Abstract
The mtDNA variation of 74 Khoisan-speaking individuals (Kung and Khwe) from Schmidtsdrift, in the Northern Cape Province of South Africa, was examined by high-resolution RFLP analysis and control region (CR) sequencing. The resulting data were combined with published RFLP haplotype and CR sequence data from sub-Saharan African populations and then were subjected to phylogenetic analysis to deduce the evolutionary relationships among them. More than 77% of the Kung and Khwe mtDNA samples were found to belong to the major mtDNA lineage, macrohaplogroup L* (defined by a HpaI site at nucleotide position 3592), which is prevalent in sub-Saharan African populations. Additional sets of RFLPs subdivided macrohaplogroup L* into two extended haplogroups—L1 and L2—both of which appeared in the Kung and Khwe. Besides revealing the significant substructure of macrohaplogroup L* in African populations, these data showed that the Biaka Pygmies have one of the most ancient RFLP sublineages observed in African mtDNA and, thus, that they could represent one of the oldest human populations. In addition, the Kung exhibited a set of related haplotypes that were positioned closest to the root of the human mtDNA phylogeny, suggesting that they, too, represent one of the most ancient African populations. Comparison of Kung and Khwe CR sequences with those from other African populations confirmed the genetic association of the Kung with other Khoisan-speaking peoples, whereas the Khwe were more closely linked to non–Khoisan-speaking (Bantu) populations. Finally, the overall sequence divergence of 214 African RFLP haplotypes defined in both this and an earlier study was 0.364%, giving an estimated age, for all African mtDNAs, of 125,500–165,500 years before the present, a date that is concordant with all previous estimates derived from mtDNA and other genetic data, for the time of origin of modern humans in Africa.
Introduction
The South African Kung San were among the first human groups in which mtDNA variation was analyzed by restriction analysis (Denaro et al. 1981; Johnson et al. 1983). In these studies, the mtDNA sequences of the Kung were surveyed for variation, by means of six rare-cutting restriction endonucleases (AvaII, BamHI, HaeII, HincII, HpaI, and MspI) and Southern blotting. Although this procedure screened only 2%–3% of the mtDNA sequence for variation, it revealed that ∼90%–95% of the Kung mtDNAs were characterized by a HpaI site gain at nucleotide position (np) 3592. This marker was subsequently found at very high frequencies in mtDNAs of other sub-Saharan African populations (Scozzari et al. 1988, 1994; Soodyall and Jenkins 1992, 1993; Chen et al. 1995) but was not observed in populations of non-African origin, with a few exceptions (Cann et al. 1987; Soodyall 1993). Furthermore, the African mtDNAs defined by the HpaI np-3592 site gain formed a group of related mtDNA haplotypes (originally defined as haplogroup “L” and here redefined as “L*”), which was found to be the most divergent of those identified in human populations from around the world (Chen et al. 1995). These findings contributed to the hypothesis of an African origin of modern human mtDNAs (Johnson et al. 1983; Cann et al. 1987; Chen et al. 1995), although other interpretations of these data have also been put forward (Excoffier and Langaney 1989; Templeton 1992).
Since 1981, relatively limited data have been collected on the RFLP variation in Kung or related Khoisan-speaking populations. Those studies that were conducted on Khoisan-speaking populations continued to use low-resolution (LR)–RFLP analysis with only the six rare-cutting restriction endonucleases listed above (Soodyall and Jenkins 1992, 1993). As a consequence, these data provided additional information about Khoisan groups but could not be entirely integrated with those obtained, on other populations, by high-resolution (HR)–RFLP analysis (Cann et al. 1987; Torroni et al. 1992).
The value of using HR-RFLP analysis with African mtDNAs, for phylogenetic reconstructions of human mtDNA, has been shown primarily by two studies of African populations (Cann et al. 1987; Chen et al. 1995). In the first study, Cann et al. (1987) conducted genomic digestions of mtDNAs, by 12 enzymes (AluI, AvaII, DdeI, FnuDII, HaeIII, HhaI, HinfI, HpaI, HpaII, MboI, RsaI, and TaqI), with the resulting restriction fragments being resolved by PAGE. In the later study, the entire genome of African mtDNAs was PCR amplified in nine overlapping fragments, which were then subjected to digestion by 14 enzymes (AluI, AvaII, BamHI, DdeI, HaeII, HaeIII, HhaI, HincII, HinfI, HpaI, HpaII, MboI, RsaI, and TaqI), with the resulting restriction fragments being resolved by agarose gel electrophoresis. Both of these studies revealed that African haplotypes form the deepest branches of the human mtDNA phylogeny and that African groups are the most divergent of all world populations.
Several additional aspects of mtDNA variation in African populations were shown in the Chen et al. (1995) study. First, two types of length polymorphisms were observed in some of the central-African mtDNAs (Mbuti [eastern] and Biaka [western] Pygmies), one of which was the COII/tRNALys intergenic 9-bp deletion that formerly had been thought to occur only in Asian populations. This length polymorphism had previously been noted in African individuals but had not been assigned to a particular haplogroup or mtDNA lineage (Cann and Wilson 1983). In addition, western-African populations from Senegal were found to have haplotypes that lacked the HpaI np-3592 site gain and, thus, appeared not to belong to haplogroup L*. Because nearly all of these “non-L” haplotypes had the DdeI np-10394 site gain, a polymorphism that was also present in almost every haplogroup L* mtDNA, the data suggested that the former haplotypes derived from the latter, although the exact subhaplogroup (L1 or L2) from which these non-L haplotypes originated was not clear. Alternatively, non-L mtDNAs could have evolved from haplotypes observed in Europeans, such as those belonging to haplogroups I–K (Torroni et al. 1994a), and could have been spread into Africa through population contractions/expansions since their origin. Such ambiguities clearly indicated that further analysis of non-L haplotypes was necessary in order to understand their phylogenetic relationships to other African and non-African mtDNAs.
Similarly, until very recently, only a limited number of Kung mtDNAs had been surveyed for control region (CR) sequence variation. In earlier studies (Vigilant et al. 1989, 1991), mtDNAs from Kung individuals consistently clustered at the deepest nodes of the phylogenies constructed from CR sequences, a result that indicated that they are some of the most divergent—hence, oldest—mtDNAs present in human populations. Soodyall et al. (1996) also studied CR sequence variation in southern-African populations and found that the COII/tRNALys intergenic 9-bp deletion had occurred multiple times in African populations, independent of those appearing in Asian mtDNAs, a trend first noted by Vigilant (1990). Interestingly, deletion mtDNAs were not observed in Khoisan-speaking populations and were rare in western- and southwestern-African groups, whereas they occurred frequently in Pygmy and so-called Negroid populations from central Africa and in Bantu-speaking peoples of southern Africa (Soodyall et al. 1996). This distribution suggested that deletion mtDNAs arose in central Africa and were disseminated into southern Africa via the recent Bantu expansion.
Although studies using HR-RFLP methods of analysis have provided new insights into the mtDNA variation of African populations, there has been almost no effort to relate the data sets generated by RFLP to those generated by a CR sequence analyses. To date, only the western-African Mandelaku (Mandenka) have been subjected to both LR-RFLP and CR sequencing analyses (Graven et al. 1995), whereas the linkage between the HR-RFLP data and the CR sequence data for the African samples analyzed by Cann et al. (1987) and Vigilant et al. (1989, 1991) has not been made explicitly clear. Thus, the relationship between HR-RFLP haplotypes and CR sequences in African populations has yet to be adequately addressed. In addition, because of the different methods of RFLP haplotyping used in the Cann et al. (1987) and Chen et al. (1995) studies, there were some inconsistencies in the positioning of certain polymorphisms that were critical for phylogenetic reconstructions of the haplotypes defined in each study. Consequently, a more complete characterization of the distribution of genetic variation in coding (i.e., RFLP) versus noncoding (i.e., CR) regions of the mtDNA genome was needed.
Both to increase our knowledge of the genetic relationships between Khoisan (Kung San and Khoi) populations and other African peoples, and to better define the relationship between RFLP and CR sequence variation within these groups, we conducted HR-RFLP and CR sequence analyses of 74 Kung San and Khwe individuals from southern Africa and then compared the resulting data to those obtained for African populations in previous studies. This comparison revealed that many of the Kung mtDNAs clustered within a distinct sublineage (L1a2) of subhaplogroup L1a and that the Khwe are more closely genetically related to western-African (Bantu-speaking) populations than they are to the Kung San. Furthermore, the genetic relationships deduced from HR-RFLP analysis were found to be completely consistent with—and sometimes more detailed than—those obtained from CR sequencing alone. These findings implied the need for studies combining the two methods of analysis, in order to more fully understand the genetic relationships among human populations.
Subjects and Methods
Subjects
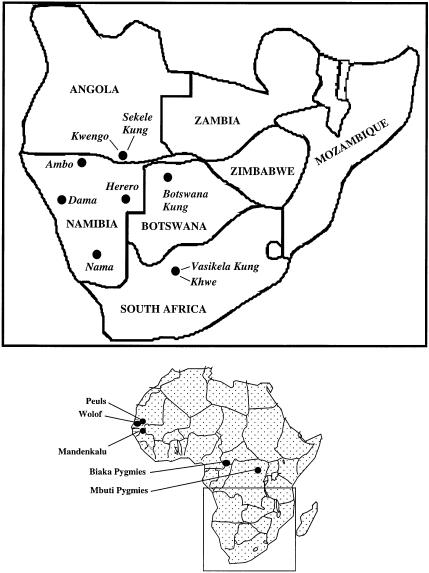
Khoisan-speaking populations can generally be divided into two distinct groups, the San and the Khoi. San populations consist of 10 different Kung groups (“!Xu” is pronounced “Kung”), as well as the //au//en, Nharo, G/wi, G//ana, !Xo and G!aokxíte, who also speak click-languages. The Khoi consist of five populations—two Topanaar groups and the Kede, Hei//om, and Nama. A third set of southern-African populations, which coexist with Khoisan-speaking groups, are the so-called Negroids, who are largely Bantu-speakers. These include the Kwisi, Kwadi, Cimba, Dama, Kgalagadi, and Denasena (Nurse et al. 1985). Although the two Khwe (Kwengo) groups physically appear to be “Negroid,” they speak a Khoisan language (de Almeida 1965). Consequently, they have been called “black Bushmen.” The geographic locations of the populations analyzed in this study, as well as the populations to which they are compared, are shown in figure 1.
Figure 1.
Map of southern Africa, and geographic locations of Khoisan and non-Khoisan populations surveyed for mtDNA variation.
A total of 181 Khoisan-speaking individuals of the northwestern Kalahari Desert in southern Africa formed the sample group for this study. Of this group, 144 were identified as Kung (Vasikela Kung), 37 as Khwe (also known as “Barakwena”). Of the 181 southern-African mtDNA samples, 74 (43 Kung and 31 Khwe) were analyzed for mtDNA sequence variation.
The individuals involved in this study were classified according to the ethnicity listed on either their identity documents or birth certificates. In the few rare cases in which this information was not available, both parents were independently questioned about their own ancestry before their child was classified as belonging to either of the two groups. Fortunately, in all cases, the family history furnished by both parents was identical, simplifying the classification of their children. Interpreters were used in the interviews and counseling sessions, and care was taken to verify the data and information gathered. Even though these two groups have been known to intermarry, no such instance was recorded for these particular individuals.
Sample Acquisition and Preparation
After informed consent was obtained, two 10-ml tubes of blood were collected from Kung and Khwe individuals, by venipuncture, in acid citrate–dextrose vacutainer tubes and then were stored at 4°C until they were shipped to Emory University. At Emory University, the blood samples from each individual were separated into their constituent cellular fractions, by low-speed centrifugation. The platelets were subsequently collected from the plasmas by centrifugation in 15-ml Corning tubes, at 5,000 g and 10°C for 20 min; all the mtDNAs used in the RFLP and CR sequence analyses were then extracted from these platelet pellets (Torroni et al. 1992).
Molecular Genetic Analysis
The entire mtDNA of each sample was subjected to HR-RFLP analysis using the primer pairs and PCR amplification conditions described by Torroni et al. (1992, 1993). This HR-RFLP analysis defined the complete haplotype for each individual. The evolutionary relationships among the Kung and Khwe mtDNAs were further differentiated by the sequencing of both hypervariable segments (HVS-I and HVS-II) of the CR of each individual, by methods described by Schurr et al. (1999). Both hypervariable segments were PCR amplified by use of heavy-strand primer H15704 (np 15704–15723) and light-strand primer L770 (np 770–751), whereas two different sets of primers—H15978 (np 15978–15997) and reverse primer L16501 (np 16501-16483) for HVS-I, H1 (np 1–19) and L429 (np 429–412), for HVS-II—were used for sequencing. The resulting data were analyzed by SEQUENCHER 3.0 software (Gene Codes).
