, molecular genetic markers, to help reveal affinities of the cosmopolitan peoples of north-central Mexico.
Subjects and Methods
Sample Collection and Preparation
Two hundred twenty-three samples of blood, hair, or placenta were collected, with informed consent, from the vicinities of Juárez (n=123) and Ojinaga (n=100), Chihuahua, in north-central Mexico (fig. 1). All individuals were of Mexican origin and resided in the vicinity of collection.
Figure 1.
Locations of sample collection: Ciudad Juárez and Ojinaga, Chihuahua, Mexico.
Extraction of DNA from whole blood was accomplished by a Super Quick-Gene DNA isolation kit (Analytical Genetic Testing Center), according to the supplier's protocol. Extraction of DNA from placenta samples followed the method of Hillis and Davis (1986). DNA extraction from hair was accomplished by techniques described by Higuchi et al. (1988).
PCR, Restriction Digestion, and DNA Sequencing
Amplification of target gene regions was accomplished by standard PCR methods, with primers listed in table 1. Restriction enzymes were obtained from Amersham, and PCR products were digested according to the protocol of the manufacturer. Sequencing templates were isolated in low melting temperature agarose and were purified with Promega's Wizard PCR Prep system. Templates were then sequenced with an Applied Biosystems Prism 377 DNA sequencer, with primers listed in table 1.
Table 1.
Markers Used for Haplogroup Identification for Individuals of North-Central Mexico
|
Primerb |
||||
| Haplogroup | Nucleotide(s)a | Length Variation | Designation | Sequence(5′→3′) |
| Native American: | ||||
| A | 16290T, 16319A | HaeIII+663 | L396 | CCAGCCTAACCAGATTTCAA |
| H1067 | GGGGTATCTAATCCCAGTTT | |||
| B | 16189C, 16217C | 9-bp Deletion | L8192 | GCTCTGAAATCTGTGGAG |
| H8361 | TATTTAGTTGGGGCATTTCAC | |||
| C | 16298C, 16327T | AluI+13262 | L12978 | CAAGCCTCACCCCACTACTA |
| H13773 | GGTAGAGGGGGATTGTTGTT | |||
| D | Not available | AluI–5176 | L5042 | CCCACATAGGATGAATAATA |
| H5442 | GCGATGAGTGTGGGGAGGAA | |||
| X | 16223T, 16278T | DdeI–1715 | L1689 | GCTAAACCTAGCCCCAAA |
| H1782 | TTTCATCTTTCCCTTGCGGTAC | |||
| European: | ||||
| H | Not available | AluI–7025 | L6886 | GACTCGCCACACTCCA |
| H7275 | GCTGTTAGAGAAATGAATGA | |||
| J | 16069T, 16126C | Not included | ||
| K | 16224C, 16311C | Not included | ||
| U | 16270T | Not included | ||
| V | 16298C | Not included | ||
| African: | ||||
| L1 | 16390G | HpaI+3592 | L3522 | CTACCATCACCCTCTACATC |
| H3650 | CACCCTGATCAGAGGATTG | |||
| L2 | 16390A | HpaI+3592 | ||
| L3 | Published sequence comparison | Not included | ||
Identified by use of HV1 PCR and sequencing primers L15988 (5′-TCTTTAACTCCACCATTAGCA-3′), H16425 (5′-GATATTGATTTCACGGAGGAT-3′), and internal sequencing primer L16123 (5′-CTGCCAGCCACCATGAATAT-3′).
Used for length variation analyses.
Haplotype Analysis
A combination of strategies was used to determine the geographic region of origin for individual haplotypes. Length variation analysis included both screening for the 9-bp deletion and RFLP analysis, to identify known diagnostic Native American, European, and African markers (table 1). A HaeIII site at nucleotide position (np) 16517 was used to confirm independent acquisition of the 9-bp deletion in a subset of haplogroup A samples. The HaeIII site is associated with haplogroup B and is dimorphic in other Native American haplogroups (Torroni et al. 1994a). Samples were screened for other European haplogroups (T, I, and J), which are characterized by control region hypervariable segment 1 (HV1) polymorphisms (Torroni et al. 1996, 1998). Restriction sites that define other haplogroups were not analyzed. Haplotypes with the presence of the HpaI site at np 3592, and with the presence of a G at np 16390, were presumed to be members of African haplogroup L1. Identification of African haplogroup L2 was established by the presence of an A at np 16390 (Chen et al. 1995). The presence or absence of this character state was determined by nucleotide sequence analysis. Control region HV1 sequences were obtained for 87 individuals. These sequences were compared with published African sequences, to aid in identification of African haplogroups L1, L2, and L3 (Watson et al. 1997).
