Abstract
Although several genetic forms of rare or syndromic hypertriglyceridemia have been reported, little is known about the specific chromosomal regions across the genome harboring susceptibility genes for common forms of hypertriglyceridemia. Therefore, we conducted a genomewide scan for susceptibility genes influencing plasma triglyceride (TG) levels in a Mexican American population. We used both phenotypic and genotypic data from 418 individuals distributed across 27 low-income, extended Mexican American families. For the analyses, TG values were log transformed (ln TG). We used a variance-components technique to conduct multipoint linkage analyses for localizing susceptibility genes that determine variation in TG levels. We used an ∼10–15–cM map, which was made on the basis of information from 295 microsatellite markers. After accounting for the effects of sex and sex-specific age terms, we found significant evidence for linkage (LOD = 3.88) of ln TG levels to a genetic location between the markers GABRB3 and D15S165 on chromosome 15q. This putative locus explains 39.7±7% (P=.000012) of total phenotypic variation in ln TG levels. Suggestive evidence was found for linkage of ln TG levels to two different locations on chromosome 7, which are ∼85 cM apart from each other. Also, there is some evidence for linkage of high-density lipoprotein cholesterol concentrations to a genetic location near one of the regions on chromosome 7. In conclusion, we found strong evidence for linkage of ln TG levels to a genetic location on chromosome 15q in a Mexican American population, which is prone to disease conditions such as type 2 diabetes and the insulin-resistance syndrome that are associated with hypertriglyceridemia. This putative locus appears to have a major influence on ln TG variation.
Introduction
The role of hypertriglyceridemia as an independent risk factor for coronary heart disease (CHD) has been controversial (Gotto 1998). Recently, however, increasingly strong evidence for its role as a CHD risk factor has emerged (Austin et al. 1998; Havel 1998; Jeppesen et al. 1998). Although fasting plasma triglyceride (TG) levels >200 mg/dl have been considered to increase risk, recent evidence suggests that an even lower threshold (>100 mg/dl) could be deleterious in patients with established coronary artery disease (Miller et al. 1998). Hypertriglyceridemia is commonly found in individuals with type 2 diabetes, and it is an element of the so-called metabolic syndrome (Ginsberg 1996; Howard 1996; Garg 1998; Grundy 1998). People with type 2 diabetes and those who are prediabetic have a greater risk of CHD than do individuals who are unaffected (Haffner et al. 1990; Hsueh and Law 1998).
Since the molecular basis of CHD is incompletely understood, there have been continued efforts to disentangle the genetic architecture of CHD risk factors, including fasting plasma TG levels. Numerous genetic epidemiological studies have demonstrated that the variation in TG levels is affected by both genetic and environmental factors (Iselius 1988; Rice et al. 1991; Mitchell et al. 1996). A few studies have examined major locus effects on TG concentrations by making use of segregation analysis, but the results have been inconclusive (Singh et al. 1988; MacCluer 1989). Several forms of hypertriglyceridemia, such as chylomicronemia, familial hypertriglyceridemia, and familial combined hyperlipidemia, are attributable to genetic abnormalities (Grundy 1998). Recently, a susceptibility locus for familial combined hyperlipidemia has been localized to a genetic location on chromosome 1q21-q23 (Pajukanta et al. 1998). Also, the gene for Tangier disease, which is associated with increased levels of TGs, has been assigned to chromosome 9q31 (Rust et al. 1998). However, little is known about the specific major genetic determinants of common forms of hypertriglyceridemia.
In this study, we scanned the genome for susceptibility loci influencing the plasma TG levels by making use of pedigree data from a low-income Mexican American population. In Mexican Americans, the prevalence of type 2 diabetes is three times higher than in the general U.S. population (Stern and Haffner 1990). Furthermore, Mexican Americans without diabetes have been shown to be more insulin resistant than members of the non-Hispanic white population (Haffner et al. 1986, 1990). Our genomewide scan for loci affecting plasma TG levels used a multipoint variance-components linkage approach and an ∼10–15–cM map.
Material and Methods
The San Antonio Family Diabetes Study (SAFADS) aims to identify susceptibility genes for type 2 diabetes and related phenotypes by use of data from extended pedigrees (Stern et al. 1996; Duggirala et al. 1996, 1999). A total of 579 individuals (140 with diabetes) in 32 families ascertained on a type 2 diabetic proband were examined, involving first-, second-, and third-degree relatives aged 18–98 years. Metabolic, anthropometric, demographic, and medical history information were obtained on all the examined individuals. A subset of 440 individuals (116 with diabetes) from the 27 largest pedigrees were selected for genotyping. All procedures were approved by the Institutional Review Board of the University of Texas Health Science Center at San Antonio, and all subjects gave informed consent.
Blood samples were obtained after a 12-h fast. The blood was assessed for various metabolic traits, including glucose, TG, and high-density lipoprotein cholesterol (HDL-C) concentrations, by methods described elsewhere (Stern et al. 1984). Duplicate measures for a given trait were used to assess the extent of measurement error. The technical error of measurement for HDL-C was 1.1% of the mean; for TG, it was 1.2%. Of the 440 SAFADS subjects, TG values were available for 422 individuals. TG values >800 mg/dl were excluded from the analysis because they were distinct outliers (i.e., only four individuals). For the present analysis, which is based on phenotypic information from 418 individuals, the TG values were log transformed (ln TG). These 418 individuals generated 3,379 relative pairs that were distributed across 17 categories of relative pairs (e.g., sibs, first cousins, second cousins, etc.), as shown in table 1. Glucose levels were also measured 2 h after a standardized oral glucose load. Diabetes was diagnosed according to criteria of the World Health Organization (1985).