Phylogenetic Analyses
The phylogenetic relationships between the mtDNA haplotypes observed in the Kung and Khwe and those previously reported in the other sub-Saharan African populations (Chen et al. 1995) were inferred by parsimony analysis with PAUP 3.1.1 (Swofford 1994) and PAUP 4.0.2b (Swofford 1998). The samples included complete haplotypes of 62 Senegalese (AF01–AF24, AF26–AF36, AF45–AF59, AF64–AF65, and AF70–AF79), 17 Pygmy (AF25, AF37–AF44, AF60–AF63 and AF66–AF69), and 29 Kung and Khwe (AF46, AF80-AF107). All dendrograms were rooted from a chimpanzee haplotype that was extrapolated from the whole mitochondrial genome sequence presented by Horai et al. (1995), by identification of all recognition sequences of the 14 enzymes used in the HR-RFLP analyses. The African haplotypes were also midpoint rooted without an outgroup.
Maximum parsimony (MP) trees were generated via random addition of sequences, by the tree-bisection and- reconnection (TBR) algorithm, with ⩽10 MP trees being saved for each replication. Because of the numerous terminal taxa in this data set, a large number of MP trees were obtained, with 3,000 MP trees being generated after 1,388 replications. Although shorter trees could exist, none were observed in this analysis. Strict and 50%-majority-rule consensus trees encompassing all MP trees were also obtained, to test the consistency of the branching order in the MP trees. Similarly, the data were subjected to bootstrap analysis to test the statistical support for the branch structure of the MP trees. All RFLP haplotypes were bootstrapped through 10–500 replications, with resampling of characters (i.e., RFLPs) by the TBR algorithm, with a 50%-majority-rule bootstrap consensus tree being generated from the best trees saved after each independent analysis. Genetic distance/neighbor-joining (NJ) trees were also generated by PAUP 4.0.2b, on the basis of the mean character differences of the haplotypes. These were also subjected to bootstrap analysis (Swofford 1998).
In addition, the CR sequences obtained from the 43 Kung and 31 Khwe samples were combined with the 11 distinct CR sequences from the Botswana Kung (sequences 1, 5–9, and 11–15) reported by Vigilant et al. (1989) and were subjected to phylogenetic analysis. To distinguish between the CR sequences belonging to the two different Kung San groups, the population samples described in this study were called the “Vasikela Kung,” and those analyzed by Vigilant et al. (1989) were called the “Botswana Kung.” Phylogenies of these sequences were obtained with the NJ (Saitou and Nei 1987) method present in the MEGA 1.01 statistical package (Kumar et al. 1993). The evolutionary distance between pairs of CR sequences were estimated as p distances, the proportion (p) of nucleotide sites at which the pair of sequences being compared differed, as calculated by dividing the number of nucleotide differences (nd) by the total number of nucleotides compared (n). Similar genetic distances were obtained with the other algorithms available in MEGA, but they are not presented here.
All p distances were used with the NJ method, to produce phylogenetic trees. The NJ trees generated from HVS-I sequences were rooted by use of either a chimpanzee (Horai et al. 1995) or the Neanderthal (Krings et al. 1997) sequence, and those generated from both HVS-I and HVS-II data were rooted from only the chimpanzee sequence, because no HVS-II data were available for the Neanderthal mtDNA. Unrooted NJ trees were midpoint rooted without an outgroup sequence. The branching structure of these NJ trees was, in turn, tested by bootstrap analysis, which produced bootstrap confidence-level estimates for each interior branch.
Sequence-Divergence Estimates
To calculate the genetic divergence of the southern-African populations, as well as the divergence within specific haplogroups and their sublineages, we used the iterative maximum likelihood (ML) method of Nei and Tajima (1983). The interpopulation ML estimates were obtained by taking a weighted average of the number of individuals in each population. “Corrected” interpopulation values (Corr) were obtained by taking an average of two intrapopulation estimates and subtracting that value from the uncorrected interpopulation value—that is, Corr=δxy-[(δx+δy)/2], where x and y are the two populations being analyzed. The same method was used for estimation of the divergence of haplogroups and their sublineages. When calculating the divergence times of these haplogroups and of their sublineages, we used the mtDNA evolutionary rate of 2.2%–2.9%/1 million years (Torroni et al. 1994c).
Results
Southern-African RFLP Haplotypes
A total of 29 haplotypes (AF46 and AF80–AF107) defined by 47 restriction-site polymorphisms were observed among the 74 South African individuals analyzed (Appendix A). One of these haplotypes, AF46, was described in our previous study of African mtDNA (Chen et al. 1995), whereas haplotypes AF80-AF107 were observed for the first time in African populations. A total of 18 haplotypes were detected in the Vasikela Kung, and 13 haplotypes were found in the Khwe, only 2 of which (i.e., AF85 and AF99) were shared between them (table 1). Of these southern-African haplotypes, 77% clustered into haplogroup L*, including 84% of the Vasikela Kung mtDNAs and 68% of the Khwe mtDNAs.
Table 1.
HR mtDNA Haplotypes in Southern-African Populations
|
Statusb |
No. |
||||
| HR Haplotypea | HpaI 3592 | DdeI 10394 | Kung | Khwe | Total |
| AF46 | + | + | 2 | 2 | |
| AF80 | − | + | 1 | 1 | |
| AF81 | − | + | 1 | 1 | |
| AF82 | − | + | 2 | 2 | |
| AF83 | − | + | 6 | 6 | |
| AF84 | − | − | 1 | 1 | |
| AF85 | − | + | 4 | 1 | 5 |
| AF86 | − | + | 1 | 1 | |
| AF87 | + | + | 6 | 6 | |
| AF88 | + | + | 3 | 3 | |
| AF89 | + | + | 5 | 5 | |
| AF90 | + | + | 2 | 2 | |
| AF91 | + | − | 2 | 2 | |
| AF92 | + | − | 5 | 5 | |
| AF93 | + | + | 2 | 2 | |
| AF94 | + | + | 1 | 1 | |
| AF95 | + | + | 1 | 1 | |
| AF96 | + | + | 1 | 1 | |
| AF97 | + | + | 2 | 2 | |
| AF98 | + | + | 1 | 1 | |
| AF99 | + | + | 1 | 6 | 7 |
| AF100 | + | + | 4 | 4 | |
| AF101 | + | + | 4 | 4 | |
| AF102 | + | + | 2 | 2 | |
| AF103 | + | + | 1 | 1 | |
| AF104 | + | + | 1 | 1 | |
| AF105 | + | + | 3 | 3 | |
| AF106 | + | + | 1 | 1 | |
| AF107 | + | + | 1 | 1 | |
| Total | 43 | 31 | 74 | ||
As defined by the polymorphic restriction sites shown in Appendix A.
A plus sign (+) denotes presence, and a minus sign (−) denotes absence.
Haplotypes belonging to haplogroup L* have previously been observed in the western-African Senegalese (Chen et al. 1995; Graven et al. 1995) and Biaka and Mbuti Pygmy (Chen et al. 1995) populations, at very high frequencies, as well as at similar frequencies in the Bamileke of Cameroon (Scozzari et al. 1994) and in various southern-African groups (Soodyall and Jenkins 1992, 1993). However, no comparable haplotypes have been observed in European, Middle Eastern, or Asian populations (Ballinger et al. 1992; Torroni et al. 1994a, 1994b, 1994c, 1996), with the exception of those groups with a history of contact with African populations (Bonné-Tamir et al. 1986; De Benedictis et al. 1989; Semino et al. 1989; Ritte et al. 1993). Thus, these results substantiate the African origin of this mtDNA lineage, as well as its widespread distribution, albeit at varying frequencies, in all African populations.
Consistent with a previous study of Khoisan mtDNA variation (Soodyall et al. 1996), none of the Vasikela Kung or the Khwe haplotypes exhibited any length polymorphisms in the COII/tRNALys intergenic region. These results support the conclusion that deletion haplotypes from haplogroup L* originated not in Khoisan populations but, instead, in Bantu-speaking and/or Pygmy groups of western and central Africa.
Phylogenetic Analysis of RFLP Haplotypes
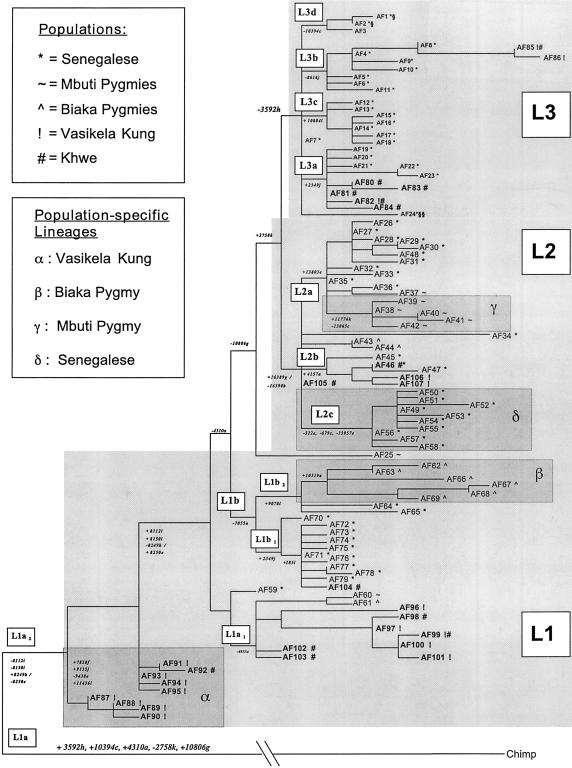
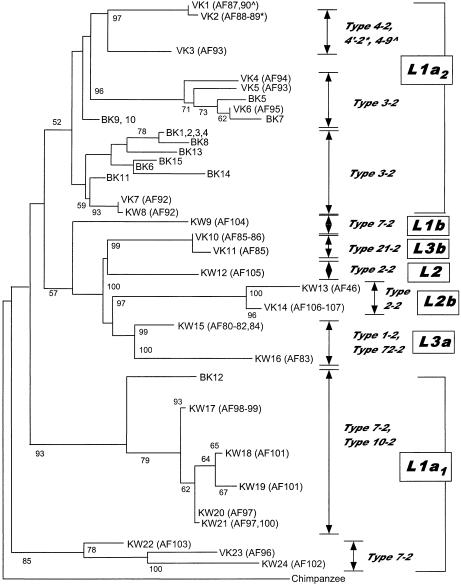
The phylogenetic relationships among the mtDNA haplotypes observed in the Vasikela Kung and Khwe and those previously reported in the other sub-Saharan African populations were assessed by both MP and genetic-distance/NJ analysis. The MP tree of African mtDNAs is presented infigure 2. The statistical robustness of the branches and subbranches of this tree are demonstrated by their preservation in 50%-majority-rule and strict consensus trees.
Figure 2.
MP tree showing evolutionary relationships among 107 haplotypes (AF1–AF107) observed in 214 sub-Saharan Africans (Chen et al. 1995; present study). The tree is 200 steps in length, has consistency and retention indices (CI and RI) of .745 and .893, respectively, and is one of 3,000 MP trees generated by the TBR algorithm. It was rooted from a chimpanzee haplotype defined by sequence analysis (Horai et al. 1995) (see text). The RFLPs that define each of the major branches of the MP tree are indicated by the number of the first nucleotide of the respective recognition sites, according to the CR sequence (Anderson et al. 1981), with the restriction enzymes being indicated by single-letter codes as given in Appendix A. The length of the horizontal branches is proportional to the number of mutations that separate the haplotypes. The three major haplogroups are denoted by the light-gray shading, whereas the subhaplotype clusters, defined in table 2, are indicated by the smaller unshaded boxes. The “AF”-prefixed numbers at the end of each branch denote the individual haplotypes, and the population(s) in which the individual haplotypes were detected are indicated by symbols as defined in the key. All population-specific lineages are encompassed by dark-gray shading and are also denoted by Greek letters as defined in the key. Haplotypes AF1 and AF2, which are aligned with European haplogroup U, are located in L3d and are indicated by a section symbol (§). Haplotype AF24, which is aligned with Asian macrohaplogroup M, is indicated by a double section symbol (§§). The bootstrap values for the major branches of the MP tree were calculated by use of 100 replicates, with resampling by the TBR algorithm. L1a2 cluster AF87–AF90 has a bootstrap value of 92%; subcluster AF88–AF90 has a bootstrap value of 100%. L1a2 cluster AF91–AF92 has a bootstrap value of 95%; but AF93 and AF94 were not resolved at bootstrap values >50%. L1a1 cluster AF97–AF101 has a bootstrap value of 100%; and subclusters AF99–AF101 and AF60–AF61 have bootstrap values of 52% and 60%, respectively; but AF60 and AF61 were not resolved at bootstrap values >50%. L1b1 cluster AF70–AF71/AF77–AF78 has a bootstrap value of 100%, as does subcluster AF71/AF77/AF78. L1b2 cluster AF62–AF69 has a bootstrap value of 100%, as do subclusters AF62–AF63 and AF67–AF69, whereas subcluster AF62–AF64/AF66–AF69 has a bootstrap value of 55%. L2 cluster AF26–AF38/AF40–AF50/AF52–AF53/AF55–AF58/AF105–AF107 has a bootstrap value of 52%. L2a cluster AF26–AF38/AF40–AF42 has a bootstrap value of 58%. L2b cluster AF46–AF47 has a bootstrap value of 100%. L2c cluster AF49–AF50/AF52–AF53/AF55–AF58 has a bootstrap value of 100%, and cluster AF106–AF107 has a bootstrap value of 52%. L3a cluster AF22–AF23 has a bootstrap value of 100%, whereas subclusters AF19–AF21/AF24 and AF80–AF84 were not resolved at bootstrap values >50%. L3b cluster AF8/AF85–AF86 has a bootstrap value of 100%, as does subcluster AF85–AF86, but cluster AF4/AF6/AF9/AF11 was not resolved at a bootstrap value of 50%. L3c cluster AF12/AF14–AF18 has a bootstrap value of 100%, as does subcluster AF14–AF18. L3d cluster AF1–AF3 has a bootstrap value of 97%.