Phylogenetic Analysis
Phylogenetic analysis of the 87 HV1 sequences was performed with PAUP, version 4.0 (Swofford 1998). A 347-bp region between np 16023 and np 16370 was analyzed. Sequences were aligned with the human reference sequence (Anderson et al. 1981). Maximum parsimony trees were generated through the simple addition of sequences, by use of the tree-bisection and reconnection branch–swapping algorithm of PAUP. A total of 2,000 most parsimonious trees were saved, and a 50% majority rule consensus tree was produced. Neandertal sequence (Krings et al. 1997) was used as outgroup. This procedure was repeated multiple times.
Results
Native American, European, or African haplotypes were identified in 221 of 223 individuals (table 2). Two of the 223 samples were not identified by the methods employed. One hundred ninety-nine (89.2%) of the 223 samples were Native American haplotypes. The remaining 22 identified samples were equivalently divided between European and African; 12 (5.4%) were identified as European, and 10 (4.5%) were identified as African. The frequencies of Native American haplotypes identified in samples collected from the two localities were not substantially different (table 2). A greater frequency of European haplotypes was observed in Juárez samples (7.3%) than in Ojinaga samples (3.0%). The frequency of African haplotypes was greater in Ojinaga samples (5.0%) than in Juárez samples (4.1%).
Table 2.
Geographic Region of Origin and Frequencies of mtDNA Haplotypes Identified for Two Cosmopolitan Populations in North-Central Mexico
| Haplotype Frequency |
||||
| CollectionArea | Native American | European | African | Unknown |
| Ojinaga (n=100) | 91 (91.0%) | 3 (3.0%) | 5 (5.0%) | 1 (1.0%) |
| Juárez (n=123) | 108 (87.7%) | 9 (7.3%) | 5 (4.1%) | 1 (<1.0%) |
| Total (n=223) | 199 (89.1%) | 12 (5.4%) | 10 (4.5%) | 2 (<.1%) |
Native American Haplotypes
Of the 223 samples studied, 199 had length variation markers diagnostic for one of the four primary Native American haplogroups: 75 (33.6%) were identified as haplogroup A; 59 (26.5%) had the 9-bp deletion, which has traditionally been used to identify haplogroup B; 52 (23.3%) were members of haplogroup C; and only 13 (5.8%) were haplogroup D. Native American haplogroup A occurred at the highest frequency, and haplogroup D occurred at the lowest frequency. This is consistent with Native American mtDNA studies in Central America (Torroni et al. 1994a; Batista et al. 1995).
Ten of these samples, identified by the presence of the HaeIII site at np 663 as haplogroup A, also possessed the 9-bp deletion. Not 1 of these 10 samples had the HaeIII site at np 16517, which is generally associated with both the deletion and haplogroup B, whereas all haplogroup B samples had the site present. Furthermore, HV1 sequence data were obtained for 7 of these 10 samples and were included in the phylogenetic analysis. The phylogenetic positions of these seven samples were within haplogroup A. These results confirm that the 9-bp deletion has arisen more than once, in two Native American haplogroups. Presence of the deletion in haplogroup A has been reported, in other studies, at low frequencies (Ballinger et al. 1992; Torroni et al. 1993, 1994c).
Of the 87 samples for which HV1 nucleotide sequence data were obtained, 63 had haplogroup A–, B–, or C–specific nucleotides at polymorphic positions, and, on the basis of restriction site analysis, one sample was identified as haplogroup D (table 3). Sixteen samples had Native American haplogroup A–specific nucleotides; Native American haplogroup B–specific nucleotides were present in 32 samples; and haplogroup C–specific nucleotides were present in 14 samples. One haplogroup B sample did not have a C at np 16189, and one haplogroup C sample did not have a C at np 16298; however, these samples were identified as being haplogroup B and haplogroup C, respectively, on the basis of the 9-bp deletion and restriction site analysis. All samples belonging to Native American haplogroups A, B, and C, which we ascertained on the basis of HV1 markers, had corresponding haplogroup restriction site markers. Samples identified as Native American did not have African or European HV1 or restriction site markers.
