Abstract
We have identified a family afflicted over multiple generations with posterior fossa tumors of infancy, including central nervous system (CNS) malignant rhabdoid tumor (a subset of primitive neuroectodermal tumors, or PNET) and choroid plexus carcinoma. Various hereditary tumor syndromes, including Li-Fraumeni syndrome, Gorlin syndrome, and Turcot syndrome, have been linked to increased risk of developing CNS PNETs and choroid plexus tumors. Malignant rhabdoid tumors of the CNS and kidney show loss of heterozygosity at chromosome 22q11. The hSNF5 gene on chromosome 22q11 has recently been identified as a candidate tumor-suppressor gene in sporadic CNS and renal malignant rhabdoid tumors. We describe a family in which both affected and some unaffected family members were found to have a germline splice-site mutation of the hSNF5 gene, leading to exclusion of exon 7 from the mature cDNA and a subsequent frameshift. Tumor tissue shows loss of the wild-type hSNF5 allele, in keeping with a tumor-suppressor gene. These findings suggest that germline mutations in hSNF5 are associated with a novel autosomal dominant syndrome with incomplete penetrance that predisposes to malignant posterior fossa brain tumors in infancy.
Germline mutations in a number of genes—including p53, (Li-Fraumeni syndrome), PATCHED, (Gorlin syndrome), and possibly APC (Turcot syndrome)—predispose to familial cancer syndromes that include the development of central nervous system (CNS) primitive neuroectodermal tumors (PNET) (Li and Fraumeni 1969; Hamilton et al. 1995; Hahn et al. 1996). PNET neoplasms of the CNS include medulloblastoma, pineoblastoma, and CNS malignant rhabdoid tumor. Truncating mutations of the hSNF5 gene on chromosome 22q11.2 (MIM 601607) were recently found in sporadic renal and CNS malignant rhabdoid tumors (Versteege et al. 1998; Biegel et al. 1999). Several infants with malignant rhabdoid tumors were noted to have germline mutations of the hSNF5 gene, but in no case could a mutation of hSNF5 be found in the parents of affected children, nor were there signs of affected individuals in other generations. Other reports have suggested the existence of familial syndromes that predispose to the development of malignant rhabdoid tumors (Lynch et al. 1983; Bonnin et al. 1984; Fort et al. 1994; Parellada et al. 1998). The hSNF5 protein is part of a multiprotein complex that functions to remodel nucleosomes and thus to regulate the access of transcription factors to a number of different promoters. The mechanism by which loss of hSNF5 leads to neoplasia is unknown.
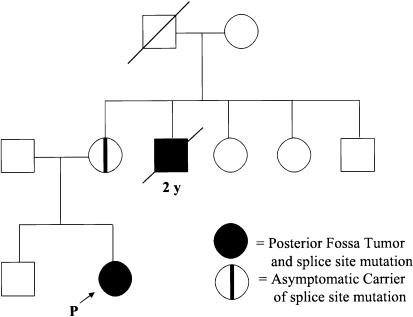
We report a family afflicted by posterior fossa brain tumors over at least two generations. The proband presented at age 18 mo with a cerebellar malignant rhabdoid tumor. Both of the parents are healthy, but the proband’s maternal uncle died at age 2 years from a posterior fossa choroid plexus carcinoma (fig. 1). The maternal grandfather’s sibling died, as an infant, from a disease process consistent with a pediatric brain tumor. Tissue from the maternal grandfather’s sibling is not available for analysis. The occurrence of two posterior fossa tumors of infancy in one family is a rare event and may suggest a germline mutation predisposing to malignancy. We hypothesized that this family might harbor a germline mutation in the hSNF5 gene, accounting for the high incidence of malignancy observed.
Figure 1.
Pedigree showing posterior fossa tumors in one family over two generations with an unaffected carrier.
Informed consent was obtained from all family members, and approval for genetic testing was obtained from the Institutional Research Ethics Board of the Hospital for Sick Children, University of Toronto. Parents of the proband received genetic counseling before and after learning the results of this study. Genomic DNA was isolated from peripheral blood leukocytes (white blood cells [wbc]) by standard techniques. Lymphocytes were separated from peripheral blood by gradient centrifugation through a percoll gradient, and RNA was extracted with Trizol (Gibco BRL). The proband’s brain tumor was flash frozen in liquid nitrogen in the operating room, and RNA and DNA were extracted with Trizol. Paraffin sections of the uncle’s tumor were digested overnight in proteinase K for extraction of genomic DNA.