From the MP tree, it is clear that the African mtDNA phylogeny forms a successive series of branches, each one occupied by clusters of related mtDNA haplotypes (fig. 2 and table 2), consistent with our previous report (Chen et al. 1995). The overall phylogeny is divided into two major parts, the lower two-thirds having the HpaI site at np 3593 and the upper one-third lacking this site. We had previously designated the mtDNAs that have the HpaI np-3592 site as “haplogroup L” (Chen et al. 1995) and had reported that it subdivides into two major branches—L1, having a HinfI site at np 10806, and L2, having a combined HinfI site gain at np 16389 and an AvaII site loss at np 16390. The expanded phylogeny infigure 2 shows that L1 is further subdivided into L1a and L1b, on the basis of the presence or absence of an AluI site at np 4310. L2 is subdivided into L2a, L2b, and L2c, on the basis of the presence of a HaeIII site at np 13803, for L2a; the presence of an AluI site at np 4157, for L2b; and the absence of HaeIII sites at np 322 and 13957 and of a DdeI site at np 69, for L2c. L1a is further subdivided into L1a1, by an AluI site loss at np 4853, and L1a2, by MspI site losses at np 8112 and 8150 and by a combined AvaII np-8249 site gain and HaeIII np-8250 site loss. L1b is also subdivided into L1b1, by a MboI np-2349 site gain, and L1b2, by a TaqI np-9070 site gain (fig. 2 and table 2).
Table 2.
mtDNA Haplogroup Distribution in African Populations
|
No. (%) in |
|||||||
| Haplogroup | Defining Polymorphisms | Kung (N=43) | Khwe (N=31) | Mandenkalu (N=60)a | Wolof (N=20)a | MbutiPygmies (N=22)a | Biaka Pygmies (N=17)a |
| L | +HpaI 3592, +DdeI 10394 | 36 (84) | 21 (68) | 44 (73) | 14 (70) | 22 (100) | 17 (100) |
| L1 | +HinfI 10806, −RsaI 2758 | 34 (79) | 16 (52) | 17 (28) | 4 (20) | 8 (36) | 15 (88) |
| L1a | +AluI 4310 | 34 (79) | 15 (48) | 1 (2) | … | 6 (27) | 4 (23) |
| L1a1 | −AluI 4853 | 12 (28) | 10 (32) | 1 (2) | … | 6 (27) | 4 (23) |
| L1a2 | −MspI 8112, −MspI 8150, +AvaII 8249, −HaeIII 8250 | 22 (51) | 5 (16) | … | … | … | … |
| L1b | −AluI 7055 | … | 1 (3) | 16 (27) | 4 (20) | … | 11 (65) |
| L1b1 | +MboI 2349 | … | 1 (3) | 13 (22) | 4 (20) | … | … |
| L1b2 | +TaqI 9070 | … | … | 3 (5) | … | … | 11 (65) |
| L2 | +HinfI 16389/−AvaII 16390 | 2 (5) | 5 (16) | 27 (45) | 10 (50) | 14 (64) | 2 (12) |
| L2a | +HaeIII 13803 | … | … | 10 (17) | 5 (25) | 14 (64) | … |
| L2b | +AluI 4157 | 2 (5) | 5 (16) | … | 3 (15) | … | … |
| L2c | −HaeIII 322, −DdeI 679, −HaeIII 13957 | … | … | 17 (28) | 2 (10) | … | … |
| L3 | −HpaI 3592 | 7 (16) | 10 (32) | 16 (27) | 6 (30) | … | … |
| L3a | +MboI 2349 | 2 (5) | 9 (29) | 4 (7) | 1 (5) | … | … |
| L3b | −MboI 8616 | 5 (12) | 1 (3) | 7 (12) | 2 (10) | … | … |
| L3c | +TaqI 10084 | … | … | 4 (7) | 3 (15) | … | … |
| L3d | −DdeI 10394 | … | … | 1 (2) | … | … | … |
Data are from Chen et al. (1995).
The upper one-third of the mtDNA phylogeny encompasses mtDNAs that lack the np-3592 HpaI site, which, in our previous study, we had designated “non-L” (Chen et al. 1995). This was subsequently redefined as “L3,” by Watson et al. (1997). When this nomenclature is adopted, L3 is subdivided into four haplogroups: L3a, associated with a MboI np-2349 site gain; L3b, associated with a MboI np-8616 site loss; L3c, associated with a TaqI np-10084 site gain; and L3d, associated with a DdeI np-10394 site loss (fig. 2 and table 2).
L1, L2, and L3 each encompass a sequence diversity that is approximately equivalent to those of other continent-specific haplogroups (Ballinger et al. 1992; Torroni et al. 1994a, 1994b; Chen et al. 1995). Therefore, it is reasonable to delineate these as haplogroups and to delineate the combination of L1 and L2, delineated by the presence of the HpaI site at np 3592, as a macrohaplogroup, “L*.” This then relegates L1a, L1b, L2a–L2c, and L3a–L3d to the level of subhaplogroups. Of our L3 subhaplogroups (i.e., L3a–L3d), L3a and L3b are identical to similarly named groupings recognized by Watson et al. (1997).
Within the African MP phylogeny (fig. 2), the southern-African Kung and Khwe samples cluster together in L1a. Within L1a, L1a2 is predominantly Kung, L1a1 is Kung and Khwe, L2b includes Kung and Khwe, and L3a is predominantly Khwe. This asymmetric distribution of haplotypes is also seen for the Biaka Pygmies, L1b2; and the Mbuti Pygmies, L2a (fig. 2).
To establish the reliability of the major branches of our African mtDNA MP tree, we subjected the MP tree to a bootstrap analysis. Because of the large number of taxa (haplotypes) and character states (restriction-site variants) in our data set, the 50%-majority-rule bootstrap tree was unable to provide confidence values for a number of the internal branches. To facilitate the analysis, we reduced the number of taxa from each of the major clusters seen infigure 2, eliminating most of the haplotype-specific restriction-site variants. This gave confidence values, for the major branches, that were in the 55%–85% range (fig. 2, legend). However, confidence values could not be obtained for a few of the subhaplogroup clusters, a common problem when bootstrap analysis is applied to large data sets containing similar taxa.
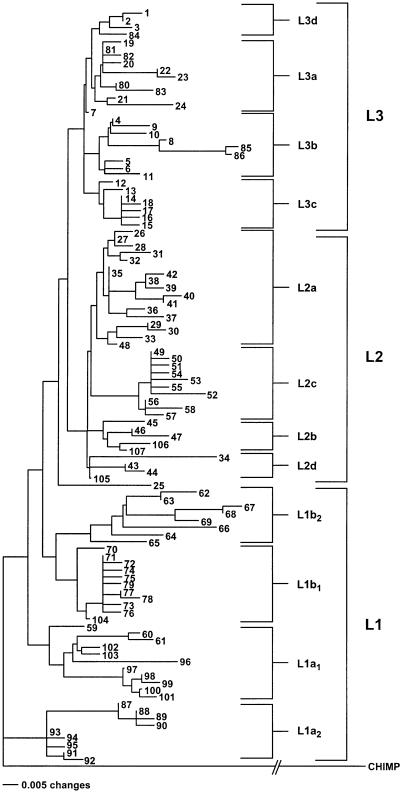
To further define the African mtDNA phylogeny, we performed a genetic distance/NJ analysis on the same data set, using PAUP 4.0.2b. The resulting NJ tree (fig. 3) is virtually identical to the MP tree shown infigure 2. The only exception is the position of haplotypes AF34, AF43, AF44, and AF105. In the MP tree, these mtDNAs are dispensed between L2a L2b, and L2c. However, in the NJ tree, they form a distinct cluster, which could be designated “L2d.”
Figure 3.
NJ tree constructed on the basis of mean character genetic differences of the RFLP haplotypes. The numbers represent the AF haplotype numbers shown in figure 2. The bootstrap values for various haplotype clusters of the NJ tree were calculated by use of 500 replicates, with resampling. The L1a2 cluster of haplotypes separated from all other African haplotypes, with a bootstrap value of 55%. L1a2 cluster AF87–AF88/AF90–AF93 has a bootstrap value of 54%; subcluster AF91–AF93 has a bootstrap value of 59%; subsubcluster AF91–AF92 has a bootstrap value of 65%; subcluster AF87–AF88/AF90 has a bootstrap value of 99%; and subsubcluster AF88–AF90 has a bootstrap value of 75%. L1b1+L1b2 cluster AF62–AF65/AF67–AF68/AF70–AF72/AF78 has a bootstrap value of 56%. L1b1 cluster AF70–AF72/AF78 has a bootstrap value of 94%; subcluster AF71–AF72/AF78 has a bootstrap value of 67%; and subsubcluster AF71, 72 has a bootstrap value of 60%. L1b2 cluster AF62–AF65/AF67–AF68 has a bootstrap value of 70%; subcluster AF67–AF68 has a bootstrap value of 100%; and subcluster AF62–AF63 has a bootstrap value of 69%. L1a1 cluster AF59–AF61/AF96–AF98/AF100–AF101/AF103 has a bootstrap value of 60%; subcluster AF60–AF61 has a bootstrap value of 90%; subcluster AF97–AF98/AF100–AF101 has a bootstrap value of 96%; subsubcluster AF97–AF98 has a bootstrap value of 72%; and subsubcluster AF100–AF101 has a bootstrap value of 67%. L2c cluster AF49–AF53/AF56/AF58 has a bootstrap value of 83%; subcluster AF49–AF53 has a bootstrap value of 53%; and subcluster AF56–AF58 of 63%. L2b cluster AF45–AF47/AF105 has a bootstrap value of 53%; subcluster AF46–AF47/AF105 has a bootstrap value of 74%; and subcluster AF46–AF47 has a bootstrap value of 76%. L2 cluster AF43–AF44 has a bootstrap value of 87%. L2a subcluster AF38–AF41 has a bootstrap value of 84%; subsubcluster AF38–AF39 has a bootstrap value of 66%; and subsubcluster AF40–AF41 has a bootstrap value of 70%. L2a subcluster AF36–AF37 has a bootstrap value of 69%; subcluster AF29–AF30 has a bootstrap value of 84%, whereas subclusters AF26–AF28 and AF33–AF35 were not resolved at bootstrap values >50%. L3d subcluster AF1–AF2 has a bootstrap value of 69%, whereas AF3 was not resolved at a bootstrap value >50%. L3c subcluster AF14–AF16 has a bootstrap value of 51%, although AF12 and AF13 were not resolved at a value of 50%. L3b cluster AF8/AF85–AF86 has a bootstrap value of 88%; and subcluster AF85–AF86 has a bootstrap value of 96%; however, clusters AF4–AF7 and AF9–AF11 were not resolved at a value of 50%. L3 cluster AF24–AF25 and L3a cluster AF80–AF83 were not resolved at a value of 50%.
To test the robustness of the African NJ phylogeny, we applied bootstrap analysis using PAUP 4.0.2b. Since the genetic distances used to construct the NJ trees represent single summaries of the genetic relationships among taxa (Saitou and Nei 1987), the number and complexity of the taxa were reduced. Consequently, bootstrap analysis of the NJ-tree genetic-distance data gave confidence values, for most branches, that were in the 51%–94% range. Again, however, some branches could not be assigned numerical values (fig. 3, legend).
Since both the MP and the genetic distance/NJ methods produced nearly identical trees, with the major branches having high bootstrap values, it is apparent that the African mtDNA phylogenies shown in figures2 and3 provide a reasonable representation of the phylogenetic relationships of African mtDNAs. Hence, the macrohaplogroups, haplogroups, and subhaplogroups shown in figures2 and3 can be used to further investigate the mtDNA genetic history of African populations.
At present, the relationship of the African mtDNA phylogeny to the mtDNA phylogenies of Europe and Asia can only partially be deduced. If included in the African phylogenetic analysis, all European mtDNAs would cluster outside haplogroups L1 and L2 and would form separate branches within haplogroup L3. This is because all European haplogroups are defined by distinct sets of RFLPs not usually present in African haplogroups (Torroni et al. 1996, 1998). Many of the L3 mtDNAs have the DdeI site at np 10394, a characteristic also seen in European haplogroups I–K; others, such as AF01, AF02, and AF03 of L3d, lack the DdeI np-10394 site, which is characteristic of European haplogroups H and T–X. Among the African mtDNAs lacking the DdeI 10394 site, haplotypes AF01 and AF02 from the Senegalese have a HinfI site at np 12308, which characterizes haplogroup U in European populations (Torroni et al. 1996). This raises the possibility that the L3d lineage may have given rise to several of the European haplogroups; however, it is also possible that the haplogroup U mtDNAs observed in these African samples may be the product of back-migrations into Africa from Europe.
