Abstract
Congenital cataracts are a common major abnormality of the eye that frequently cause blindness in infants. At least one-third of all cases are familial; autosomal-dominant congenital cataract appears to be the most-common familial form in the Western world. Elsewhere, in family ADCC-3, we mapped an autosomal-dominant cataract gene to chromosome 3q21-q22, near the gene that encodes a lens-specific beaded filament protein gene, BFSP2. By sequencing the coding regions of BFSP2, we found that a deletion mutation, ΔE233, is associated with cataracts in this family. This is the first report of an inherited cataract that is caused by a mutation in a cytoskeletal protein.
Congenital cataracts are a common major abnormality of the eye that frequently cause blindness in infants. At least one-third of all cases are familial; autosomal-dominant congenital cataract (ADCC) appears to be the most-common familial form in the Western world (Ionides et al. 1999). Thirteen distinct loci in humans have been identified for 10 phenotypically distinct forms of ADCC (Francis et al. 1999; Ionides et al. 1999).
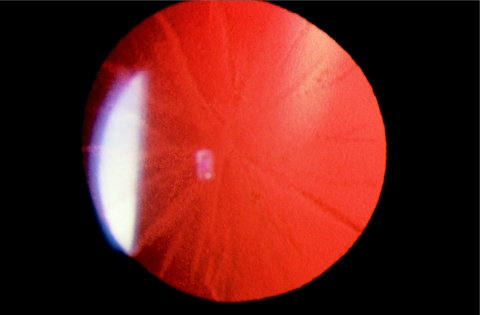
We have been studying a large family (ADCC-3) with ADCC (Kramer et al., in press). The family consists of 15 affected and more than 15 unaffected individuals across three generations. Individuals gave informed consent to participate in the study protocol, which was approved by the institutional review board of Oregon Health Sciences University. Affected members of the family had congenital nuclear, sutural, and stellate or spokelike cortical cataracts that varied in severity among different individuals. The mildest expression of the cataract was that of spokelike anterior and posterior subcapsular cortical opacities with a ground-glass appearance throughout the cataract and, most notably, the radially oriented fine vacuoles (fig. 1). The unoperated visual acuity has been as good as 20/30–20/40; however, many of those affected that are have required bilateral cataract surgery during early childhood, most often at age 3–5 years.
Figure 1.
Retroillumination slit-lamp photograph of left lens of a 12-year-old boy, demonstrating spokelike cortical opacities with radially oriented fine vacuoles. Bilateral cataracts were first noted at age 7 years, at which time the acuity was 20/40 in the left eye and 20/30 in the right eye. The acuity declined to 20/50 in the right eye; cataract surgery with intraocular lens implantation restored the acuity to 20/25. The visual acuity in the left eye is now 20/40.
Using linkage mapping in this family, we identified a new ADCC locus on chromosome 3q21.2-q22.3. This locus was mapped to a 12-cM region bounded by D3S3674 and D3S3612 (Kramer et al., in press). Hess et al. (1995) had previously localized a gene for a lens-specific beaded filament structural protein (BFSP2; also known as “CP47,” “CP49,” or “phakinin” [MIM 603212]) to chromosome 3q21-q25. Because this localization overlapped the ADCC critical region in family ADCC-3 and because expression of BFSP2 is restricted to the lens fiber cells (Ireland and Maisel 1989; Fitzgerald and Casselman 1991; Gounari et al. 1993; Hess et al. 1993), this gene became an excellent candidate gene for the site of the ADCC mutation in family ADCC-3.
To refine the localization of BFSP2, we used radiation hybrid mapping on the Genebridge-4 panel. The primers for this experiment (primers 2521 and 2438; table 1) were designed from the sequence of the 3′ UTR (Hess et al. 1995). We used the default LOD-score cutoff of 15 for linkage to the framework map. The most-likely position of BFSP2 indicated by this analysis was on chromosome 3, flanked by D3S1290 and D3S1615 (data not shown). This falls within the critical region in family ADCC-3 (Kramer et al., in press), strengthening the candidacy of BFSP2 for the disease gene in this family.
Table 1.
Primers and Conditions Used for Amplification of Exons
| Primer | Sequence | Size (nt) | Taa (°C) | Bufferb |
| 2521 | TGGAGAAACTTCCTCTTTTTC | |||
| 2438 | GACTGGAGCTCAGGAGAAAGC | 92 | 65–55 | 1 |
| Exon1/f | CACGGAAGACAGCAGACAAA | |||
| Exon1/r | GTAGNGCACCCCAGTTTCCC | 604 | 65–50 | 1 |
| Exon2/f | CTCTAGGTGGGTGAGGCAGT | |||
| Exon2/r | CTAGGGATGTGGAGGGACTG | 134 | 70–55 | 2 |
| Exon3/f | TTTTGGGCTACTCAGTTATGCTA | |||
| Exon3/r | AGCACAGGCAGACAGATGTG | 273 | 70–55 | 2 |
| Exon4/f | AATTCTATGCCATTTATTTCCC | |||
| Exon4/r | ACATTGACTAGTTTGCAGCTC | 297 | 65–50 | 1 |
| Exon5/f | CGCTCACACTGAGACCTGAC | |||
| Exon5/r | ATTAGCACAGGGACACCAGG | 202 | 65–50 | 1 |
| Exon6/f | TAGAAACGAGGCCTGGAGAA | |||
| Exon6/r | ATGGGTGCCTATGTGAGAGG | 353 | 65–50 | 1 |
| Exon7/f | CTCACCCATTGCTTCTGAC | |||
| Exon7/r | AAGCATTATTGGAAGAGGACAA | 262 | 70–55 | 2 |
“Touchdown” PCR was used: the annealing temperature Ta was decreased by 1°C/cycle for the first 10 or 15 cycles, followed by 25 cycles at the final annealing temperature. Initial and final annealing temperatures are shown.