Table 1.
Distribution of Relative Pairs by Category in a Subset of 27 SAFADS Families Containing 418 Individuals
| Relative Pair | Relationship Coefficienta | No. ofPairs |
| Parent-offspring | .5000 | 396 |
| Sibs | .5000 | 491 |
| Grandparent-grandchild | .2500 | 100 |
| Avuncular | .2500 | 633 |
| Half-sibs | .2500 | 65 |
| Great-grandparent–grandchild | .1250 | 12 |
| Grand avuncular | .1250 | 124 |
| Half-avuncular | .1250 | 66 |
| First cousins | .1250 | 656 |
| Great-grandavuncular | .0625 | 3 |
| Half-grandavuncular | .0625 | 11 |
| First cousins, once removed | .0625 | 479 |
| Half–first cousins | .0625 | 75 |
| First cousins, twice removed | .0312 | 9 |
| Half–first cousins, once removed | .0312 | 32 |
| Second cousins | .0312 | 212 |
| Second cousins, once removed | .0156 | 15 |
| Total | 3,379 |
The relationship coefficient is 2 × coefficient of kinship of two individuals.
Genotyping
A set of 295 markers was used for the multipoint linkage analysis, which provided coverage at intervals of ∼10–15 cM on chromosomes 1–22 (Duggirala et al. 1999). Primers were purchased from Research Genetics. Further details concerning oligonucleotide primer sequences and polymorphisms can be obtained from the Genome Data Base at Johns Hopkins University (Fasman et al. 1996). PCR conditions were optimized by testing a range of annealing temperatures with varying concentrations of MgCl2 so that the PCR amplification produced a specific product that could be visualized by ethidium bromide staining (0.5 μg/ml).
Genotypes were collected primarily by PCR assays with radiolabeled oligonucleotide primers. The antisense primer was 5′-radiolabeled in a standard polynucleotide kinase reaction by 3,000 Ci/mmol [γ32P]-adenosine triphosphate (ATP; from NEN) at a molar ratio of 18 [γ32P]-ATP:1 primer, as described elsewhere (O'Connell et al. 1994; Duggirala et al. 1996). Thirty cycles of PCR (denaturing at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min) were done in a 15-μl assay containing 50 ng of DNA template, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1–4 mM MgCl2, 0.1 M spermidine (Sigma), 0.1 μM of each PCR primer, 0.1 μM dNTPs (Gibco BRL), and 0.5 U of Taq polymerase (PE Biosystems). Amplified DNA was diluted 1:1 with stop solution (97% formamide, 1% EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol) and denatured at 85°C for 2 min. Three microliters of denatured DNA from each sample were loaded onto a 7% denaturing polyacrylamide gel (19:1 acrylamide/bis-acrylamide) containing 32% formamide and 34% urea and fractionated by gel electrophoresis for 2.5 h at 60 W. Gels were transferred to filter paper (Whatman 3MM; from W. R. Balston) covered with plastic wrap, equilibrated with 20% methanol–20% acetic acid solution, and dried. Dried gels were exposed to X-ray film (Fuji Photo Film). As reported elsewhere (Duggirala et al. 1999), data for some of the markers were collected by means of fluorescent-labeled primers, purchased from Research Genetics. These were PCR amplified, as described above, and were loaded onto an Applied Biosystems model 373 sequencer, and the data were analyzed by Applied Biosystems GENOTYPER software.
The genotypic data used for this study were analyzed for discrepancies (i.e., violations of Mendelian inheritance) by the program INFER (Dyke 1996). Before conducting linkage analyses, the SAFADS pedigree structures were verified with information from ∼50 polymorphic loci, including red blood cell antigens. The discrepancies were checked for mistyping in the laboratory, and a few instances that could not be resolved were presumed to be due to factors such as sample mix-up, nonparentage, or mutations. These unresolved cases were treated as missing data. Pedigree information was rechecked with family members, and blood samples were redrawn from relevant individuals whenever possible. Checking and rechecking the pedigree resulted in eight discrete pedigree revisions.
The genotypic information for SAFADS participants was routinely verified, and discrepancies were checked in the laboratory for mistyping. Markers for discrepant individuals were either corrected or blanked out prior to analysis. The average percentage of genotypes blanked (i.e., the number of genotypes blanked divided by the total number of genotypes) is ∼.06%. Thus far, >400 markers have been typed, and all markers were used for two-point linkage analysis. Since our multipoint linkage approach yields optimum results when similar numbers of individuals are genotyped at all loci, markers with <80% of the sample genotyped were not considered for multipoint analysis unless their absence would result in a map gap of 20 cM (Duggirala et al. 1999). This arbitrary requirement excluded ∼25% of the markers. Thus, the multipoint analyses are made on the basis of a total of 295 markers. In addition, ∼4% of the total SAFADS marker data set could not be used for the multipoint analyses because of genotyping problems (i.e., markers failed to map to their expected positions or led to map expansion).