The Asian mtDNA phylogeny is subdivided into two macrohaplogroups, one of which is M. M is delineated by a DdeI site at np 10394 and an AluI site of np 10397. The only African mtDNA found to have both of these sites is the Senegalese haplotype AF24. This haplotype branches off African subhaplogroup L3a (figs.2 and3), suggesting that haplogroup M mtDNAs might have been derived from this African mtDNA lineage; however, it is also possible that this particular haplotype is present in Africa because of back-migration from Asia.
Interestingly, the MboI np-2349 site gain is shared by the haplogroup L3a mtDNAs in the Vasikela Kung and the Khwe populations and a subset of subhaplogroup L1b mtDNAs from Senegalese populations, implying a possible linkage between these population groups. Unlike subhaplogroup L1b mtDNAs, however, these L3a mtDNAs do not have either the HinfI np-10806 site gain or the AluI np-7055 and RsaI np-2758 site losses that define haplogroup L1b. Thus, it appears that the haplogroup L3a mtDNAs acquired the MboI np-2349 site independently of the subhaplogroup L1b mtDNAs.
Haplogroup Distribution in African Populations
The overall distribution of mtDNA haplogroups in African populations, including the Vasikela Kung and the Khwe, is shown in table 2. Both the Mbuti and Biaka Pygmy populations have 100% macrohaplogroup L* mtDNAs. In addition, macrohaplogroup L* haplotypes constitute the large majority of the mtDNAs of the Vasikela Kung, Khwe, and Senegalese groups. However, these groups also encompass significant frequencies of L3 haplotypes, with subhaplogroups L3a–L3d being nonuniformly distributed among these populations. Furthermore, there is some degree of geographic partitioning of the clusters within macrohaplogroup L* (table 2), a trend also noted by Watson et al. (1997). The distribution of haplogroups among these African populations implies that they have experienced quite different genetic histories and that macrohaplogroup L* has undergone considerable regional diversification in Africa.
The Vasikela Kung and the Khwe encompass different arrays of mtDNA haplotypes. Of those present in the Vasikela Kung, 79% and 5% are found in haplogroups L1 and L2, respectively, with these haplotypes belonging to subhaplogroups L1a and L2b (table 2). In addition, Vasikela Kung subhaplogroup L1a mtDNAs segregate into two distinct lineages: L1a2, which encompass >51% of their mtDNAs (AF87–AF91 and AF93–AF95), and L1a1, which encompass 28% of their mtDNAs (AF96–AF97 and AF99–AF101) (figs.2 and3 and table 2). L1a2 is found almost exclusively in the Vasikela Kung, while lineage L1a1 includes Mbuti and Biaka Pygmy mtDNAs (haplotypes AF60–AF61), as well as Khwe mtDNAs; thus L1a1 is not Kung specific.
Among the Khwe, 52% and 16% of the mtDNAs are found in haplogroups L1 and L2, respectively (table 2). With the exception of AF104, which is located in subhaplogroup L1b, within a cluster of related western-African mtDNAs, all the Khwe mtDNAs from haplogroup L1 cluster within subhaplogroup L1a. Within L1a, 32% of the Khwe mtDNAs (AF98–AF99 and AF102–AF103) belong to lineage L1a1, and only 16% occur in lineage L1a2. Lineage L1a1 is also present in the Vasikela Kung and Biaka Pygmies, but L1a2 is found almost exclusively in the Vasikela Kung, with only one Khwe haplotype (AF92) being found in this cluster. Hence, the presence of a Khwe haplotype in lineage L1a2 is most likely due to admixture with Khoisan-speakers.
Both the Vasikela Kung and the Khwe possess haplotypes belonging to subhaplogroup L2b (table 2). The two Vasikela Kung haplotypes from this subhaplogroup (i.e., AF106 and AF107) are closely related. The two Khwe haplotypes include one (AF46) that is the same as that occurring in three Wolof from Senegal (Chen et al. 1995) and another (AF105) that is found in three Khwe and that occupies the nodal position in the haplogroup L2; hence, AF105 could be the founding haplotype for this subhaplogroup. This distribution raises the possibility that the L2b haplotypes found in the Vasikela Kung were acquired by admixture with the Khwe.
Both southern-African populations also have haplogroup L3 mtDNAs (table 2). Two of the L3 haplotypes in the Kung (i.e., AF85 and AF86) are quite distinctive and cluster on a distant branch of haplogroup L3b, haplotype AF85 also occurring in the Khwe. Khwe haplotypes AF80–AF84 all cluster together within haplogroup L3a, with AF82 being shared with the Kung. This pattern of haplotype sharing suggests that the L3 haplotypes in the Vasikela Kung and the Khwe are probably of Khwe origin. Hence, it seems probable that lineage L1a2 mtDNAs originated in the Vasikela Kung and were disseminated into the Khwe, whereas the L2 and L3 mtDNAs originated in the Khwe and were passed on to the Vasikela Kung.
HR-RFLP analysis also reveals several distinct sublineages that appear to be population specific (figs.2 and3 and table 3). Among Khoisan-speakers, the Vasikela Kung have sublineage α within subhaplogroup L1a, the Biaka Pygmies have sublineage β within subhaplogroup L1b, the Mbuti Pygmies have sublineage γ within subhaplogroup L2a, and the Senegalese have sublineage δ within subhaplogroup L2c (fig. 2 and table 3). The presence of such population-specific sublineages within macrohaplogroup L* further demonstrates the extensive regional sequence divergence of African mtDNAs.
Table 3.
Sequence Divergence and Divergence Time of Population-Specific Lineages
| Population-Specific Haplogroup | Primary Defining Polymorphism(s)a | Haplotypes | No. of (%) Subjects | Sequence Divergence(%) | Divergence Time(YBP) |
| Kung: α | −8112 MspI, −8150 MspI, +8249 AvaII, −8250 HaeIII | AF87–AF95b | 26 (59)b | .119 | 41,000–54,100 |
| Biaka Pygmies: β | +10319 AluI | AF62–63, AF66–AF69 | 11 (65) | .225 | 77,600–102,000 |
| Mbuti Pygmies: γ | +11776 RsaI, −13065 DdeI | AF38–AF42 | 10 (45) | .042 | 14,500–19,100 |
| Senegalese: δ | −322 HaeIII, −679 DdeI, −13957 HaeIII | AF49–AF58 | 22 (22) | .052 | 17,900–23,200 |
All population-specific lineages listed are also defined by HpaI 3592 and DdeI 10394 site gains.
Haplotype AF92 was the only haplotype α lineage that was observed in the Khwe; its presence in the Khwe is attributed to genetic admixture with the Vasikela Kung, since five other Vasikela Kung individuals also have AF92 mtDNAs.
Sequence Divergence of African Haplogroups
On the basis of intrapopulation ML calculations, we estimated the divergence times for all major clusters of mtDNAs thus far observed in African populations (table 4 and Appendix B). The sequence-divergence value for all African HR haplotypes is 0.364%, an estimate that gives a maximum age, for the most recent common ancestor (MRCA), of ∼126,000–166,000 years before the present (YBP). Similarly, macrohaplogroup L* shows a sequence divergence of 0.356%, which gives a divergence time of 123,000–162,000 YBP. These findings are concordant with earlier estimates of African mtDNA divergence, which gave coalescence values of 110,000–160,000 YBP (Chen et al. 1995; Graven et al. 1995; Horai et al. 1995; Vigilant et al. 1989, 1991; Watson et al. 1997), and confirm that macrohaplogroup L* is the most divergent of all haplogroups in human populations (Chen et al. 1995). These data further support the hypothesis that, in being the oldest mtDNAs in human populations, these African haplotypes form the root of the modern human mtDNA phylogeny (figs.2 and3).
Table 4.
Sequence Divergence and Divergence Time of African mtDNA Haplogroups
| Haplogroup | No. of Haplotypes | No. of Subjects | Sequence Divergence (%) | Divergence Time(YBP) |
| All African | 107 | 214 | .364 | 125,500–165,500 |
| L | 76 | 164 | .356 | 122,800–161,800 |
| L1 | 40 | 98 | .328 | 113,100–149,100 |
| L1a | 20 | 60 | .265 | 91,400–120,500 |
| L1a1 | 10 | 32 | .166 | 57,200–75,500 |
| L1a2 | 9 | 27 | .119 | 41,000–18,600 |
| L1b | 19 | 36 | .214 | 73,800–97,300 |
| L1b1 | 11 | 21 | .041 | 14,100–18,600 |
| L1b2 | 8 | 15 | .246 | 84,800–111,800 |
| L2 | 36 | 66 | .171 | 59,000–77,700 |
| L2a | 17 | 29 | .113 | 39,000–51,400 |
| L2b | 5 | 9 | .072 | 24,800–32,700 |
| L2c | 10 | 22 | .052 | 17,900–23,600 |
| L3 | 31 | 50 | .227 | 78,300–103,200 |
| L3a | 10 | 17 | .120 | 41,400–54,500 |
| L3b | 9 | 17 | .225 | 77,600–102,300 |
| L3c | 7 | 9 | .082 | 28,300–37,300 |
| L3d | 3 | 5 | .051 | 17,600–23,200 |
Within macrohaplogroup L*, the major clusters of African mtDNAs show varying levels of sequence diversity (table 4). Haplogroup L1 is nearly as diverse as macrohaplogroup L*, whereas haplogroup L2 is half as diverse. This indicates that haplogroup L1 originated well before the emergence of haplogroup L2. Among the smaller haplotype clusters within macrohaplogroup L*, subhaplogroup L1a has the largest divergence value, whereas subhaplogroup L2c has the smallest value, implying that there are considerable differences in times of origin for these haplotype clusters within African populations. Haplogroup L3 is less divergent than L1, and the subhaplogroups of L3 are generally less divergent than those of L1. Only haplogroup L3b is comparable, in terms of sequence diversity, to the oldest subhaplogroups of L1 and L2; however, the L3b value could be inflated, because of the inclusion of the highly divergent haplotypes AF85 and AF86, since these southern-African haplotypes are at least five mutations different from any other haplotypes (AF04–AF06 and AF08–AF11) of this Senegalese-specific subhaplogroup (Chen et al. 1995). Therefore, it appears that the L3 subhaplogroups arose long after the radiation of haplogroup L1 and approximately at the time that haplogroup L2 split from haplogroup L1.
Sequence-divergence estimates were also calculated for each of the population-specific sublineages (table 3). These results show that sublineages found in the Mbuti Pygmies (γ) and the Senegalese (δ) have approximately comparable divergence values (0.042%–0.052%). By contrast, the Vasikela Kung lineage (α) is twice as diverse (0.119%), and the Biaka Pygmy (β) is four times as diverse (0.225%), as the Mbuti Pygmies and Senegalese.
These diversity levels suggest that the Biaka Pygmies of sublineage β may be one of the oldest distinct African populations and, hence, one of the oldest human populations in the world, although other demographic factors could have contributed to this level of diversity. The Vasikela Kung sublineage α may be the other truly ancient African mtDNA cluster. Not only is sublineage α of the Kung the second most diverse, but it is positioned at the deepest root of the African phylogenetic tree (figs.2 and3), suggesting that the Kung San became differentiated very early during human radiation.
Genetic Divergence of African Populations
The intra- and interpopulation sequence divergences within and between African populations are shown in table 5. The Biaka Pygmies have the highest intrapopulation sequence divergence (0.342%), followed by the Vasikela Kung (0.320%) and then the Khwe (0.277%). In addition, all the interpopulation sequence-divergence values between Vasikela Kung and other African populations are higher than those between the Khwe and other African populations. Thus, the Vasikela Kung are more divergent from other African populations than are the Khwe. This finding is consistent both with the Vasikela Kung being genetically distinct from the Khwe (Nurse et al. 1977) and with the Khwe being more closely related to the Negroid populations of South Africa, who, in turn, show greater affinities with populations from western and central Africa.
Table 5.
Sequence Divergence Within and Between African Populations[Note]
|
Sequence Divergence(%) |
||||||
| Kung(N=43) | Khwe(N=31) | Mandenkalu(N=60) | Wolof(N=20) | Mbuti Pygmies (N=22) | Biaka Pygmies (N=17) | |
| Kung | .320 | .334 | .366 | .368 | .378 | .400 |
| Khwe | .036 | .277 | .300 | .298 | .315 | .353 |
| Mandenkalu | .069 | .024 | .274 | .275 | .294 | .332 |
| Wolof | .076 | .028 | .007 | .263 | .283 | .369 |
| Mbuti Pygmies | .098 | .056 | .036 | .032 | .241 | .371 |
| Biaka Pygmies | .068 | .043 | .024 | .067 | .080 | .342 |
Note.— Intrapopulation sequence-divergence values are on the diagonal (underlined), whereas the raw and corrected interpopulation sequence-divergence values are above and below the diagonal, respectively.