Table 3.
Native American Haplogroups and HV1 Sequence Variation Observed for Individuals of North-Central Mexico
| Variable Nucleotide Position |
|
| 11111111111111111111111111111111111111111111111116666666666666666666666666666666666666666666666666001111111111111222222222222222222233333333333333359112224677888801122233344567779990001122234556661213679542617894782383491866048048149193575267025 | |
| Reference Sequencea | ATCATAGGATCACCTGTCCCCACCACCCCGCCCTCTATGTTCATTTCTC |
| Haplogroup A: | |
| C1 | ..T................T.........A.T......A......C.C. |
| 524I | ...................T.........A.T......A........C. |
| C68 | ..TT...............T.........A.T......A........C. |
| C70 | ..T................T.........A.T......A........C. |
| D36 | ..T................T.........A.T......A........C. |
| D37 | ..T................T.........A.T......A........C. |
| O8 | ..T...........C....T.........A.T......A........C. |
| P13 | ..T................T..TT.......TT.....A........C. |
| P9 | ..T................T...T.......T......A........C. |
| P4 | ..T................T...........T......A........C. |
| 5291 | ..T................T...........T......A........C. |
| C12 | ..T.........T......T...........T......A........C. |
| C43 | ..T................T...........T......A........C. |
| C47 | ..T................T...........T......A........C. |
| C56 | ..T................T...........T......A...G...... |
| O21 | ..T................T...........T......A........C. |
| Haplogroup B: | |
| O23 | ..............C.C....G........................T.. |
| C4 | ..............C.C.............................T.. |
| P14 | ..............C.C.............................T.. |
| 6032 | ..T...........C.C...T.......T.................?.. |
| C30 | .CT...........C.C...T....?..T.........A.......?.. |
| P20 | ..T...........C.C...T....?....................T.. |
| C67 | ......A.......C.C........T..T.................... |
| C42 | ..............C.C.............................?.. |
| N11 | ......AA....T.C.C......................C......... |
| P15 | ......AA....T.C.C......................C......... |
| N64 | ..............C.C................................ |
| BB1 | ..............C.C........?....................?.. |
| BB2 | ..............C.C.............................?.. |
| C39 | ..T...........C.C.......................C...C.... |
| P8 | ..T...........C.C................................ |
| O32 | ..T...........C.C...?....?....................?.. |
| P12 | ..............C.C................................ |
| P19 | ..............C.C.............T.................. |
| 524IV | ..............C.C...?.........................?.. |
| 5292 | ..............C.C........?.......C............?.. |
| C31 | ..............C.C........?....................... |
| C62 | ......A.......C.C........T..T.................... |
| D30 | ..............C.C.........T....................C. |
| C33 | ..............C.C................C.............C. |
| C53 | ..............C.C...........?.................?.. |
| C73 | ..............C.C...?....?.T..................?.. |
| D16 | ..............C.C...?....?....................... |
| C64 | ......A.......C.C........T..T.................... |
| D20 | ...........G..C.C..............T................. |
| D52 | ..............C.C........?..?.................... |
| O39 | ..............C.C................................ |
| C8 | ........G.....C.C..................C....C........ |
| D27 | ........G.......C................................ |
| Haplogroup C: | |
| BB3 | .C........T......T.T.............C......CT....... |
| P11 | .C........T......T.T.............C.......T....... |
| O4A | .C........T......T.T.............C......CT....... |
| O4 | .....T.............T.............C..G...CT....... |
| P3 | ....C..............T.............C......CT....... |
| 5299 | ...................T.............C......CT....... |
| O12 | ......A..C.........T.............C...C..CT....... |
| C9 | G..................T.............C......CT....... |
| P2 | ......A............T.............C...C..CT....... |
| O2A | ...................T.............C......CT......T |
| O3A | ...................T.............C......CT...C... |
| O9 | ...................T.............C......CT....... |
| P5 | ...................T.............C......CT....... |
| C10 | G............T.A...T....................CT.....C. |
| Haplogroup D: | |
| C75 | ...................T..G.G.........T........C...C. |
Source: Anderson et al. (1981).