Statistical Genetic Analysis
We used a variance-components approach to test for linkage of a genetic location with ln TG levels by means of maximum-likelihood methods (Amos 1994; Blangero and Almasy 1997; Almasy and Blangero 1998). The variance-components method uses information from all possible biological relationships simultaneously in an attempt to disentangle the genetic architecture of a quantitative trait. This method specifies the expected genetic covariances between relatives as a function of their identity-by-descent (IBD) relationships at a marker locus (which is hypothesized to be linked to a locus influencing the quantitative trait [QTL]). It allows for locus-specific effects (h2q is heritability attributed to the QTL), residual additive genetic effects (h2 is heritability attributed to the residual genetic effects), covariate effects (e.g., age and sex), and individual-specific random environmental factors [e2=1–(h2q+h2)].
In this study, in addition to the variance components, mean and SD of the phenotype and covariate effects (i.e., sex-specific age, and age2 and sex terms) were simultaneously estimated by maximum-likelihood techniques. Hypothesis testing was performed by the likelihood ratio tests. The hypothesis of no linkage (i.e., additive genetic variance due to the QTL=0) was tested by comparing the likelihood of this restricted model with that of a model in which the additive genetic variance due to the QTL is estimated. Twice the difference in ln likelihoods of these two models yields a test statistic that is asymptotically distributed as a ½:½ mixture of a χ21 and a point mass at zero (Self and Liang 1987). LOD scores were obtained by converting the ln likelihood values into values of log10. After obtaining locus-specific IBD information for pairs of relatives by the computer program SOLAR (Almasy and Blangero 1998), the multipoint mapping strategy proposed by Fulker et al. (1995) was extended and modified (Almasy and Blangero 1998) to perform multipoint variance-components linkage analysis. These procedures were implemented in the SOLAR program.
Results
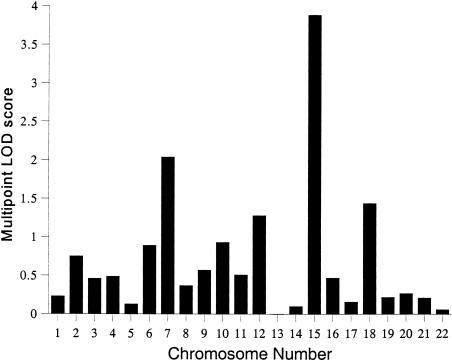
The characteristics of the subjects used for this study are reported in table 2. All genetic analyses included sex and sex-specific age and age2 terms as covariates of ln TG values. After accounting for the significant covariate effects, the overall polygenic heritability (h2) for ln TG values was estimated to be 33.6±9%, with high statistical significance (P<.0001). The covariates explained ∼14% of total phenotypic variation in ln TG. Following this, the multipoint linkage analysis was conducted, and the results of our genome scan (i.e., peak LOD score by chromosome) are summarized in figure 1. All LOD scores reported below relate to multipoint analysis, and only multipoint LOD scores near 2 or above are discussed.
Table 2.
Characteristics of SAFADS Subjects Distributed across 27 Families Included in the Genotyping Set
| Variable | Mean ± SD or % |
| Age at examination (years) | 43.3 ± 17.3 |
| Men | 41.2% |
| Women | 58.8% |
| Diabetics | 26.4% |
| Body mass index (kg/m2) | 30.0 ± 6.7a |
| TGs (mg/dl) | 173.6 ± 109.2b |
| ln TGs | 5.0 ± .6c |
| HDL-C (mg/dl) | 38.0 ± 10.2d |
Subjects with diabetes, 31.9±6.6; subjects without diabetes, 29.4±6.6.
Subjects with diabetes, 212.4±116.4; subjects without diabetes, 160.3±103.5.
Subjects with diabetes, 5.2±.5; subjects without diabetes, 4.9±.5.
Subjects with diabetes, 37.9±11.0; subjects without diabetes, 38.0±9.9.
Figure 1.
Summary of the multipoint linkage analyses of ln TG: peak multipoint LOD score by chromosome.
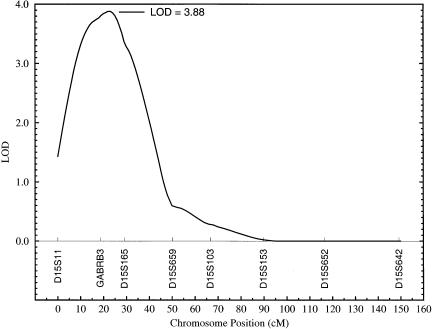
The highest LOD score that we observed was 3.88, which is highly statistically significant and corresponds to a susceptibility locus for ln TG on chromosome 15q between the markers GABRB3 (gamma-aminobutyric acid A receptor, beta-3) and D15S165 (fig. 2). The multipoint LOD scores obtained for ln TG were plotted against map positions on chromosome 15 (fig. 2). As can be seen from this figure, the markers GABRB3 and D15S165 cover a region of ∼10 cM, which appears to harbor a major susceptibility locus for hypertriglyceridemia. The heritability attributable to this putative locus (h2q) was estimated to be 39.7±7% (P=.000012). Although the QTL heritability (h2q) is greater than the overall polygenic heritability (h2), it is within the estimated SE around the overall polygenic heritability (i.e., h2=33.6±9%). Also, since the polygenic model was rejected in favor of the model including the QTL effect, the latter gives a more unbiased estimate of the heritability.