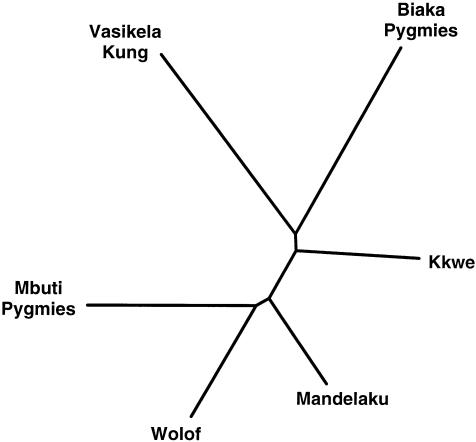
These same relationships are seen quite clearly in the NJ tree based on these ML estimates (fig. 4 and Appendix B). In this tree, the Khwe clearly cluster more closely to other Bantu-speaking Senegalese groups—the Mandelaku and Wolof—than to the Vasikela Kung. The Vasikela Kung, on the other hand, form a distant outlier between the Biaka and Mbuti Pygmy populations. In addition, the Mbuti Pygmies are closer to western-African Bantu-speaking groups than to the Biaka Pygmies, a relationship that reflects the considerably different haplotype compositions of the two populations. The marked divergence between the two Pygmy groups raises the possibility that these two Pygmy populations had independent origins.
Figure 4.
NJ tree of ML genetic distances for major African populations (table 5).
Distribution of CR Sequences in Southern-African Populations
A total of 640 nucleotides from HVS-I and HVS-II of the CRs for all 74 southern-African individuals were sequenced. The sequence data from the Vasikela Kung and Khwe are shown in table 6. A total of 24 distinct CR sequences based on 65 variable nucleotides were detected. Of the 65 variable nucleotides found in this study, only 31 had previously been reported in the study of CR sequence variation in the Botswana Kung (Vigilant et al. 1989). The vast majority (84%) of these nucleotide substitutions are transitions, both in the HVS-I and in the HVS-II, a result that is consistent with patterns of variation observed in previous CR-sequencing studies of human populations (Horai and Hayasaka 1990; Vigilant et al. 1991).
Table 6.
Kung and Khwe CR Sequences
|
No. in |
Positions in |
||||
| CR | Haplotype(s) (Haplogroup) | Kung | Khwe | HVS-1 | HVS-2 |
| 1 | AF87, AF90 (L1a) | 8 | … | 16167T, 16187T, 16189C, 16223T, 16230G, 16234T, 16242T, 16243C, 16311C | 73G, 146C, 152C, 195C, 198T, 247A, 315C+ |
| 2 | AF88–AF89 (L1a) | 8 | … | 16167T, 16187T, 16189C, 16223T, 16230G, 16234T, 16242T, 16243C, 16311C | 73G, 146C, 152C, 195C, 198T, 247A, 309C+, 315C+ |
| 3 | AF93 (L1a) | 1 | … | 16153A, 16187T, 16189C, 16223T, 16230G, 16243C, 16294T, 16311C | 73G, 146C, 152C, 195C, 247A, 315C+ |
| 4 | AF94 (L1a) | 1 | … | 16129A, 16187T, 16189C, 16230G, 16234T, 16243C, 16266A, 16311C | 73G, 146C, 152C, 199C, 247A, 315C+ |
| 5 | AF93 (L1a) | 1 | … | 16129A, 16187T, 16189C, 16230G, 16234T, 16243C, 16264T, 16266G, 16311C | 73G, 146C, 152C, 195C, 199C, 247A, 315C+ |
| 6 | AF95 (L1a) | 1 | … | 16093C, 16129A, 16187T, 16189C, 16230G, 16234T, 16243C, 16266G, 16311C | 73G, 146C, 152C, 195C, 199C, 247A, 315C+ |
| 7 | AF91 (L1a) | 2 | … | 16129A, 16179T, 16187T, 16189C, 16223T, 16230G, 16243C, 16311C | 73G, 146C, 152C, 188G, 195C, 247A, 315C+ |
| 8 | AF92 (L1a) | 5 | 16129A, 16179T, 16187T, 16189C, 16223T, 16230G, 16243C, 16311C | 73G, 146C, 152C, 188G, 195C, 247A, 309C+, 315C+ | |
| 9 | AF104 (L1b) | … | 1 | 16126C, 16187T, 16189C, 16223T, 16264T, 16270T, 16278T, 16311C | 73G, 151T, 195C, 247A, 263A, 315C+ |
| 10 | AF85–AF86 (L3b) | 5 | … | 16124C, 16183C, 16189C, 16193C+, 16223T, 16278T, 16304C, 16311C | 73G, 152C, 195C, 263G, 309C+, 315C+ |
| 11 | AF85 (L3b) | … | 1 | 16124C, 16183C, 16189C, 16193C+, 16223T, 16278T, 16304C, 16311C | 73G, 152C, 263G, 315C+ |
| 12 | AF105 (L2) | … | 3 | 16189C, 16192T, 16223C, 16278T, 16294T, 16309G, 16390A | 73G, 146C, 152C, 195C, 263G, 309C+, 315C+ |
| 13 | AF46 (L2b) | … | 2 | 16114A, 16129A, 16213A, 16223T, 16278T, 16284G, 16355T, 16362C, 16390A | 73G, 150T, 151T, 152C, 182T, 195C, 198T, 204C, 263G |
| 14 | AF106–107 (L2b) | 2 | … | 16114A, 16129A, 16213A, 16223T, 16278T, 16354T, 16390A | 73G, 150T, 151T, 152C, 182T, 195C, 198T, 204C, 263G |
| 15 | AF80–AF82, AF84 | 2 | 3 | 16172C, 16189C, 16223T, 16320T | 73G, 150T, 152C, 195C, 263G, 315C+ |
| 16 | AF83 (L3a) | … | 6 | 16185T, 16209C, 16223T, 16327T | 73G, 150T, 152C, 189G, 195C, 200G, 207A, 263G, 309C+, 315C+ |
| 17 | AF98–AF99 (L1a) | 1 | 7 | 16166C, 16172C, 16187T, 16189C, 16209C, 16214T, 16223T, 16230G, 16278T, 16291G, 16311C | 73G, 146C, 152C, 189G, 195C, 198T, 247A, 315C+ |
| 18 | AF101 (L1a) | 2 | … | 16166C, 16172C, 16187T, 16189C, 16209C, 16214T, 16223T, 16230G, 16278T, 16291G, 16311C | 73G, 146C, 152C, 189G, 195C, 198T, 204C, 207A, 247A, 309C+, 315C+ |
| 19 | AF101 (L1a) | 2 | … | 16166C, 16172C, 16189C, 16209C, 16214T, 16223T, 16230G, 16278T, 16291G, 16311C | 73G, 146C, 152C, 189G, 195C, 198T, 204C, 207A, 247A, 309C+, 315C+ |
| 20 | AF97 (L1a) | 1 | … | 16166C, 16172C, 16187T, 16189C, 16209C, 16214T, 16223T, 16230G, 16278T, 16291G, 16311C | 73G, 146C, 152C, 189G, 195C, 198T, 207A, 247A, 309C+, 315C+ |
| 21 | AF97, AF100 (L1a) | 5 | … | 16166C, 16172C, 16187T, 16189C, 16209C, 16214T, 16223T, 16230G, 16278T, 16291G, 16311C | 73G, 146C, 152C, 189G, 195C, 198T, 207A, 247A, 315C+ |
| 22 | AF103 (L1a) | … | 1 | 16129A, 16148T, 16172C, 16187T, 16188G, 16189C, 16223T, 16230G, 16311C, 16320T | 73G, 95C, 152C, 189G, 207A, 236C, 247A, 263G, 309C+, 315C+ |
| 23 | AF96 (L1a) | 1 | … | 16129A, 16148T, 16169T, 16172C, 16187T, 16188G, 16189C, 16223T, 16230G, 16278T, 16311C | 93G, 146C, 152C, 185A, 189G, 195C, 198T, 207A, 236C, 247A, 263G, 315C+ |
| 24 | AF102 (L1a) | … | 2 | 16129A, 16148T, 16168T, 16172C, 16187T, 16188G, 16189C, 16223T, 16230G, 16278T, 16293G, 16311C, 16320T, 16344T | 93G, 95C, 185A, 189G, 236C, 247A, 263G, 315C+ |
The distribution of CR sequences in the Vasikela Kung and the Khwe show them to be quite distinctive from each other; indeed, there is virtually no overlap of their CR sequences (table 6). Therefore, each CR sequence has been given a two-letter prefix indicating its probable population of origin, as shown in figure 5. The only CR sequence that the Vasikela Kung and the Khwe populations have in common is CR 17, which is associated with haplotype AF99. Since this mtDNA is found in seven Khwe but in only one Vasikela Kung, we have given it a Khwe designation, “KW17.” Similarly, two very similar CR sequences—10 and 11—share the same haplotype, AF85, that is found in five Kung but in only one Khwe. Hence, the AF85 CR sequence 11 that appears in the Khwe is most likely of Vasikela Kung origin and therefore has been given the designation “VK11.”
Figure 5.
NJ tree based on HVS-I and HVS-II sequences from Vasikela Kung and Khwe (present study) and Botswana Kung (Vigilant et al. 1989). The CR sequence numbers are positioned at the end of each branch; VK = Vasikela Kung sequence, KW = Khwe sequence, and BK = Botswana Kung sequences; and the associated HR haplotypes are in parentheses. LR types are indicated in boldface italic, and the square brackets indicate all CR sequences associated with them. The major haplogroups and subhaplogroups to which these mtDNAs belong are shown within the boxes. Bootstrap values for each major branch of the tree are shown at their corresponding positions. This tree was rooted from a chimpanzee CR sequence (Horai et al. 1995).
Phylogenetic Analysis of CR Sequences
The phylogenetic relationships among the CR sequences of the Vasikela Kung, Khwe, and Botswana Kung (Vigilant et al. 1989) are shown in figure 5 and table 6. In general, there is a very good correspondence between the groupings identified by CR sequences and those identified by HR-RFLP haplotypes (figs.2 and3). A similar correlation between CR sequences and LR-RFLP types was seen, by Graven et al. (1995), in the Mandenka of Senegal. This correspondence is strongest when only HVS-I sequences are used in the comparison, since the addition of HVS-II sequences creates minor difficulties in the resolution of certain branching relationships among these mtDNAs, a trend noted in previous studies of CR sequence variation (Horai and Hayasaka 1990; Vigilant et al. 1991). In addition, when only HVS-I sequences are compared, there is no major difference, in the branching order or structure of the NJ tree, irrespective of whether the chimpanzee or Neanderthal mtDNA is used to root the tree, or whether midpoint rooting is used to construct the tree.
The CR sequences of all L1a subhaplogroup sequences—both those in L1a1 and those in L1a2—cluster together with one another and apart from those belonging to haplogroups L2 and L3 (fig. 5). The CR sequences also split lineages L1a1 and L1a2 into distinct branches, encompassing distinct subclusters, including ones that are constituted almost solely of population-specific mtDNAs. By contrast, the CR sequences of haplogroups L2 and L3 cluster closer to each other, as they do in the RFLP phylogeny (figs.2 and3), while also showing some degree of genetic substructure.
Although there is a good correspondence between the RFLP haplotypes and CR sequences, specific RFLP haplotypes have multiple CR sequences associated with them (e.g., AF93 and CR sequences VK3 and VK5). In other cases, the same CR sequence is associated with multiple RFLP haplotypes (e.g., CR sequence KW15 with AF80–82 and AF84). These observations indicate that both HR-RFLP and CR sequence data provide valuable phylogenetic information about human mtDNAs and are more valuable when used together than when employed separately.
With regard to the population-specific clusters, the CR sequences for the Kung San populations generally segregate into distinct clusters of related types. With one exception (BK12), all the Botswana Kung CR sequences (BK01–BK11 and BK13–BK15) cluster together within lineage L1a2 (fig. 5), along with similar Vasikela Kung CR sequences (VK01–VK07). The large majority of the CR sequences in the Botswana Kung cluster within a single group to which virtually no Vasikela Kung mtDNAs belong. The remainder cluster more closely with CR sequences in the Vasikela Kung that correspond to sublineage α. This pattern of CR sequence diversity within subhaplogroup L1a, for both the Botswana Kung and the Vasikela Kung, suggests a significant degree of genetic differentiation of the Kung San populations, although the perceived difference between the Botswana Kung and Vasikela Kung could simply reflect limited or regional sampling (Vigilant et al. 1989, 1991).
Discussion
The Origins of African Populations
Studies of African mtDNA sequence variation that have used HR-RFLP analysis (Chen et al. 1995; present study) have confirmed the ubiquity of macrohaplogroup L* in African populations. Moreover, we have documented that macrohaplogroup L* is the oldest African and human mitochondrial lineage, encompassing a sequence diversity of 0.364% and having an estimated age ∼126,000–165,000 YBP; as such, it must be considered the root of the human mtDNA phylogeny, from which all other haplogroups evolved. Macrohaplogroup L* is further subdivided into two major haplogroups: L1 and L2 (figs.2 and3 and table 2). Haplogroup L1 is the most ancient of African haplogroups, and it is further subdivided into subhaplogroups L1a and L1b, with L1a being the older of the two (table 4). L1a is further subdivided into lineages L1a1 and L1a2 (table 2). Lineage L1a2 contains the core population-specific haplotypes (α) of the Vasikela Kung, and lineage L1b2 contains the population-specific haplotypes (β) of the Biaka Pygmies, the two most ancient and distinct African populations known (figs.2 and3 and tables 2 and 3).
Haplogroup L2 is considerably younger than haplogroup L1, being approximately half the age of the latter. It is subdivided into three subhaplogroups: L2a, L2b, and L2c. L2a contains the core population-specific haplotypes (γ) of the Mbuti Pygmies, whereas L2c contains the core haplotypes (δ) of the Bantu-speaking Senegalese (figs.2 and3 and tables 2 and 3).