Non–Native American Haplotypes
Twenty-four samples did not possess Native American mtDNA markers. Twelve of 24 non–Native American samples were identified as European haplotypes (table 4). The European haplogroup H RFLP marker was identified in five samples (C5, O36, P16, D41, and D60). Samples C2 and C25 had diagnostic HV1 markers that characterize European haplogroup K (Torroni et al. 1996). Samples C80 and D58 were identified as European haplogroup J, on the basis of HV1 markers (Torroni et al. 1996). Sample D40 had the HV1 marker that characterizes subhaplogroup U6 of haplogroup U (Macaulay et al. 1999). Samples D33 and O6 were identified as European haplogroup V (Torroni et al. 1998).
Table 4.
Non–Native American Haplotypes and HV1-Sequence Variation Observed for Individuals from North-Central Mexico
|
Variable Nucleotide Position |
||
| 111111111111111111111111111111111111666666666666666666666666666666666666001111111111122222222222222233333333691222466788911222356677899900122566931469502279247234461508614848103602 | Haplogroup | |
| Reference sequencea | CTCTTGGAATCTCCTCCTCCCACCCCCTTTTCTTCT | |
| European: | ||
| C5 | ..............C..................C.. | H |
| O36 | .................................... | H |
| P16 | .................................... | H |
| D41 | ..............C..................C.. | H |
| D60 | .C....................T............. | H |
| C80 | T...C.A..C.....T....T............... | J |
| D58 | T...C............................... | J |
| C2 | ...........C.....C............C..... | K |
| C25 | ...........C.....C............C..... | K |
| O6 | ...............T...........C........ | V |
| D33 | ...........................C....C... | V |
| D40 | .C.................T..T..T...C...... | U |
| African: | ||
| O2S | (Sequence was not obtained) | L |
| C66 | .....A...CTC....T....C.TG.T...C...T. | L1 |
| D47 | .....A...CTC....T....C.TG.T...C...T. | L1 |
| N18 | ...........CT...T......T..T........C | L2 |
| P1 | ..TC........T...TC.....T............ | L2 |
| N16 | ................T......T............ | L2 |
| C59 | ...C............T......T...........C | L3 |
| O17 | ................T......T...........C | L3 |
| C78 | .........C...T..T..............T.... | L3 |
| C27 | .....A..GC.C................C.C....C | L3 |
| Unknown: | ||
| D42 | ..................T................. | … |
| AL3 | .......G..........T................. | … |
Source: Anderson et al. (1981).
Of 24 non–Native American samples, 10 were identified as African haplotypes (table 4). Six samples (C66, D47, N18, P1, N16, and O2S) had the HpaI site present at np 3592. Three of those samples (P1, N16, and N18) had an A at np 16390 and belong to haplogroup L2, thus suggesting that other samples (C66 and D47) are part of haplogroup L1. Of the 87 samples sequenced, only P1, N16, and N18 had an A at np 16390. Sequence data for sample O2S were not obtained. Samples identified here as haplogroup L1 or haplogroup L2 shared HV1-sequence polymorphisms with many African samples belonging to cluster L1 and cluster L2, respectively, reported by Watson et al. (1997).
Four samples (C59, O17, C78, and C27) identified as African did not have the African haplogroup L restriction site or an A at np 16390; however, these samples were identical or similar to many published African sequences. Sample C59 had HV1 sequence identical to that of 10 African samples belonging to cluster L3 as reported by Watson et al. (1997). Of the cluster L3 sequences described by Watson et al. (1997), 2 were identical to, and 12 were 1 bp different from, sample O17. Sample C78 had HV1 sequence that differed by two nucleotides from five African samples identified as cluster L3, and it shared three of four variable positions (C at np 16172, T at np 16223, and T at np 16320) with 10 African samples (cluster L3) reported by Watson et al. (1997). Sample C27 shared five of seven variable positions with L3 sequences reported by Watson et al. (1997).
Samples identified as African did not share definitive HV1 sequence or restriction site polymorphisms with Native American or European samples. Of 10 haplotypes identified as African, 7 shared T's at np 16223 and np 16278; a single African sample had the T only at np 16223. These character states have been associated with haplogroup X in concurrence with a DdeI site loss at np 1715 (Forster et al. 1996; Torroni et al. 1996); however, all African, European, and Native American samples lacked this haplogroup X marker.