Figure 2.
Linkage of ln TG to a quantitative trait locus between markers GABRB3 and D15S165 on chromosome 15.
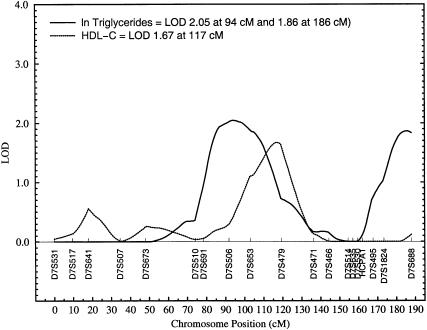
Two regions on chromosome 7 are suggestively linked (i.e., a multipoint LOD score near 2 or above) to ln TG levels: a genetic location between markers D7S506 and D7S653 (LOD 2.05) and a location between markers D7S1824 and D7S688 (LOD 1.86). This is shown in figure 3. The former peak is relatively broad and covers a region of ∼11 cM between the sequential markers D7S506 and D7S653. The two suggestive susceptibility loci on chromosome 7 are ∼85 cM apart from each other. No other multipoint LOD scores near 2 or above were observed in our genomewide scan. Given the known correlation between TGs and HDL-C, only one of the three chromosomal regions of interest for ln TGs in this study appears to be also linked with HDL-C levels—namely, the region near marker D7S479 (i.e., between markers D7S653 and D7S479) on chromosome 7 (fig. 3). Although the LOD curves peak at different locations for these phenotypes (ln TGs at 94 cM and HDL-C at 117 cM), the trait-specific linkage curves exhibit appreciable overlapping. The region on chromosome 15q, however, failed to show any evidence for linkage to HDL-C (LOD score = 0).
Figure 3.
Plot of LOD scores obtained for ln TG and HDL-C concentrations against map positions on chromosome 7.
We conducted two independent analyses after our original findings were concluded to verify whether our results were sensitive to ascertainment bias or to the diabetic status of the subjects. Since the SAFADS families were ascertained on type 2 diabetic probands, we reran the analysis, as a conservative approach, by correcting for the ascertainment by conditioning on likelihood of observing the ln TG of the proband with diabetes. We also reran the analysis after accounting for the effect of type 2 diabetes on ln TG values. However, the differences between the three analyses were trivial.
Discussion
Hypertriglyceridemia is an important component of the risks associated with CHD, type 2 diabetes, and the metabolic syndrome. Increased concentrations of TG are associated with several types of dyslipidemias. Fasting plasma TG levels are surrogates for TG-rich lipoproteins (e.g., chylomicrons and very-low-density lipoproteins; Grundy 1998). Hypertriglyceridemia often correlates with hyperinsulinemia in the general population (Steiner and Lewis 1996) and is associated with increased concentrations of small, dense, low-density lipoproteins (Mykkänen et al. 1997).
A number of studies have examined population associations between various candidate genes related to lipoprotein metabolism and plasma TG levels. For example, studies have shown associations between variation in or near the lipoprotein lipase (LPL) gene (chromosome 8p22) and plasma TG levels (Gerdes et al. 1995; Humphries et al. 1998). Also, TG levels have been shown to be correlated with variants in the promotor region of the LPL gene (Ehrenborg et al. 1997; Wittrup et al. 1997; Talmud et al. 1998). Recently, a meta-analysis was conducted to assess the effect of various mutations at the LPL locus (i.e., Gly188Glu, Asp9Asn, Asn291Ser, and Ser447Ter substitutions) in the heterozygous state on lipid metabolism and risk of ischemic heart disease in whites (Wittrup et al. 1999). Individuals who carry the Gly188Glu, Asp9Asn, and Asn291Ser substitutions, compared with individuals who do not carry these substitutions, were shown to have an atherogenic lipoprotein profile, whereas individuals who carry the Ser447Ter substitution were found to have a protective lipoprotein profile. There is evidence for an association between TG levels and variation in the APO CIII (apolipoprotein CIII) gene (chromosome 11q23) or in its promotor (Zeng et al. 1995; Surguchov et al. 1996; Hegele et al. 1997). Our failure to find linkage in genetic regions containing loci such as the LPL and the APO CIII in Mexican Americans may be due to the possible heterogeneity of common forms of hypertriglyceridemia across the populations.
Population association studies have limitations attributable to problems such as misclassification and population stratification, and they are more likely to generate false-positive results (Khoury et al. 1993). Also, it is usually not possible to evaluate the magnitude of the genetic effects exerted by such loci on the basis of association analysis; linkage approaches, on the other hand, that make use of data gathered from members of families can be helpful in determining the magnitude of genetic effects (Cox and Bell 1989). Recently, by making use of linkage analysis, susceptibility genes for familial combined hyperlipidemia (Pajukanta et al. 1998) and Tangier disease (Rust et al. 1998), which are associated with elevated TG levels, have been localized. However, our failure to find evidence for linkage in regions corresponding to familial combined hyperlipidemia and Tangier disease may be related to their distinct etiologies, which could play a minor role in determining variation in TG levels in Mexican Americans.