Overlapping primer pairs were used to amplify portions of the hSNF5 cDNA by PCR, and the products were analyzed by agarose gel electrophoresis, to assess for the presence of aberrant transcripts. Primer SSNF5-5R (CGC GGA TCT TCT TCT CCAT) was used to sequence normal and mutant transcripts amplified from tumor and control cDNA. Primers (ssnf57-2f: GAG TTT GTC ACC ACC ATC G) and (ssnf5-7-2R: GAG CAA ACA CAC AGA CC) were used to amplify exon 7 and surrounding intronic sequences from genomic DNA. PCR products were sequenced by cycle sequencing using Thermo Sequenase (USB). In addition, the entire coding region of hSNF5 (mutant and wild-type transcripts) was amplified from maternal cDNA with use of the forward primer (5′-ccggaatccgccgcctctgccgccgcaatg-3′) and the reverse primer (5′-ccggaattcccaggccgggcccgtgttggcaagacg-3) and cloned into the EcoR1 site of pBluescript. Full-length and mutant clones were sequenced on a Li-Cor automated sequencer with the T7 and T3 primers.
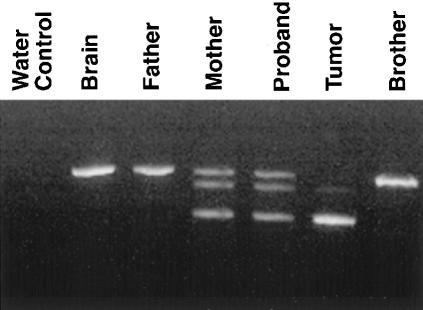
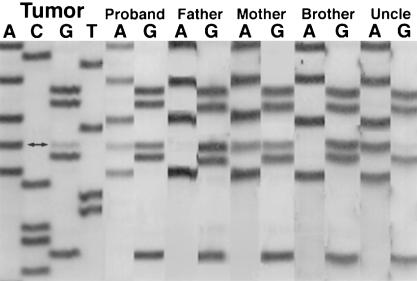
Analysis of the reverse transcriptase (RT)-PCR products from one of the hSNF5 primer pairs revealed an aberrant transcript in the proband, her tumor, and maternal cDNA (fig. 2). This product was seen in neither paternal cDNA nor cDNA from normal brain tissue from an unrelated individual. Although lymphocyte cDNA from the proband and her mother show both the expected wild-type product and the aberrant transcript, tumor cDNA shows only the aberrant product with loss of wild-type product, consistent with the role of hSNF5 as a tumor-suppressor gene. Sequencing of wild-type and truncated transcripts amplified by RT-PCR revealed the truncated product to be hSNF5 but to lack the portion of the cDNA corresponding to exon 7 (data not shown). This leads to joining of exon 6 to exon 8 with a subsequent frameshift and generation of a premature stop codon. RT-PCR of the full-length hSNF5 gene from maternal cDNA revealed two products: the expected wild-type 1200-bp product and a truncated 1,000-bp product. Other than the lack of exon 7, the sequence of this hSNF5 allele contained no other mutations (data not shown). Sequencing of exon 7 and surrounding intronic sequences from genomic DNA revealed a G→A mutation in the donor splice site of exon 7. This mutation alters the conserved GT sequence at the beginning of the intron and thus violates the GT rule for splice-site recognition. This mutation was seen in genomic DNA from the proband, her tumor, maternal DNA, and DNA from the uncle’s tumor (fig. 3). Although lymphocyte samples show heterozygosity with preservation of the wild-type allele, both tumor specimens show only the mutant allele, consistent with loss of heterozygosity (LOH) at this locus, typical of a tumor-suppressor gene.
Figure 2.
RT-PCR of a portion of the hSNF5 cDNA, showing the expected wild-type product from normal brain and from paternal and sibling (brother) wbc cDNA. Maternal and proband wbc cDNA shows wild-type and mutant transcripts, with an intervening heteroduplex. Tumor cDNA shows only the mutant transcript, with loss of the wild-type allele.
Figure 3.
Sequencing of exon 7 of hSNF5, showing a G→A mutation (double arrow) in the proband’s tumor, proband’s germline, and mother’s germline DNA. Leukocyte DNA from proband’s father and brother show the wild-type sequence. The proband’s tumor and her uncle’s tumor (Uncle) show the same G→A mutation.