Given that only a select number of restriction site markers were analyzed, we were unable to identify the geographic region of origin for samples D42 and Al3. These samples did not have the Native American, European, or African restriction sites or specific HV1 markers screened for here. These two samples differed in sequence by one and two nucleotides, respectively, from HV1 of the European reference sequence (table 4; Anderson et al. 1981). Both samples shared a T at np 16234; however, this site has not been shown to characterize a specific region of origin.
Phylogenetic Analysis
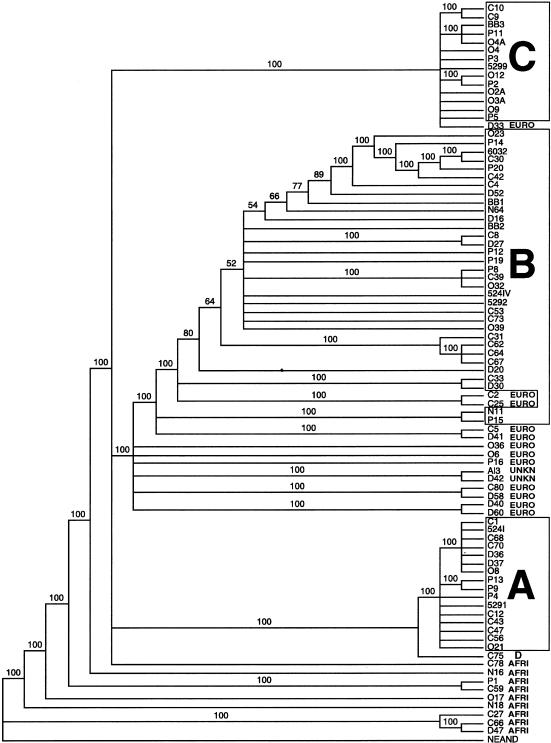
Phylogenetic analysis produced a 50% majority rule consensus tree generated from 2,000 maximum parsimony trees (fig. 2). Tree length was 146 steps, with a consistency index, excluding uninformative characters, of 0.47. Native American haplogroups were primarily monophyletic. Most European sequences formed a paraphyletic group at the base of haplogroup B. The single haplogroup D sample was the sister to haplogroup A. African sequences formed a basal, paraphyletic group. Native American haplogroup A was monophyletic. Two European haplotype lineages, C2 and C25, originated within the Native American haplogroup B clade. The positions of two others, C5 and D41, were unresolved in relationship to haplogroup B. Samples C2 and C25 had a C at np 16189, which is a definitive HV1 site for haplogroup B. Samples C5 and D41 both had the other haplogroup B definitive site, a C at np 16217. The position of European haplotype D33 was unresolved but was allied with Native American haplogroup C, because of the presence of one definitive HV1 marker, a C at np 16298. Other than these few exceptions, Native American clades were monophyletic and European lineages originated basal to haplogroup B. The phylogenetic position of the two unknown samples, D42 and Al3, was among the European samples. It is likely that these two samples are of European origin; however, further analysis is needed to confirm identity.
Figure 2.
Majority rule consensus tree of 2,000 maximum parsimony trees generated from control region sequence data with a heuristic search with the tree bisection reconnection branch swapping algorithm of PAUP, rooted by using Neandertal as outgroup. Numbers on branches indicate the percentage of 2,000 trees with a depicted clade. This analysis was calculated multiple times, with the same result. Tree length is 146 steps; the consistency index (excluding uninformative characters) is 0.47. Boxes indicate Native American haplogroups. D = Native American haplogroup D; EURO = European haplotypes; AFRI = African haplotypes; and UNKN = unknown haplotypes. Because of high homoplasy, consistent character state changes do not define most major clades, yet this shortest, rooted network provides resolution of clades that is concordant with previous studies that made use of various genetic systems and with results of the restriction site analysis presented in table 1.
Discussion
A low frequency of African haplotypes in northern Mexico might be expected because of possible rare contact with African Americans from the United States. The historical and present African American population of the west Texas–Mexican border region is small, as has been African American emigration to Mexico from the United States. The discovery of a proportion of African haplotypes roughly equivalent to the proportion of European haplotypes cannot be explained by recent admixture of African Americans from the United States. This is especially the case for the Ojinaga area, which presently is, and historically has been, largely isolated from U.S. African Americans. In the Ojinaga sample set, the frequency of African haplotypes was higher than that of European haplotypes (table 2). The findings of a basal phylogenetic position and paraphyly of African haplotypes were generally concordant with other studies (e.g., Cann et al. 1987; Vigilant et al. 1991; Nei and Roychoudhury 1993; Chen et al. 1995).