Knowledge of the major determinants of common forms of hypertriglyceridemia in humans is limited. We conducted a genomewide search for susceptibility genes influencing ln TG levels in a Mexican American population by using pedigree data and a variance-components linkage approach; we found significant evidence for a major susceptibility locus influencing TG concentrations on chromosome 15q between markers GABRB3 (15q11.2) and D15S165 (15q12-q13.1). Several criteria have been proposed to verify a claim of significant linkage at the level of a genomewide scan (Lander and Kruglyak 1995; Elston 1998; Morton 1998). For example, in reference to the allele-sharing methods, such as the one used in this study, Lander and Kruglyak (1995), by making use of simulations, proposed the term “significant linkage” to refer to LOD scores for various types of relative pairs in the range of ∼3.3–3.8. Following this suggestion, the observed multipoint LOD of 3.88 at this location on chromosome 15q corresponds to a significant linkage.
Since a multivariate normal distribution is assumed in our linkage approach, there may be some concerns about violations of this assumption (Elston 1998; Allison et al. 1999). Although the variance-component approach has been shown to be robust to such violations (Beaty et al. 1985; Amos 1994), recently, sib-pair–based data simulations have shown that some types of nonnormality (e.g., leptokurtosis) can lead to inflated type I error rates and that the degree of such an inflation rate seems to be directly related to the residual sib correlation (Allison et al. 1999). In this connection, the significance of the present finding is less likely to be spuriously inflated for the following reasons. After accounting for the effects of covariates on ln TG levels, the violation of nonnormality of the residuals used in this study appears, on the basis of the coefficients for skewness (.37) and kurtosis (.02), to be minor. Also, the extent of proportion of variance explained by the major QTL in our study implies the low residual sibling correlation.
There are no obvious candidate genes for hypertriglyceridemia in this region of chromosome 15q, although it is worth noting that the Prader-Willi syndrome (MIM 176270) and the Angelman syndrome (MIM 105830) map to the chromosomal region 15q11-13, which is close to the area of interest in our study. The hepatic lipase (HL) gene (MIM 151670), which has a role in TG metabolism, has been localized to chromosomal region 15q21-23 (Sparkes et al. 1987). The HL gene is located ∼25–30 cM centromeric to the genetic location (i.e., peak) of interest in the present study. However, given our recent finding, in another set of Mexican American families, of significant linkage between a genetic location near the HL gene and a component of HDL-C—namely, unesterified HDL2a-C (Almasy et al. 1999)—we are planning to screen the HL gene to assess its relation to the present finding.
Two different locations on chromosome 7 are suggestively linked to susceptibility loci influencing ln TG levels. The first susceptibility locus (LOD 2.05) is between markers D7S506 and D7S653. A candidate gene close to marker D7S653 is the collagen and thrombospondin receptor CD36 (MIM 173510), which maps to 7q11.2 (Fernández-Ruiz et al. 1993). Recently, Aitman et al. (1999) identified CD36 as an insulin-resistance gene that underlies defective fatty acid metabolism and hypertriglyceridemia in the spontaneously hypertensive rat. These authors suggested a possible role for CD36 in the etiology of the insulin-resistance syndrome in humans. Earlier, in a sibship-based analysis, we reported evidence for linkage of insulin precursors (intact and 32,33 split proinsulin) to a genetic location near marker D7S479 on 7q (Duggirala et al. 1996), which is ∼16 cM telomeric to the region of interest (i.e., the region between markers D7S506 and D7S653) in the present study. As discussed earlier, there is some evidence for linkage of HDL-C levels to a genetic location near marker D7D479, and the LOD curves for both ln TG and HDL-C exhibit appreciable overlap, suggesting the possibility that a locus is influencing both phenotypes. Other genes of interest in this region of 7q are paraoxonase genes and plasminogen activator inhibitor 1 (Klinger et al. 1987; Mochizuki et al. 1998; Sanghera et al. 1998). Recently, we have found strong evidence for linkage (LOD 3.6) of HDL-C levels to a region on chromosome 9p near markers D9S925 and D9S1121 (Arya et al. 1999), although this region failed to show evidence for linkage to ln TG. These findings are consistent with those of an earlier study involving the Mexican American population—that TG and HDL-C concentrations are influenced by both shared genes and that each of these phenotypes appears to be influenced independently by unshared genes as well (Mahaney et al. 1995).
The second susceptibility locus on chromosome 7q for ln TG is near marker D7S688. In a previous sibship-based analysis, we found evidence for a susceptibility locus for various obesity-related traits, which is linked to a genetic location near marker D7S495 (i.e., between markers D7S495 and D7S688) on chromosome 7q (Duggirala et al. 1996). It is possible that our earlier and present results may be related to the same susceptibility locus. Given the known association between pancreatitis and TG levels, it is interesting to note that the gene for hereditary pancreatitis (MIM 167800) maps to chromosome 7q35 (Whitcomb et al. 1996; Gorry et al. 1997). The chromosomal region between markers D7S495 and D7S688 has been shown to encompass the hereditary pancreatitis locus (Pandya et al. 1996). Our second susceptibility locus on chromosome 7 lies in the same region, and it is 2 cM centromeric to marker D7S688.