This splice mutation is predicted to cause a truncation of the protein and likely contributes to the high incidence of posterior fossa brain tumors seen in this family. The proband’s tumor shows LOH for chromosome 22, as has been described in malignant rhabdoid tumors (Biegel et al. 1990, 1996). This particular mutation in hSNF5 shows incomplete penetrance, because the proband’s mother is completely unaffected. Whether the mother has an increased risk of developing a malignancy in the future is uncertain; however, both choroid plexus carcinoma and CNS malignant rhabdoid tumors are uncommon in adults. Although hSNF5 mutations have been described in renal rhabdoid tumors, only CNS tumors were seen in this kindred. Whether germline hSNF5 mutations predispose specifically to CNS tumors requires description of further families with germline mutations. The region of the hSNF5 protein deleted in this family includes the second highly conserved repeat region as well as the C terminal coiled-coil region (Muchardt et al. 1995; Morozov et al. 1998). The c-myc oncogene is amplified in many medulloblastomas, and c-myc has been shown to bind to the hSNF5 protein (Bigner et al. 1990; Cheng et al. 1999). This interaction would be predicted to be preserved in the family described in the present report. Malignant rhabdoid tumors are a subset of CNS PNETs only recently differentiated from medulloblastoma (Rorke et al. 1996). PNETs and choroid plexus carcinoma are tumor types known to occur in the Li-Fraumeni syndrome. Although this family does not meet the criteria for a diagnosis of either Li-Fraumeni syndrome or Li-Fraumeni–like syndrome, other such families in which a p53 mutation is not found may have germline hSNF5 mutations.
In conclusion, we describe a family affected over at least two generations with posterior fossa brain tumors of infancy, in whom we have characterized a germline mutation in the hSNF5 gene. The absence of symptoms in the proband’s unaffected mother suggests that this mutation can show incomplete penetrance. Families with an increased incidence of pediatric brain tumors may harbor germline mutations in the hSNF5 gene.
Acknowledgments
We are indebted to the family described for their assistance in the performance of this study. M.D.T. and T.G.M. are Terry Fox Fellows of the National Cancer Institute of Canada with funds from the Terry Fox Run. J.T.R. is a career scientist of the Medical Research Council of Canada. This work was supported in part by BRAINCHILD and the National Cancer Institute of Canada.
Electronic-Database Information
Accession number and URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/ (for hSNF5 [OMIM 601607])
References
- Biegel JA, Allen CS, Kawasaki K, Shimizu N, Budarf ML, Bell CJ (1996) Narrowing the critical region for a rhabdoid tumor locus in 22q11. Genes Chromosomes Cancer 16:94–105 [DOI] [PubMed]
- Biegel JA, Rorke LB, Packer RJ, Emanuel BS (1990) Monosomy 22 in rhabdoid or atypical tumors of the brain. J Neurosurg 73:710–714 [DOI] [PubMed]
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B (1999) Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res 59:74–79 [PubMed]
- Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD (1990) Amplification of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Res 50:2347–2450 (erratum [1990] Cancer Res 50:3809) [PubMed]
- Bonnin JM, Rubinstein LJ, Palmer NF, Beckwith JB (1984) The association of embryonal tumors originating in the kidney and in the brain: a report of seven cases. Cancer 54:2137–2146 [DOI] [PubMed]
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV (1999) c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet 22:102–105 [DOI] [PubMed]
- Fort DW, Tonk VS, Tomlinson GE, Timmons CF, Schneider NR (1994) Rhabdoid tumor of the kidney with primitive neuroectodermal tumor of the central nervous system: associated tumors with different histologic, cytogenetic, and molecular findings. Genes Chromosomes Cancer 11:146–152 [DOI] [PubMed]
- Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, et al (1996) Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85:841–851 [DOI] [PubMed]
- Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, et al (1995) The molecular basis of Turcot's syndrome. N Engl J Med 332:839–847 [DOI] [PubMed]
- Li FP, Fraumeni JF Jr (1969) Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst 43:1365–1373 [PubMed]
- Lynch HT, Shurin SB, Dahms BB, Izant RJ Jr, Lynch J, Danes BS (1983) Paravertebral malignant rhabdoid tumor in infancy: in vitro studies of a familial tumor. Cancer 52:290–296 [DOI] [PubMed]
- Morozov A, Yung E, Kalpana GV (1998) Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc Natl Acad Sci USA 95:1120–1125 [DOI] [PMC free article] [PubMed]
- Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M (1995) A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res 23:1127–1132 [DOI] [PMC free article] [PubMed]
- Parellada JA, Gupta P, Rose W (1998) Rhabdoid tumor of the kidney with primitive neuroectodermal tumor of the central nervous system. Eur J Radiol 28:219–221 [DOI] [PubMed]
- Rorke LB, Packer RJ, Biegel JA (1996) Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood: definition of an entity. J Neurosurg 85:56–65 [DOI] [PubMed]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, et al (1998) Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203–206 [DOI] [PubMed]