We found that the frequencies of European haplotypes were lower than published estimates of European admixture in Mestizo populations (cosmopolitan communities) found, throughout Mexico, by classical genetic systems (Lisker et al. 1996). Other estimates ranged from 22.0% in the city of Tlaxcala to 86.3% in Monterrey (Crawford et al. 1974; Cerda-Flores and Garza-Chapa 1989). European-admixture estimates in our study are likely lower because of the maternal inheritance of mtDNA, as historically more Spanish males than Spanish females colonized Mexico.
The Mexican east coast is known to have greater African admixture, as is evident today in the physical appearance and culture of the populations residing there. This has also been seen in blood group marker studies, with east coast Mestizo communities having African-admixture estimates ranging from 21.7% in Parafso to 40.5% in Tamiahua (Lisker et al. 1996). By contrast, the rest of Mexico exhibits lower African-admixture estimates, ranging from 0% in Monterrey to 18.1% in Cuanalan (Crawford et al. 1976; Cerda-Flores and Garza-Chapa 1989). In this context, the low frequency of African mtDNA haplotypes reported here is likely related to disproportionate emigration by male slaves.
Prior to the Spanish conquest, Mexico was inhabited largely by peoples who crossed the Bering Land Bridge from Asia at the end of the Pleistocene Epoch. Some historians have estimated that 4–5 million Native Americans inhabited Mexico before the Spanish arrived, and other estimates are as high as 25 million (Mörner 1967). The presence of a large proportion of Native American haplotypes in north-central Mexico would be expected, given that a large number of Native Americans inhabited Mexico for thousands of years. A drastic decline in the Native American population occurred following the Spanish settlement of the 1500s (Valdés 1987; Stern 1994). European diseases caused widespread epidemics, such as smallpox, measles, and typhus, which took a toll on the native population, especially during the late 1500s (Valdés 1987; Stern 1994). With the Spanish invasion, European genetic lineages increased in frequency, and thus the presence of European mtDNA haplotypes in Mexican populations should be expected.
With the drastic reduction of the Native American population, a large number of African slaves were brought to Mexico during the late 16th and early 17th centuries, to replace native labor (Beltrán 1944; Palmer 1976; Valdés 1987; Stern 1994). Importation of African slaves into Mexico by the Spanish during the 16th–18th centuries is documented historically yet was often denied and largely forgotten (Beltrán 1944; Muhammad 1995). Records from this period are sketchy at best, since many slaves were brought into the country illegally and since African ancestry was often denied by mulattos in census data (Beltrán 1944; Tjarks 1978; Muhammad 1995). It has been estimated that, in Mexico during the late 16th and early 17th centuries, the slave population was both greater than that in any other country in the Americas and larger than the European population of Mexico (Beltrán 1944; Muhammad 1995). Denial of African origin improved social status and thereby increased opportunities and privileges (Beltrán 1945, 1970; Muhammad 1995). The denial and dilution of African ancestry during the past 500 years has deprived the contemporary Mexican peoples of knowledge of a significant portion of their ethnic heritage.
Today, the number of Mexicans with African heritage is not known; however, some suggest that as much as 75% of the modern Mexican population has some African ancestry (Muhammad 1995). Estimation of African contribution to the genetic heritage of Mexico may now be accomplished through identification of population of origin by use of diagnostic, discrete character, molecular genetic markers. Such analyses will ultimately provide a more accurate understanding of Mexican history and culture.
On the basis of both our results and the historical evidence (Beltrán 1945; Palmer 1976; Meyer and Sherman 1991), it appears that a greater proportion of African slaves and European colonizers in Mexico were males rather than females. When paternally inherited genetic markers in the nonrecombining portion of the Y chromosome are studied in the cosmopolitan Mexican peoples, we predict that, compared with maternally inherited markers, a significantly larger proportion will be found to be of African and European origin. When both maternal and paternal markers are studied, and when other regions of Mexico are sampled, a more complete picture of the African contribution to Mexico will be revealed.