The existence of four distinct L3 subhaplogroups (Chen et al. 1995) also has been confirmed. Of these, L3a and L3b are the oldest, whereas L3c and L3d are of more recent origin. In addition, L3 mtDNAs have been found to be much more closely related to those from L2 than either is to L1, by both RFLP and CR sequence analysis (figs.2 and3 and table 2). These findings suggest that haplogroups L2 and L3 have a more recent origin than does haplogroup L1 and that the split of L3 from L2 happened relatively soon after L2 split from L1 (figs.2 and3 and table 4).
An unexpected result is the marked difference, in haplotype distribution, between the Mbuti and Biaka Pygmy populations. Not only do the mtDNAs of these two populations form distinct clusters by parsimony analysis (figs.2 and3), but they branch separately in the NJ tree of RFLP genetic distances (fig. 4). The distinction between the two Pygmy populations is further demonstrated by the significantly greater sequence diversity of the Biaka Pygmies (0.342%) relative to that of the Mbuti Pygmies (0.241%). This difference suggests that the Biaka Pygmies arose before the Mbuti Pygmies (table 3) and represent one of the oldest African populations, whereas the Mbuti Pygmies appear to have arisen independently and more recently. This conclusion is consistent with the nuclear genetic data reported by Cavalli-Sforza et al. (1994) and with the different linguistic affiliations of the two populations, with the Mbuti speaking a Nilo-Saharan language and with the Biaka speaking a Niger-Kordofarian language (Greenberg 1963).
The Vasikela Kung mtDNA data also suggest a relatively ancient origin for the Kung San. The population-specific haplotypes of the Biaka Pygmies (β) and the Vasikela Kung (α) are both located in the ancient African haplogroup L1. Moreover, the Vasikela Kung sublineage α haplotypes form the deepest root of the African tree, relative to the chimpanzee outgroup haplotype (figs.2 and3). Thus, the Kung must also represent one of the most ancient modern human populations. The antiquity of the Vasikela Kung is further emphasized by the high intragroup sequence diversity, 0.320%, of this population, a value only slightly smaller than that of the Biaka Pygmies (0.342%) (table 5). Thus, our data suggest that the Biaka Pygmies and the Vasikela Kung represent the earliest modern human populations, whereas the Mbuti Pygmies and the Bantu-derived Senegalese have emerged more recently.
The Relationship between the Kung and the Khwe
The Vasikela Kung and the Khwe both speak Khoisan languages. However, the Khwe have a physical appearance that is more similar to that seen in many Bantu-derived populations than to that of the Vasikela Kung. Therefore, the Khwe may have originated from the Bantu migration into southern Africa and subsequently may have adopted the Khoisan language. This possibility is supported both by the striking differences between the population-specific haplotypes of the Kung San (α) and those of the Bantu-speaking Senegalese (δ) and by the greater similarities between the Khwe and Senegalese haplotypes than between the Kung and the Khwe haplotypes.
To most effectively address this question, we have compared the mtDNA variation in all of the southern-African populations that have been studied thus far. However, with the exception of our current and previous (Chen et al. 1995) studies, all African mtDNA RFLP variation has been analyzed by LR-RFLP methods rather than by HR-RFLP methods. As noted above, LR-RFLP analysis typically employs the five rare-cutting restriction endonucleases—HpaI, BamHI, HaeII, MspI, and AvaII—and, in some instances, HincII also has been included (Johnson et al. 1983; Scozzari et al. 1988; Soodyall and Jenkins 1992, 1993). Fortunately, the most informative restriction-site polymorphisms for African mtDNAs are recognized by HpaI, MspI, and AvaII, which are included in both LR- and HR-RFLP analysis. Hence, once nomenclatural differences between LR- and HR-RFLP haplotypes have been resolved, a constructive population comparison can be made for African populations, by use of both types of data.
LR-RFLP analysis involves examination of restriction-fragment patterns by use of Southern blot analysis, with a unique restriction-endonuclease pattern being called a morph and being given a specific morph number. The sum of all of the individual morphs for a particular mtDNA is defined as its mtDNA type. The LR-RFLP types are numbered and described by listing the morph numbers of the five restriction endonucleases, in the order HpaI-BamHI-HaeII-MspI/HpaII-AvaII. Thus, type 1 is defined by morph-2 for HpaI, morph-1 for BamHI, morph-1 for HaeII, morph-1 for MspI/HpaII, and morph-1 for AvaII, which can be written, in shorthand form, as “2-1-1-1-1.” Later, when HincII was added to the enzyme set, its morph was appended to the type number, such that a type 1 mtDNA with a HincII morph-2 became type 1-2. Most of the variation represented by these morphs can now be equated with the presence or absence of specific restriction sites identified by HR-RFLP analysis. Thus, the LR-RFLP types can be equated with HR-RFLP haplogroups, at the restriction site—and, thus, at the nucleotide level. These associations can be further refined by use of CR sequence data.
HpaI morph-3 is the most common African HpaI restriction pattern. By contrast, HpaI morph-2 delineates all L3 African mtDNAs and is found in all European and Asian mtDNAs. HpaI morph-3 differs from morph-2 by having the HpaI site gain at np 3592. Thus HpaI np-3592 morph 3 defines macrohaplogroup L*, and distinguishes L* from L3 and the remaining global mtDNAs. Accordingly, all African LR-RFLP types that start with a 3 (e.g., 3-1-1-1-1-2 or type 7-2) can be considered L*(L1 or L2) mtDNAs, while those which start with a 2 (e.g., 2-1-1-1-1-2 or Type 1-2) can be considered L3 mtDNAs (table 7).
Table 7.
Distribution of LR-RFLP Types in Khoisan Populations and in Neighboring Groups
|
No. in |
|||||||||
| LR Type (Designation) [HR Haplogroup]a | Vasikela Kung (N=43) | Botswana Kungb (N=34) | Sekele Kungc (N=49) | Namad (N=46) | Khwee (N=41) | Bantub (N=40) | Hereroc (N=54) | Damac (N=43) | Amboc (N=22) |
| 2-1-1-1-1-2 (1-2) [L3a] | … | 2.9 | 2.0 | 6.5 | 26.8 | 25.0 | 33.3 | 20.9 | 13.6 |
| 2-1-1-1-2-2 (21-2) [L3b] | 11.6 | … | 4.1 | 13.0 | 2.4 | … | 50.0 | 32.6 | 4.5 |
| 2-1-1-1-12-2 (72-2) [L3a] | 4.6 | … | … | … | … | … | … | … | … |
| 3-1-1-1-3-2 (2-2) [L2] | 4.7 | 2.9 | … | … | 17.1 | 32.5 | … | … | 13.6 |
| 3-1-1-2-2-2 (3-2) [L1a2] | 14.0 | 26.5 | 14.3 | 54.3 | 17.1 | 7.5 | 5.6 | 2.3 | 4.5 |
| 3-1-1-3-2-2 (4-2) [L1a2] | 20.9 | 26.5 | 40.8 | … | … | … | … | … | … |
| 3-1-1-3-2-9 (4-9) [L1a2] | 4.7 | … | 2.0 | … | … | … | … | … | … |
| 3-1-1-3′-2-2 (4′-2) [L1a2] | 11.6 | … | … | … | … | … | … | … | … |
| 3-1-1-2-5-2 (5-2) [L1a2] | … | 20.6 | 2.0 | 6.5 | … | … | … | … | … |
| 3-1-1-1-1-2 (7-2) [L1] | 18.6 | 5.9 | 20.4 | 6.5 | 36.6 | 10.0 | … | 2.3 | 54.5 |
| 3-1-1-1-2-2 (10-2) [L1a1] | 9.3 | 5.9 | 12.2 | 6.5 | … | 5.0 | 1.9 | … | … |
| 3-1-1-2-4-2 (14-2) [L1] | … | 5.9 | … | … | … | … | … | … | … |
| 3-1-1-5-1-2 (32-2) [L1] | … | 2.9 | … | … | … | 2.5 | … | … | … |
| 3-1-1-12-1-2 (113-2) [L1] | … | … | 2.0 | … | … | … | … | … | … |
| 3-1-1-1-6-2 (140-2) [L1] | … | … | … | 2.2 | … | … | … | … | … |
| 7-1-6-13-1-2 (139-2) [?] | … | … | … | 4.3 | … | … | … | … | … |
HR haplogroups were assigned to LR types according to the RFLPs that define these types. The LR designations are according to the nomenclature that was developed by Johnson et al. (1983) and that subsequently was elaborated in many subsequent studies (Bonné-Tamir et al. 1986; De Benedictis et al. 1989; Scozzari et al. 1988, 1994; Semino et al. 1989; Soodyall and Jenkins 1992, 1993; Graven et al. 1995). The LR haplotypes are defined by restriction-endonuclease morphs, in the order HpaI-BamHI-HaeII-MspI-AvaII-HincII; those types defined by Johnson et al. (1983) would have an “#” in place of the HincII morph number, since this enzyme was not used to characterize mtDNA variation; however, since the vast majority of mtDNAs from various world populations have HincII morph-2, it is assumed to be the default morph for this enzyme unless additional data show otherwise (see text). The following restriction-site polymorphisms identify each LR-RFLP haplotype: 1 = no polymorphisms relative to CR sequence; 2 = +3592h, −16390b; 3 = +3592h, −8112i, −8150i, +8249b; 4 = +3592h, −8112i, −8150i, +8249b, +11436i; 4-9 = +3592h, −8112i, −8150i, +8249b, +11436I, +13259o; 4′-2 = +3592h, −8112i, −8150i, +8249b, +11436i, +16494i; 5 = +3592h, −8112i, −8150i, +8249b; 7 = +3592h; 10 = +3592h, +8249b; 14 = +3592h, −8112i, −8150i, +8249b, +15890b; 21-2 = +8249b; 32 = +3592h, −8112i, −8150i, +13070i, +8249b; 72-2 = −12629b; 113-2 = +3592h, +8469i; 140-2 = +3592h, +15882b; and 139-2 = −12406h, +9250n, +11164i.
Data are from Johnson et al. (1983).
Data are from Soodyall and Jenkins (1992).
Data are from Soodyall and Jenkins (1993).
The data obtained in the present study were combined with those reported, by Soodyall and Jenkins (1992), for the related Kwengo.
Similarly, the MspI/HpaII morphs delineate major subsets of African mtDNAs. These are represented by the fourth number in the LR-RFLP type designation (table 7). MspI-2 is defined by site losses at np 8112 and np 8150. These are the primary site changes that define the HR-RFLP lineage L1a2 and the Kung San-specific sublineage α (figs.2 and3). Similarly, MspI-3 is defined by the site losses of MspI-2, as well as by an additional site gain at np 11436. This new polymorphic site further subdivides lineage L1a2, into two major subgroups (figs.2 and3). Finally, MspI -3′ has the same site changes as does MspI-3, together with an additional site gain at np 16494. This further subdivides sublineage-α mtDNAs. Thus, all mtDNA types having MspI morphs 2, 3, and 3′ correspond to the Vasikela Kung–specific sublineage α in L1a2 (figs.2 and3 and table 7).
Several of the AvaII morphs also correspond to RFLPs that define major sublineages of the African phylogeny. The first is AvaII-2, which is defined by a site gain at np 8249. This site appears in haplotypes from lineage L1a2 and from the Vasikela Kung–specific sublineage α (figs.2 and3 and table 7). AvaII-3 is defined by a site loss at np 16390, and AvaII-5 is defined by both a site gain at np 8249 and a site loss at np 16390. Thus, AvaII-5 could derive from either AvaII-2 or AvaII-3.
Therefore, the LR-RFLP sites for HpaI-3, for MspI -2, -3, and –3′, and for AvaII-2 all help to define the lineage L1a2, which encompasses the Kung-specific mtDNA sublineage α. Consequently, mtDNA types 3-2, 4-2, 4-9, 4′-2, 5-2, 14 –2, and 32-2 (all HpaI-3 and MspI-2 or 3) can be considered to be of Kung San origin (table 7). As a result, these mtDNA types can be used to determine the relative representation of Kung San–derived mtDNAs in other African populations that have been analyzed by LR-RFLP analyses (table 7).
By contrast, mtDNAs that are HpaI-2 and MspI-1 belong to haplogroup L3, with type 1-2 (i.e., 2-1-1-1-1-2) belonging to subhaplogroup L3a and with type 21-2 (i.e., 2-1-1-1-2-2) belonging to subhaplogroup L3b. Since the Senegalese have a much higher frequency of L3 types than does the Kung population, the presence of these mtDNAs are more indicative of a Bantu origin than of a Kung origin (table 7).
Using these distinctions, we can now compare our current data on the Vasikela Kung and Khwe with those from other LR-RFLP studies of the Kung San and Bantu populations (table 7). The three Kung San groups that have been studied are the Vasikela Kung, the Botswana Kung, and the Sekele Kung. The single Khoi group that has been examined is the Nama, whereas the Bantu-speaking groups hat have been analyzed include the Bantu, Herero, Dama, and Ambo.