In conclusion, on the basis of a genomewide scan, we found evidence for a major susceptibility locus on chromosome 15q influencing ln TG levels in a Mexican American population. This putative locus appears to have substantial influence on the phenotypic variation in ln TG levels. Also, two other susceptibility loci on chromosome 7 appear to have minor influences on variation in ln TG levels. One of these two loci on chromosome 7 may have a common influence on both ln TG and HDL-C. These findings may be unique to the Mexican Americans, a population prone to diseases such as obesity, insulin-resistance syndrome, and type 2 diabetes. However, as noted earlier, the findings appear not to be influenced by the diabetic condition. Given the possibility of heterogeneity across populations, generalization of our observations would require evidence for influence of the same loci on variation in TG levels in other populations. For precise localization of these susceptibility loci, especially the one on chromosome 15q, we plan to conduct fine-structure mapping with single nucleotide polymorphisms and linkage disequilibrium mapping techniques.
Acknowledgments
We wish to thank Dr. Mary Pat Moyer and Ms. Florence Wall for establishing some of the lymphoblastoid cell lines. We also wish to thank Rajeswari Cheruvu, Edgardo Benavides, Stefenie Fleming, Michelle Zavala, and Bonnie Reus for technical assistance. We warmly thank members of the SAFADS families for their support and participation. This research was supported by grants from the National Institute of Diabetes, Digestive and Kidney Diseases (RO1 DK42273, RO1 DK47482, and RO1 DK53889), and by grants GM18897 and MH59490 from the National Institutes of Health.
Electronic-Database Information
Accession numbers and URL for data in this article are as follows:
References
- Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, et al (1999) Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet 21:76–83 [DOI] [PubMed]
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci-mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed]
- Almasy L, Hixson JE, Rainwater DL, Cole S, Williams JT, Mahaney MC, VandeBerg JL, et al (1999) Human pedigree-based quantitative-trait-locus mapping: localization of two genes influencing HDL-cholesterol metabolism. Am J Hum Genet 64:1686–1693 [DOI] [PMC free article] [PubMed]
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed]
- Arya R, Duggirala R, Almasy L, Stern MP, O'Connell P, Blangero J (1999) Strong evidence for linkage of high-density lipoprotein cholesterol (HDL-C) concentrations to a genetic location on chromosome 9p in Mexican Americans. Am J Hum Genet Suppl 65:A2 [Google Scholar]
- Austin MA, Hokanson JE, Edwards KL (1998) Hypertriglyceridemia as a cardiovascular risk factor. Am J Cardiol 81(4A):7B–12B [DOI] [PubMed]
- Beaty TH, Self SG, Liang KY, Connolly MA, Chase GA, Kwiterovich PO (1985) Use of robust variance components models to analyze triglyceride data in families. Ann Hum Genet 49:315–328 [DOI] [PubMed]
- Blangero J, Almasy L (1997) Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol 14:959–964 [DOI] [PubMed]
- Cox NJ, Bell GI (1989) Disease associations: chance, artifact, or susceptibility genes? Diabetes 38:947–950 [DOI] [PubMed]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, P. O'Connell, et al (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed]
- Duggirala R, Stern MP, Mitchell BD, Reinhart LJ, Shipman PA, Uresandi OC, Chung WK, et al (1996) Quantitative variation in obesity-related traits and insulin precursors linked to the OB gene region on human chromosome 7. Am J Hum Genet 59:694–703 [PMC free article] [PubMed]
- Dyke B (1996) PEDSYS: a pedigree data management system: version 2.0: user's manual (PGL Technical Report no. 2), Population Genetics Laboratory, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio [Google Scholar]
- Ehrenborg E, Clee SM, Pimstone SN, Reymer PWA, Benlian P, Hoogendijk CF, Davis HJ, et al (1997) Ethnic variation and in vivo effects of the −93t→g promoter variant in the lipoprotein lipase gene. Arterioscler Thromb Vasc Biol 17:2672–2678 [DOI] [PubMed]
- Elston RC (1998) Methods of linkage analysis—and the assumptions underlying them. Am J Hum Genet 63:931–934 [DOI] [PMC free article] [PubMed]
- Fasman KH, Letovsky SI, Cottingham RW, Kingsbury DT (1996) Improvements to the GBDTM Human Genome Data Base. Nucleic Acids Res 24:57–63 [DOI] [PMC free article] [PubMed]
- Fernández-Ruiz E, Armesilla AL, Sánchez-Madrid F, Vega MA (1993) Gene encoding the collagen type I and thrombospondin receptor CD36 is located on chromosome 7q11.2. Genomics 17:759–761 [DOI] [PubMed]