Acknowledgments
This work was supported, in part, by a research-enhancement grant and other research funds from Sul Ross State University. We thank Dr. Lou Densmore for graciously allowing us use of his laboratory at Texas Tech University before ours was fully functional. We thank Dr. Clifton Pearce and Dr. Fernando Gallegos for their invaluable assistance in sample collection and Dr. Eppie Rael of the University of Texas at El Paso for generously providing the Juárez DNA samples. We thank Dr. Marilyn Brady for helpful discussion. We also thank all those who donated samples for DNA extraction. We thank Dr. A. Michael Powell and the late Dr. Jim V. Richerson for support and encouragement. We thank Lee Greer for contributions made in the early stages of this study, and we thank two anonymous reviewers for comments that improved this article.
References
- Anderson S, Bankier AT, Barriel BG, De Bruin MHL, Coulson AR, Drouin J, Eperon IC, et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed]
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH, et al (1992) Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient Mongoloid migrations. Genetics 130:139–152 [DOI] [PMC free article] [PubMed]
- Batista O, Kolman CJ, Bermingham E (1995) Mitochondrial diversity in the Kuna Amerinds of Panama. Hum Mol Genet 4:921–929 [DOI] [PubMed]
- Beltrán AG (1944) The slave trade in Mexico. Hispanic Am Hist Rev 24:412–431 [Google Scholar]
- ——— (1945) Races in 17th century Mexico. Phylon 6:212–218 [Google Scholar]
- ——— (1970) The integration of the Negro into the national society of Mexico. In: Mörner M (ed) Race and class in Latin America. Columbia University Press, New York, pp 11–27 [Google Scholar]
- Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 [DOI] [PubMed]
- Cerda-Flores RM, Garza-Chapa R (1989) Variation in the gene frequencies of three generations of humans from Monterrey, Nuevo León, Mexico. Hum Biol 61:249–261 [PubMed]
- Chen YS, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC (1995) Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet 57:133–149 [PMC free article] [PubMed]
- Crawford MH, Leyshon WC, Brown K, Lees F, Taylor L (1974) Human biology in Mexico. II. A comparison of blood group, serum and red cell enzyme frequencies, and genetic distances of the Indian populations of Mexico. Am J Phys Anthropol 41:251–268 [DOI] [PubMed]
- Crawford MH, Lisker R, Pérez-Briceño R (1976) Genetic microdifferentiation of two transplanted Tlaxcaltecan populations. In: Crawford M (ed) The Tlaxcaltecans: prehistory, demography, morphology and genetics. Publications in anthropology 7. University of Kansas, Lawrence, pp 169–175 [Google Scholar]
- Curtin PD (1969) The Atlantic slave trade. University of Wisconsin Press, Milwaukee [Google Scholar]
- Forster P, Harding R, Torroni A (1996) Origin and evolution of Native American mtDNA: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed]
- Higuchi R, von Berolding C, Sensabaugh GF, Erlich H (1988) DNA typing from single hairs. Nature 332:543–546 [DOI] [PubMed]
- Hillis DM, Davis SK (1986) Evolution of ribosomal DNA: fifty million years of recorded history in the frog genus Rana. Evolution 40:1275–1288 [DOI] [PubMed] [Google Scholar]
- Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, Tajima K (1993) Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol 10:23–47 [DOI] [PubMed]
- Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S (1997) Neandertal DNA sequences and the origin of modern humans. Cell 90:19–30 [DOI] [PubMed]
- Lisker R, Babinsky V (1986) Admixture estimates in nine Mexican Indian groups and five east coast localities. Rev Invest Clin 38:14–149 [PubMed] [Google Scholar]
- Lisker R, Pérez-Briceño R, Granados J, Babinsky V (1988) Gene frequencies and admixture estimates in the state of Puebla, Mexico. Am J Phys Anthropol 76:331–335 [DOI] [PubMed]
- Lisker R, Pérez-Briceño R, Granados J, Babinsky V, De Rubens J, Armendares S, Buentello L (1986) Gene frequencies and admixture estimates in a Mexico City population. Am J Phys Anthropol 71:203–207 [DOI] [PubMed]
- Lisker R, Ramírez E, Babinsky V (1996) Genetic structure of autochthonous populations of Meso-America: Mexico. Hum Biol 68:395–404 [PubMed]