The Kung-associated mtDNA types (3-2, 4-2, 4-9, 4′-2, 5-2, 14-2, and 32-2) constitute 51% of the Vasikela, 88% of the Botswana, and 59% of the Sekele Kung mtDNAs (table 7). Of these, types 4-2, 4-9, and 4′-2 are Kung specific, whereas types 3-2 and 7-2 are present in all southern-African populations, although at generally higher frequencies in the Kung and Khoi (Nama) (table 7). By contrast, the Khwe lack virtually all the Kung San–specific L1a2 types and, instead, exhibit 29% of the Bantu-associated types 1-2 and 21-2 of haplogroup L3. This frequency of L3 mtDNAs in the Khwe is comparable to the 18%–83% seen in the Bantu-derived groups but is quite different from the 3%–12% seen in the Kung San populations (table 7). Thus, on the basis of these data, as well as on the basis of genetic-distance analysis (fig. 4 and table 5), the Khwe appear to be much more closely related to Bantu-derived populations than to the other Khoisan-speaking populations (Nurse et al. 1985).
Since nearly all of the haplotypes in Bantu-speaking populations from western Africa and in the Pygmy populations from eastern and central Africa are MspI-1 (Chen et al. 1995; Graven et al. 1995; authors' unpublished data), MspI-2 must be of Khoisan origin. This further supports the conclusion that the MspI-2 mtDNAs in the Bantu-speaking populations have been acquired through genetic admixture with Khoisan-speaking populations since the Bantu expansion into southern Africa.
The population distribution of the individual LR-RFLP morphs can also permit deduction of their interrelatedness; for example, AvaII-5 is defined by the np-8249 site gain of AvaII-2 and by the np-16390 site loss of AvaII-3. Therefore, it could have derived from either of these morphs. However, both AvaII-2 and AvaII-5 occur primarily in the Kung San, whereas AvaII-3 does not. Therefore, it is most likely that AvaII-5 derived from AvaII-2 within the Kung population, through the loss of the np-16390 site. A parallel and independent mutation must then have created the np-16390 site loss seen in AvaII-3. This conclusion is supported by the fact that the np-16390 site is located in the hypervariable CR at the very end of HVS-I and, thus, is prone to repeated mutational events.
Correlation between RFLP and CR Sequence Data
In most respects, the phylogenetic relationships between the CR sequences of the Kung and Khwe correlated well with those seen for the combined LR- and HR-RFLP haplotype data (fig. 5 and table 7). The CR sequences of haplogroup L1 mtDNAs were clearly delineated from those of L2 and L3, and the four subhaplogroups of L3 were also distinctive. In a number of cases, the CR sequence provided more information than did HR-RFLP analysis. Indeed, the CR sequence data revealed several distinct subdivisions within these RFLP groupings; for example, lineage L1a1 was divided into two major subbranches by CR sequences (AF97–AF99 and AF100-AF101 in one branch and AF96, AF102, and AF103 in a distinct second branch) (fig. 5). Moreover, some HR-RFLP haplotypes have more than one CR sequence (AF93 with VK3 and VK5). By contrast, in other instances the CR sequences provided less resolution. CR sequences were unable to clearly distinguish between L2 and L3, such that, in the absence of supporting RFLP data, the L2 and L3 CR sequences could be interpreted as belonging to the same haplogroup (fig. 5). Furthermore, certain CR sequences were found to be present in several different RFLP haplotypes (CR 15 is common to haplotypes AF80–AF82 and AF84) (table 6). Only when both RFLP and CR sequence data are used can the optimal phylogenetic relationships between African mtDNAs be resolved.
The conclusion that the deepest root of the African mtDNA tree occurs at the Vasikela Kung sublineage α—and that the Kung and the Biaka and Mbuti Pygmies harbor distinctive groups of mtDNA types—is supported by both RFLP and CR sequence data (figs.2,3, and 5). This result is fully consistent with earlier analyses, by Vigilant et al. (1989, 1991), of African and non-African CR sequences and with our own RFLP data (Chen et al. 1995). Thus, all of these studies place the Kung San among the oldest African and extant human populations.
The importance of macrohaplogroup L*, with its component haplogroups L1 and L2, as opposed to haplogroup L3 (non-L), was also confirmed in another study, by Watson et al. (1997), of African mtDNA CR sequence variation. In that study, a total of 407 African samples, including Kung San, Mbuti and Biaka Pygmies, and multiple Bantu-derived populations, were analyzed, by CR sequencing and screening, for both the presence of the HpaI np-3592 site (delineating L*) and the absence of the AvaII np-16390 site (equal to the presence of the HinfI site at np 16389 and delineating L2) (figs.2 and3 and table 2). This analysis confirmed the distinctive nature of haplogroups L1–L3, which we previously had described (Chen et al. 1995), and also revealed that haplogroup L3 has three distinct sublineages: L3a, L3b, and L3c (Watson et al. 1997).
The primary question addressed by Watson et al. (1997) is the number and nature of African population expansions. To address this question, they examined the sequence diversity of these samples, partitioning their CR sequences into those which were observed in more than one population (87%) versus those which were found in only one population (13%). The multipopulation CR sequences were then used to construct a median network, which resulted in the separation of these sequences into haplogroups L1 (a and b), L2, and L3 (a–c). Estimation of the distribution of pairwise differences of and between haplogroups L1a, L1b, L2, L3a, and L3b revealed all of them to have unimodal distributions, a finding that is consistent with each haplogroup being associated with a different population expansion. The remaining population-specific CR sequences (termed “isolated lineages”) were then combined into a new category termed “L1i.” A median network of the L1i sequences converged on a MCRA having CR transition mutations at np 16129, 16187, 16189, 16223, 16230, 16278, and 16311; however, this L1i network was not starlike, and the pairwise distribution of CR sequences was not unimodal.
The nature of Watson et al. (1997)’s L1i haplogroup can be more fully explained if it is compared with our data (Chen et al. 1995; present study). By placing all of the population-specific CR sequences into L1i, Watson et al. (1997) combined mtDNAs from the Kung San–specific sublineage α from L1a, the Biaka Pygmy sublineage β from L1b, the Mbuti Pygmy sublineage γ from L2a, and the Senegalese sublineage δ from L2a. In addition, a comparison of the variant sites of their MRCA with the data from our study shows that it has the greatest similarities to our CR sequences 4, 5, 7, and 8 of lineage L1a2, sequence 9 of subhaplogroup L1b, and sequences 22–24 of lineage L1a1 (fig. 5 and table 6). Not surprisingly, therefore, L1i coalesces to the deepest root of the African phylogeny, at subhaplogroup L1a, because it encompasses widely different clusters of haplotypes from both haplogroup L1 and haplogroup L2; for this same reason, it follows that L1i is not starlike or unimodal, since it subsumes several different population-specific sublineages.
Our estimates of the age of the African population and its haplogroups are also comparable to estimates made by other investigators. In our studies, the antiquity of African mtDNAs has been demonstrated by means of a sequence-divergence time of 125,500–165,500 YBP (Chen et al. 1995; present study). Similarly, the coalescence time for the MRCA, as calculated by Watson et al. (1997) on the basis of their L1i cluster, was 111,000 ± 5,700 YBP; that estimated by Horai et al. (1995) on the basis of whole mtDNA sequences was 143,000 ± 18,000 YBP; and that estimated by Cann et al. (1987) on the basis of RFLP haplotype data was 120,000 YBP. By contrast, the age of haplogroup L2 was much younger, being 59,000–77,700 YBP by our analysis, 56,000 ± 3,000 YBP according to Watson et al. (1997), and 70,333 ± 25,710 YBP according to Graven et al. (1995). Finally, the age of haplogroup L3 is 78,300–103,200 YBP by our analysis, 60,000 ± 2,400 YBP according to Watson et al. (1997), and 66,321 ± 24,965 YBP according to Graven et al. (1995). Thus, all of the most recent studies show consistent trends in the diversity and ages of the major clusters of mtDNAs in African populations.
The other interesting question raised both by our analyses (Chen et al. 1995; present study) and by that of Watson et al. (1997) is the origin and dispersal of haplogroup L3 mtDNAs. These haplotypes are widely dispersed in eastern Africa but are less prevalent in western and southern Africa (table 2; also see Watson et al. 1997; Passarino et al. 1998). Moreover, there are several different subhaplogroups within L3 (figs.2 and 4), and these appear to be differentially distributed in these populations (table 2). On the basis of the continental distribution and age that they reported, Watson et al. (1997) proposed that a subset of L2 and L3 mtDNAs moved out of eastern Africa to found the Eurasian populations ∼60,000 YBP.
Since the subhaplogroups of L3 are the most likely precursors of modern European and Asian mtDNA haplotypes (Chen et al. 1995; Watson et al. 1997), their sequence variation and age are of considerable importance in the determination of the timing and process by which these mtDNAs were dispersed out of Africa. In this regard, subhaplogroups L3a and L3b appear to be the oldest of the L3 subhaplogroups, dating to 41,400–54,500 YBP and 77,600–102,300 YBP, respectively. These estimates are somewhat similar to the 60,000 ± 3,200 YBP and 44,000 ± 3,000 YBP coalescence times that Watson et al. (1997) calculated for the analogous subhaplogroups. In contrast, subhaplogroups L3c and L3d have somewhat younger divergence times—28,300–37,300 YBP and 17,600–23,200 YBP, respectively (table 4)—suggesting that they emerged after the evolution of L3a and L3b.
We have also observed that subhaplogroup L3d lacks the DdeI np-10394 site, a polymorphism that it has in common with the vast majority of European haplogroups (H and T–X). Although our estimate of the age of subhaplogroup L3d is younger than those of most European haplogroups (Torroni et al. 1994a, 1996, 1998), the relatively young age of L3d in the present study might be due to the limited number of L3d mtDNAs that we sampled. Since two of the L3d haplotypes (i.e., AF01 and AF02) identified in our study possess the HinfI np-12308 site-gain marker for haplogroup U (Torroni et al. 1996), haplotype U could have arisen in Africa and migrated into Europe. Consistent with this hypothesis, the third haplotype in this subhaplogroup (i.e., AF03) lacks this haplogroup U marker but clusters with the haplogroup U mtDNAs. Hence, AF03 could be an African precursor to haplogroup U; alternatively, the haplogroup U mtDNAs in our sample may have been introduced into Africa by a back-migration/flow of European mtDNAs. Additional L3d mtDNAs, from other African populations, will need to be analyzed to further clarify the relationship of African haplogroup L3 and L3d mtDNAs to European mtDNA haplogroups.
Similarly, L3a was found to have a close affinity to haplotype AF24, a mtDNA that has the DdeI np-10394 and AluI np-10397 site gains characteristic of Asian macrohaplogroup M (figs.2 and3). Therefore, it is possible that subhaplogroup L3a was the progenitor of Asian mtDNAs belonging to M. Although the age of subhaplogroup L3a is somewhat less than our estimate for the age of Asian haplogroup M (Torroni et al. 1994b; Chen et al. 1995), the differences could be due to the limited number of L3a mtDNAs in our African sample. Alternatively, AF24 may have been introduced from Asia into Africa more recently.
On the basis of these observations, it is possible that mtDNA subhaplogroups L3a and L3d arose in sub-Saharan Africa and then moved upward into eastern Africa and out of eastern Africa into the Middle East, to yield Asian macrohaplogroup M and European haplogroup U. Such a hypothesis is supported by recent studies of eastern-African populations, which have revealed an unusually high percentage of L3 mtDNAs (Watson et al. 1997; Passarino et al. 1998). Interestingly, a number of these mtDNAs also have the DdeI np-10394 and AluI np-10397 site gains characteristic of “Asian” macrohaplogroup M (Passarino et al. 1998). Therefore, it is possible that subhaplogroups L3a and L3d radiated out of eastern Africa, to give rise to European and Asian mtDNAs. If so, then eastern Africa may still harbor the progenitor haplotypes from which European and Asian mtDNAs were derived.
Acknowledgments
The authors would like to thank Lorri Griffin and the Clinical Research Center of the Emory University School of Medicine, supported by National Institutes of Health (NIH) grant M01-RR-00039, for their assistance in the processing of blood samples. This work was supported by NIH grants AG13154, NS 21328, and HL45572 (to D.C.W.).
Appendix A
Table A1.