- Fulker DW, Cherny SS, Cardon LR (1995) Multipoint interval mapping of quantitative trait loci, using sib pairs. Am J Hum Genet 56:1224–1233 [PMC free article] [PubMed]
- Garg A (1998) Treatment of diabetic dyslipidemia. Am J Cardiol 81(4A):47B–51B [DOI] [PubMed]
- Gerdes C, Gerdes LU, Hansen PS, Faergeman O (1995) Polymorphisms in the lipoprotein lipase gene and their associations with plasma lipid concentrations in 40-year-old Danish men. Circulation 92:1765–1769 [DOI] [PubMed]
- Ginsberg HN (1996) Diabetic dyslipidemia: basic mechanisms underlying the common hypertriglyceridemia and low HDL cholesterol levels. Diabetes 45(Suppl 3):S27–S30 [DOI] [PubMed] [Google Scholar]
- Gorry MC, Gabbaizedeh D, Furey W, Gates LK Jr, Preston RA, Aston CE, Zhang Y, et al (1997) Mutations in the cationic trypsinogen gene are associated with recurrent acute and chronic pancreatitis. Gastroenterology 113:1063–1068 [DOI] [PubMed]
- Gotto AM (1998) Triglyceride: the forgotten risk factor. Circulation 97:1027–1028 [DOI] [PubMed]
- Grundy SM (1998) Hypertriglyceridemia, atherogenic dyslipidemia, and the metabolic syndrome. Am J Cardiol 81(4A):18B–25B [DOI] [PubMed]
- Haffner SM, Stern MP, Hazuda HP, Mitchell BD, Patterson JK (1990) Cardiovascular risk factors in confirmed prediabetic individuals: does the clock for coronary heart disease start ticking before the onset of clinical diabetes? JAMA 263:2893–2898 [DOI] [PubMed]
- Haffner SM, Stern MP, Hazuda HP, Pugh JA, Patterson JK (1986) Hyperinsulinemia in a population at high risk for non–insulin-dependent diabetes mellitus. N Engl J Med 315:220–224 [DOI] [PubMed]
- Havel RJ (1998) Plasma triglycerides and the clinician: time for reassessment. J Am Coll Cardiol 31:1258–1259 [DOI] [PubMed]
- Hegele RA, Connelly PW, Hanley AJG, Sun F, Harris SB, Zinman B (1997) Common genomic variation in the APOC3 promoter associated with variation in plasma lipoproteins. Arterioscler Thromb Vasc Biol 17:2753–2758 [DOI] [PubMed]
- Howard BV (1996) Macrovascular complications of diabetes mellitus. In: LeRoith D, Taylor SI, Olefsky JM (eds) Diabetes mellitus. Lippincott-Raven, Philadelphia, pp 792–797 [Google Scholar]
- Hsueh WA, Law RE (1998) Cardiovascular risk continuum: implications of insulin resistance and diabetes. Am J Med 105(1A):4S–14S [DOI] [PubMed]
- Humphries SE, Nicaud V, Margalef J, Tiret L, Talmud PJ, for the EARS (1998) Lipoprotein lipase gene variation is associated with a paternal history of premature coronary artery disease and fasting and postprandial plasma triglycerides: the European Atherosclerosis Research Study (EARS). Arterioscler Thromb Vasc Biol 18:526–534 [DOI] [PubMed]
- Iselius L (1988) Genetic epidemiology of common diseases in humans. In: Weir BS, Eisen EJ, Goodman MM, Namkoong G (eds) Proceedings of the second international conference on quantitative genetics. Sinauer, Sunderland, MA pp 341–352 [Google Scholar]
- Jeppesen J, Hein HO, Suadicani P, Gyntelberg F (1998) Triglyceride concentration and ischemic heart disease: an eight-year follow-up in the Copenhagen male study. Circulation 97:1029–1036 [DOI] [PubMed]
- Khoury MJ, Beaty TH, Cohen BH (eds) (1993) Fundamentals of genetic epidemiology. New York, Oxford University Press [Google Scholar]
- Klinger KW, Winqvist R, Riccio A, Andreasen PA, Sartorio R, Nielsen LS, Stuart N, et al (1987) Plasminogen activator inhibitor type 1 gene is located at region q21.3-q22 of chromosome 7 and genetically linked with cystic fibrosis. Proc Natl Acad Sci USA 84:8548–8552 [DOI] [PMC free article] [PubMed]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed]
- MacCluer JW (1989) Statistical approaches to identifying major locus effects on disease susceptibility. In: Lusis AJ, Sparkes SR (eds) Genetic factors in atherosclerosis: approaches and model systems. Karger, Basel, pp 50–78 [Google Scholar]
- Mahaney MC, Blangero J, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW (1995) Plasma HDL cholesterol, triglycerides, and adiposity: a quantitative genetic test of the conjoint trait hypothesis in the San Antonio Family Heart Study. Circulation 92:3240–3248 [DOI] [PubMed]
- Miller M, Seidler A, Moalemi A, Pearson TA (1998) Normal triglyceride levels and coronary artery disease events: the Baltimore Coronary Observational Long-Term Study (COLTS). J Am Coll Cardiol 31:1252–1257 [DOI] [PubMed]
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, et al (1996) Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans: the San Antonio Family Heart Study. Circulation 94:2159–2170 [DOI] [PubMed]
- Mochizuki H, Scherer SW, Xi T, Nickle DC, Majer M, Huizenga JJ, Tsui L-C, et al (1998) Human PON2 gene at 7q21.3: cloning, multiple mRNA forms, and missense polymorphisms in the coding sequence. Gene 213:149–157 [DOI] [PubMed]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62:690–697 [DOI] [PMC free article] [PubMed]