- Lisker R, Rameriz E, Gonzalez-Villapando C, Stern MP (1995) Racial admixture in a Mestizo population from Mexico City. Am J Hum Biol 7:213–216 [DOI] [PubMed] [Google Scholar]
- Lisker R, Ramirez E, Penaloza R, Salamanca F (1994) Red-cell acid phosphatase types and GC polymorphisms in Merida, Oaxaca, Leon, and Saltillo, Mexico. Hum Biol 66:1103–1109 [PubMed]
- Lisker R, Ramírez E, Pérez-Briceño R, Granados J, Babinsky V (1990) Gene frequencies and admixture estimates in four Mexican urban centers. Hum Biol 62:791–801 [PubMed]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, et al (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed]
- Meyer MC, Sherman WL (1991) The course of Mexican history, 4th ed. Oxford University Press, New York [Google Scholar]
- Mörner M (1967) Race mixture in the history of Latin America. Little & Brown, Boston [Google Scholar]
- Muhammad JS (1995) Mexico and Central America. In: Minority Rights Group (ed) No longer invisible: Afro-Latin Americans today. Minority Rights Publications, London, pp 163–180 [Google Scholar]
- Nei M, Roychoudhury AF (1993) Evolutionary relationships of human populations on a global scale. Mol Biol Evol 10:927–943 [DOI] [PubMed]
- Palmer CA (ed) (1976) Slaves of the white god. Harvard University Press, Cambridge, MA [Google Scholar]
- Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, et al (1996) Paleolithic and Neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59:185–203 [PMC free article] [PubMed]
- Schanfield MS, Fudenberg HH, Crawford MH, Turner KR (1978) The distribution of immunoglobulin allotypes in two Tlaxcaltecan populations. Ann Hum Biol 5:577–590 [DOI] [PubMed]
- Schurr TG, Ballinger SW, Gan Y, Hodge JA, Merriwether DA, Lawrence DN, Knowler WC, et al (1990) Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet 46:613–623 [PMC free article] [PubMed]
- Stern P (1994) Gente de color quebrado: Africans and Afromestizos in colonial Mexico. Colonial Latin Am Hist Rev 3:185–204 [Google Scholar]
- Swofford DL (1998) PAUP: phylogenetic analysis using parsimony, release 4.0b1. Sinauer Associates, Sunderland, MA [Google Scholar]
- Tiburcio V, Romero A, De Garay AL (1978) Gene frequencies and racial intermixture in a Mestizo population from Mexico City. Ann Hum Biol 5:131–138 [DOI] [PubMed]
- Tjarks AV (1978) Demographic, ethnic and occupational structure of New Mexico, 1790. Americas 35:45–88 [Google Scholar]
- Torroni A, Bandelt HJ, DíUrbano L, Lahermo P, Moral P, Sellitto D, Rengo C, et al (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northwestern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed]
- Torroni A, Chen YS, Semino O, Santachiara-Benerecetti AS, Scott CR, Lott MT, Winter M, et al (1994a) Mitochondrial DNA and Y-chromosome polymorphisms in four Native American populations from southern Mexico. Am J Hum Genet 54:303–318 [PMC free article] [PubMed]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, et al (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed]
- Torroni A, Lott MT, Cabell MF, Chen YS, Lavergne L, Wallace DC (1994b) mtDNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet 55:760–776 [PMC free article] [PubMed]
- Torroni A, Neel JV, Barrantes R, Schurr TG, Wallace DC (1994c) Mitochondrial DNA “clock” for the Amerinds and its implications for timing their entry into North America. Proc Natl Acad Sci USA 91:1158–1162 [DOI] [PMC free article] [PubMed]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith SG, et al (1993) Asian affinities and continental radiation of the four founding Native American mitochondrial DNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed]
- Valdés DN (1987) The decline of slavery in Mexico. Americas 44:167–194 [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed]
- Ward RH, Frazier BL, Dew-Jager K, Pääbo S (1991) Extensive mitochondrial diversity within a single Amerindian tribe. Proc Nat Acad Sci USA 88:8720–8724 [DOI] [PMC free article] [PubMed]
- Watson E, Forster P, Richards M, Bandelt HJ (1997) Mitochondrial footprints of human expansion in Africa. Am J Hum Genet 61:691–704 [DOI] [PMC free article] [PubMed]