Polymorphic Sites Defining RFLP Haplotypes in Southern-African Populations
|
Haplotype (AF)b |
|
| Polymorphic Sitea | 000000000000000000000111111114888888888899999999990000000060123456789012345678901234567 |
| 102e | 00000000000000000000000100000 |
| 185l | 00000000000000000000000001000 |
| 762k | 00000000000000000000000000100 |
| 1715c | 11111100111111101111111111111 |
| 2349j | 01111100000000000000000010000 |
| 2758k | 11111111000000000000000000111 |
| 3592h | 11000000111111111111111111111 |
| 4157a | 10000000000000000000000000011 |
| 4310a | 11111111111111111111111110000 |
| 4341g | 11111111111111111011111111111 |
| 4464k | 11111111111111111011111111111 |
| 4583a | 11111111111111111000000001111 |
| 4769a* | 00000000000000000000000000000 |
| 4901j | 00000000000000000100000000000 |
| 5164k | 00000000000000000100000000000 |
| 5176a | 11111100111111111111111111111 |
| 5811g | 00000000000000000011111000000 |
| 6610g | 00000000000000000000000000011 |
| 7025a* | 11111111111111111111111111111 |
| 7055a | 11111111111111111111111110111 |
| 7828f | 00000000111100000000000000000 |
| 7967c | 00000000011100000000000000000 |
| 8112i | 11111111000000000111111111111 |
| 8150i | 11111111000000000111111111111 |
| 8249b/8250e | 00000011111111111000001000000 |
| 8519g | 00000000000000000100000000000 |
| 8587k | 11111111111111111111111101111 |
| 8616j | 11111100111111111111111111111 |
| 8858f* | 11111111111111111111111111111 |
| 8921k | 00000000000000000001100000000 |
| 8994e | 11111111111111111111000111111 |
| 9135j | 00000000111100000000000000000 |
| 9246e | 00001000000000000000000000000 |
| 9438e | 11111111000011111111111111111 |
| 10394c | 11111011111100111111111111111 |
| 10806g | 0000010011111111111111111000 |
| 11436i | 00000000111100000000000000000 |
| 11641e | 00000000000000000100000110000 |
| 12629b, j | 11101111111111111111111111111 |
| 12802a | 00000000000000001000000000000 |
| 13259o | 11111111111011111111111111111 |
| 13702e* | 00000000000000000000000000000 |
| 14149o* | 00000000000000000000000000000 |
| 14268g* | 11111111111111111111111111111 |
| 14368g* | 00000000000000000000000000000 |
| 14406c | 00000000000000000000000000010 |
| 15208k | 00000011000000000000000000000 |
| 15357j | 11111100111111111111111111111 |
| 16208k | 11110100111111111100000111111 |
| 16208k | 11110100111111111100000111111 |
| 16275k | 00000011000000000000000000000 |
| 16373e | 00000000000000100000000000000 |
| 16389g/16390b | 10000000000000000000000000111 |
| 16501i | 00000000001000000000000000000 |
| 16517e | 00110101111111110111111111101 |
Restriction enzymes used in the analysis are designated by single-letter code, as follows: a =AluI; b = AvaII; c = DdeI; e = HaeIII, HhaI; g = HinfI; h = HpaII; i = MspI; j = MboI; k = RsaI; l = TaqI; m = BamHI; n = HaeII; and o = HincII. Sites separated by a slash (/) indicate either (a) simultaneous site gains or site losses for two different enzymes or (b) a site gain for one enzyme and a site loss for another, because of a single inferred nucleotide substitution; in the parsimony analysis and sequence-divergence calculations, these sites are considered to be only one restriction-site polymorphism. Sites marked with an asterisk (*) were found to be present or absent in all samples, contrary to the published sequence.
A “1” indicates the presence of a site; and a “0” indicates the absence of a site. Sites are numbered from the first nucleotide of the recognition sequence, according to the sequence published by Anderson et al. (1981).
Appendix B
Table B1.
Percent Sequence Divergence Within and Between African Haplogroups
|
Sequence Divergencea(%) |
|||||||||||||||||
| L | L1 | L2 | L3 | L1a | L1b | L2a | L2b | L2c | L3a | L3b | L3c | L3d | L1a1 | L1a2 | L1b1 | L1b2 | |
| L | .356 | .360 | .331 | .362 | .357 | .355 | .343 | .353 | .351 | .355 | .373 | .357 | .356 | .348 | .365 | .343 | .370 |
| L1 | .018 | .328 | .357 | .362 | .315 | .322 | .352 | .343 | .364 | .341 | .369 | .339 | .334 | .309 | .322 | .305 | .347 |
| L2 | .068 | .108 | .171 | .264 | .335 | .288 | .165 | .172 | .163 | .214 | .251 | .199 | .185 | .277 | .310 | .231 | .274 |
| L3 | .071 | .084 | .065 | .227 | .350 | .306 | .262 | .249 | .270 | .213 | .243 | .221 | .220 | .309 | .336 | .254 | .320 |
| L1a | .047 | .019 | .118 | .104 | .265 | .323 | .328 | .305 | .341 | .315 | .347 | .300 | .286 | .252 | .252 | .281 | .337 |
| L1b | .070 | .051 | .096 | .086 | .083 | .214 | .289 | .272 | .303 | .260 | .333 | .259 | .243 | .279 | .313 | .165 | .249 |
| L2a | .108 | .132 | .023 | .092 | .139 | .125 | .113 | .141 | .162 | .204 | .266 | .180 | .155 | .271 | .311 | .214 | .301 |
| L2b | .140 | .144 | .051 | .100 | .137 | .129 | .099 | .072 | .120 | .196 | .286 | .193 | .160 | .243 | .267 | .175 | .343 |
| L2c | .147 | .174 | .051 | .130 | .182 | .170 | .100 | .101 | .052 | .206 | .274 | .176 | .128 | .283 | .325 | .221 | .320 |
| L3a | .117 | .117 | .069 | .039 | .122 | .093 | .197 | .147 | .138 | .120 | .134 | .119 | .071 | .259 | .294 | .163 | .321 |
| L3b | .082 | .092 | .053 | .017 | .102 | .114 | .155 | .132 | .101 | .239 | .225 | .248 | .052 | .318 | .333 | .271 | .401 |
| L3c | .138 | .134 | .073 | .067 | .127 | .111 | .148 | .183 | .146 | .158 | .104 | .082 | .098 | .239 | .253 | .161 | .320 |
| L3d | .152 | .144 | .074 | .081 | .128 | .111 | .093 | .140 | .127 | .140 | .226 | .128 | .051 | .215 | .208 | .125 | .309 |
| L1a1 | .088 | .063 | .108 | .113 | .037 | .090 | .218 | .149 | .169 | .214 | .259 | .155 | .101 | .166 | .267 | .215 | .298 |
| L1a2 | .127 | .099 | .165 | .163 | .060 | .146 | .305 | .220 | .258 | .293 | .228 | .216 | .141 | .221 | .119 | .239 | .329 |
| L1b1 | .144 | .120 | .126 | .120 | .128 | .038 | .141 | .144 | .210 | .084 | .087 | .120 | .114 | .090 | .161 | .041 | .217 |
| L1b2 | .069 | .060 | .065 | .084 | .081 | .019 | .168 | .169 | .125 | .195 | .380 | .156 | .114 | .218 | .202 | .012 | .246 |
Intrahaplogroup sequence-divergence values are shown on the diagonal (underlined), whereas the raw and corrected interhaplogroup values are shown above and below the diagonal, respectively.
References
- Anderson S, Bankier AT, Barrell BG, De Bruijn MHL, Coulson AR, Drouin J, Eperon IC, et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH, et al (1992) Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient Mongoloid migrations. Genetics 130:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonné-Tamir B, Johnson MJ, Natali A, Wallace DC, Cavalli-Sforza LL (1986) Mitochondrial DNA types in two Israeli populations—a comparative study at the DNA level. Am J Hum Genet 38:341–351 [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 [DOI] [PubMed] [Google Scholar]
- Cann RL, Wilson AC (1983) Length mutations in human mitochondrial DNA. Genetics 104:699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Chen Y-S, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC (1995) Analysis of mtDNA variation in African populations identifies the most ancient of all human continent-specific haplogroups. Am J Hum Genet 57:133–149 [PMC free article] [PubMed] [Google Scholar]
- De Almeida A (1965) Bushmen and other non-Bantu peoples of Angola. Witwatersrand University Press for the Institute for the Study of Man in Africa, Johannesburg [Google Scholar]
- De Benedictis G, Rose G, Passarino G, Quagliarello C (1989) Restriction fragment length polymorphism of human mitochondrial DNA in a sample population from Apulia (south Italy). Ann Hum Genet 53:311–318 [DOI] [PubMed] [Google Scholar]
- Denaro M, Blanc H, Johnson MJ, Chen KH, Wilmsen E, Cavalli-Sforza LL, Wallace DC (1981) Ethnic variation in HpaI endonuclease cleavage patterns of human mitochondrial DNA. Proc Natl Acad Sci USA 78:5768–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Langaney A (1989) Origin and differentiation of human mitochondrial DNA. Am J Hum Genet 44:73–85 [PMC free article] [PubMed] [Google Scholar]
- Graven L, Passarino G, Semino O, Boursot P, Santachiara-Benerecetti AS, Langaney A, Excoffier L (1995) Evolutionary correlation between control region sequence and restriction polymorphisms in the mitochondrial genome of a large Senegalese Mandenka sample. Mol Biol Evol 12:334–345 [DOI] [PubMed] [Google Scholar]
- Greenberg JH (1963) The languages of Africa. Indiana University Press, Bloomington [Google Scholar]
- Horai S, Hayasaka K (1990) Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet 46:828–842 [PMC free article] [PubMed] [Google Scholar]
- Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N (1995) Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc Natl Acad Sci USA 92:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MJ, Wallace DC, Ferris SD, Rattazzi MC, Cavalli-Sforza LL (1983) Radiation of human mitochondrial DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol 19:255–271 [DOI] [PubMed] [Google Scholar]
- Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S (1997) Neandertal DNA sequences and the origin of modern humans. Cell 90:19–30 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (1993) MEGA: molecular evolutionary genetics analysis, version 1.01. Pennsylvania State University, University Park [Google Scholar]
- Nei M, Tajima F (1983) Maximum likelihood estimation of the number of nucleotide substitutions from restriction site data. Genetics 105:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse GT, Botha MC, Jenkins T (1977) Sero-genetic studies of the San of south west Africa. Hum Hered 27:81–98 [DOI] [PubMed] [Google Scholar]
- Nurse GT, Weiner JS, Jenkins T (1985) Research monographs on human population biology. Vol 3: The peoples of southern Africa and their affinities. Clarendon Press, Oxford [Google Scholar]
- Passarino G, Semino O, Quintana-Murici L, Excoffier L, Hammer M, Santachiara-Benerecetti AS (1998) Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet 62:420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte U, Neufeld E, Prager FM, Gross M, Hakim I, Khatib A, Bonné-Tamir B (1993) Mitochondrial DNA affinities of several Jewish communities. Hum Biol 65:359–385 [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya EB, Wallace DC (1999) Mitochondrial DNA diversity in Koryaks and Itel’men: population replacement in the Okhotsk Sea - Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–40 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Torroni A, Semino O, Cruciani F, Spedini G, Santachiara Benerecetti AS (1994) Genetic studies in Cameroon: mitochondrial DNA polymorphisms in Bamileke. Hum Biol 66:1–12 [PubMed] [Google Scholar]
- Scozzari R, Torroni A, Semino O, Sirugo G, Brega A, Santachiara Benerecetti AS (1988) Genetic studies on the Senegal population. I. Mitochondrial DNA polymorphisms. Am J Hum Genet 43:534–544 [PMC free article] [PubMed] [Google Scholar]
- Semino O, Torroni A, Scozzari R, Brega A, De Benedictis G, Santachiara-Benerecetti AS (1989) Mitochondrial DNA polymorphisms in Italy. III. Population data from Sicily: a possible quantitation of maternal African ancestry. Ann Hum Genet 53:193–202 [DOI] [PubMed] [Google Scholar]
- Soodyall H (1993) Mitochondrial DNA polymorphisms in Southern African populations. Ph.D. thesis, University of the Witwatersand, Johannesburg [Google Scholar]
- Soodyall H, Jenkins T (1992) Mitochondrial DNA polymorphisms in Khoisan populations from southern Africa. Ann Hum Genet 56:315–324 [DOI] [PubMed] [Google Scholar]
- Soodyall H, Jenkins T (1993) Mitochondrial DNA polymorphisms in Negroid populations from Namibia: new light on the origins of the Dama, Herero, and Ambo. Ann Hum Biol 20:477–485 [DOI] [PubMed] [Google Scholar]
- Soodyall H, Vigilant L, Hill AV, Stoneking M, Jenkins T (1996) mtDNA control-region sequence variation suggests multiple independent origins or an “Asian-specific” deletion in sub-Saharan Africans. Am J Hum Genet 58:595–608 [PMC free article] [PubMed] [Google Scholar]
- Swofford D (1994) Phylogenetic analysis using parsimony (PAUP), version 3.1.1. Illinois Natural History Survey, Champaign [Google Scholar]
- Swofford D (1998) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0. Sinauer Associates, Sunderland, MA [Google Scholar]
- Templeton AR (1992) Human origins and analysis of mitochondrial DNA sequences. Science 255:737 [DOI] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, D'Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, et al (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu P, et al (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Lott MT, Cabell MF, Chen Y-S, Lavergne L, Wallace DC (1994a) Mitochondrial DNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet 55:760–776 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Miller JA, Moore LG, Zamudio S, Zhuang J, Droma T, Wallace DC (1994b) Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol 93:189–199 [DOI] [PubMed] [Google Scholar]
- Torroni A, Neel JV, Barrantes R, Schurr TG, Wallace DC (1994c) A mitochondrial DNA “clock” for the Amerinds and its implications for timing their entry into North America. Proc Natl Acad Sci USA 91:1158–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, et al (1993) Asian affinities and continental radiation of the four founding Native American mitochondrial DNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang C-C, Szathmary EJE, Williams RC, Schanfield MS, Troup GA, et al (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L (1990) Control region sequences from African populations and the evolution of human mitochondrial DNA. PhD thesis, University of California, Berkeley [Google Scholar]
- Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC (1989) Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci USA 86:9350–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt H-J (1997) Mitochondrial footprints of human expansions in Africa. Am J Hum Genet 61:691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]