- Mykkänen L, Haffner SM, Rainwater DL, Karhapää P, Miettinen H, Laasko M (1997) Relationship of LDL size to insulin sensitivity in normoglycemic men. Arterioscler Thromb Vasc Biol 17:1447–1453 [DOI] [PubMed]
- O'Connell P, Leach RJ, Rains D, Taylor T, Garcia D, Ballard L, Holik P, et al (1994) A PCR-based genetic map for human chromosome 3. Genomics 24:557–567 [DOI] [PubMed]
- Pajukanta P, Nuotio I, Terwilliger JD, Porkka KVK, Ylitalo K, Pihlajamäki J, Suomalainen AJ, et al (1998) Linkage of familial combined hyperlipidaemia to chromosome 1q21-q23. Nat Genet 18:369–373 [DOI] [PubMed]
- Pandya A, Blanton SH, Landa B, Javaheri R, Melvin E, Nance WE, Markello T (1996) Linkage studies in a large kindred with hereditary pancreatitis confirms mapping of the gene to a 16-cM region on 7q. Genomics 38:227–230 [DOI] [PubMed]
- Rice T, Vogler GP, Laskarzewski PM, Perry TS, Rao DC (1991) Familial aggregation of lipids and lipoproteins in families ascertained through random and nonrandom probands in the Minnesota Lipid Research Clinic Family Study. Hum Biol 63:419–439 [PubMed]
- Rust S, Walter M, Funke H, von Eckardstein A, Cullen P, Kroes HY, Hordijk R, et al (1998) Assignment of Tangier disease to chromosome 9q31 by a graphical linkage exclusion strategy. Nat Genet 20:96–98 [DOI] [PubMed]
- Sanghera DK, Aston CE, Saha N, Kamboh MI (1998) DNA polymorphisms in two paraoxonase genes (PON1 and PON2) are associated with the risk of coronary heart disease. Am J Hum Genet 62:36–44 [DOI] [PMC free article] [PubMed]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Singh CF, Boerwinkle E, Moll PP, Templeton AR (1988) Characterization of genes affecting quantitative traits in humans. In: Weir BS, Eisen EJ, Goodman MM, Namkoong G (eds) Proceedings of the second international conference on quantitative genetics. Sinauer, Sunderland, MA, pp 250–269 [Google Scholar]
- Sparkes RS, Zollman S, Klisak I, Kirchgessner TG, Komaromy MC, Mohandas T, Schotz MC, et al (1987) Human genes involved in lipolysis of plasma lipoproteins: mapping of loci for lipoprotein lipase to 8p22 and hepatic lipase to 15q21. Genomics 1:138–144 [DOI] [PubMed]
- Steiner G, Lewis GF (1996) Hyperinsulinemia and triglyceride-rich lipoproteins. Diabetes 45(Suppl 3):S24–S26 [DOI] [PubMed] [Google Scholar]
- Stern MP, Duggirala R, Mitchell BD, Reinhart LJ, Shivakumar S, Shipman PA, Uresandi OC, et al (1996) Evidence for linkage of regions on chromosomes 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Res 6:724–734 [DOI] [PubMed]
- Stern MP, Haffner SM (1990) Type II diabetes and its complications in Mexican Americans. Diabetes Metab Rev 6:29–45 [DOI] [PubMed]
- Stern MP, Rosenthal M, Haffner SM, Hazuda HP, Franco LP (1984) Sex difference in the effects of sociocultural status on diabetes and cardiovascular risk factors in Mexican Americans: the San Antonio Heart Study. Am J Epidemiol 120:834–851 [DOI] [PubMed]
- Surguchov AP, Page GP, Smith L, Patsch W, Boerwinkle E (1996) Polymorphic markers in apolipoprotein C-III gene flanking regions and hypertriglyceridemia. Arterioscler Thromb Vasc Biol 16:941–947 [DOI] [PubMed]
- Talmud PJ, Hall S, Holleran S, Ramakrishnan R, Ginsberg HN, Humphries SE (1998) LPL promoter −93T/G transition influences fasting and postprandial plasma triglycerides response in African-Americans and Hispanics. J Lipid Res 39:1189–1196 [PubMed]
- Whitcomb DC, Preston RA, Aston CE, Sossenheimer MJ, Barua PS, Zhang Y, Wong-Chong A, et al (1996) A gene for hereditary pancreatitis maps to chromosome 7q35. Gastroenterology 110:1975–1980 [DOI] [PubMed]
- Wittrup HH, Tybjaerg-Hansen A, Nordestgaard BG (1999) Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease: a meta-analysis. Circulation 99:2901–2907 [DOI] [PubMed]
- Wittrup HH, Tybjaerg-Hansen A, Steffensen R, Deeb SS, Brunzell JD, Jensen G, Nordestgaard BG (1997) A common mutation T(−93)G in the promoter of the lipoprotein lipase gene is associated with elevated plasma triglycerides in both genders and with ischemic heart disease in men: the Copenhagen City Heart Study. Circulation 96 Suppl 8:I-336 [Google Scholar]
- World Health Organization Expert Committee (1985) Diabetes mellitus: report of a WHO study group. World Health Organization Technical Report Series 727, Geneva, pp 1–113 [PubMed] [Google Scholar]
- Zeng Q, Dammerman M, Takada Y, Matsunaga A, Breslow JL, Sasaki J (1995) An apolipoprotein CIII marker associated with hypertriglyceridemia in Caucasians also confers increased risk in a west Japanese population. Hum Genet 95:371–375 [DOI] [PubMed]