1 = Standard PE Biosystems/Cetus buffer with 1.5 mM Mg++; 2 = Kogan buffer: 16.6 mM ammonium sulfate, 67 mM Tris-Cl- (pH 8.8 at 25°C), 6.7 mM MgCl2, 10 mM β-mercaptoethanol, 6.7 μM EDTA, 170 μg BSA per ml, and 10% dimethylsulfoxide (Kogan et al. 1987).
To screen the coding regions of BFSP2 for mutations, it was first necessary to localize exon-intron boundaries so that intronic primers that flank exons could be designed. To accomplish this, partial genomic sequences of BFSP2 (GenBank accession numbers AF195044 and AF195849) were compared with the coding sequence (GenBank accession number U48224). With the exception of exon 1, primer pairs (table 1) were designed from noncoding sequences within introns or UTRs. In the case of exon 1, a reliable sequence from the downstream intron was unavailable, so we designed a reverse primer (exon 1/r in table 1) that was complementary to nucleotides (nt) 441–460 of the coding sequence (Hess et al. 1996). The product of this PCR included all but the last 49 nucleotides of exon 1. Using the primers listed in table 1, we amplified and directly sequenced 96% of the coding sequence from an affected individual and also from an unaffected sibling in family ADCC-3. All sequences were normal, except for exon 3 in the affected individual, which had a 3-bp deletion of nt 696–698 (GAA) in the coding sequence (data not shown). This deletion is predicted to cause an in-frame deletion of glutamic acid residue 233 (ΔE233) from the polypeptide encoded by the mutant gene. The presence of this mutation was confirmed by the sequencing of two cloned exon 3 PCR products from the affected family member (data not shown).
Allele-specific oligonucleotides (ASOs) specific for mutant (CP49mut: TGAAAGAACTTGGCTCT) and wild-type (CP49wt: AAAGAAGAACTTGGCTC) sequences were labeled with digoxygenin and were used to screen members of family ADCC-3 and unrelated unaffected controls, as described elsewhere (Litt et al. 1998). Dot blots of PCR products from exon 3 of the family members were probed with the mutant ASO. All 12 family members that were tested and were heterozygous for the disease-bearing chromosome, as determined by linkage analysis, gave signals with this ASO. None of the 19 family members that were tested and that lacked this chromosome gave signals. When a duplicate blot was probed with the wild-type ASO, all family members gave signals. All 149 unrelated unaffected individuals tested by this method hybridized only with the wild-type ASO (data not shown).
Although all 149 unaffected unrelated individuals that were screened lacked the ΔE233 alteration, this sample size is too small to rule out the possibility that the alteration could occur in 0.5%–1% of the normal population. We do, however, believe that the ΔE233 alteration is the causative mutation in ADCC-3, rather than just a rare polymorphism in strong linkage disequilibrium with the causative mutation. The affected gene product, phakinin, and its assembly partner, filensin, are both members of the intermediate filament (IF) family of proteins (Gounari et al. 1993; Hess et al. 1996, 1998). Both proteins localize to a cytoskeletal element referred to as the beaded filament (Ireland and Maisel 1984, 1989; Fitzgerald and Gottlieb 1989; Fitzgerald 1990). The precise function of the beaded filament in the lens fiber cell is unknown; however, (1) both proteins are restricted in expression to the lens fiber cell, (2) homologues to both proteins are found in lenses of at least five vertebrate orders, and (3) the beaded filament has been described only in the lens fiber cell (Fitzgerald and Casselman 1991; Gounari et al. 1993; Hess et al. 1993, 1995). Collectively, this suggests a role that is critical and unique to lens fiber cell biology.
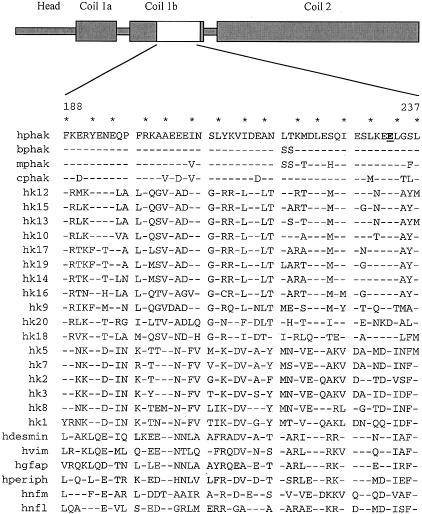
Phakinin shows most of the secondary structural features that are conserved among IF proteins. Among these is a central rod domain dominated by runs of alpha helix. These runs of alpha helix exhibit a heptad repeat pattern in which the 1 and 4 positions of the heptad are dominated by hydrophobic residues (fig. 2). This pattern results in a hydrophobic “stripe” along the helix, which aligns with the corresponding stripe in its assembly partner, resulting in a coiled-coil dimer, the first stage of intermediate filament assembly (reviewed by Parry and Steinart 1992). IF proteins are generally tolerant of conservative amino-acid mutations in the central rod domain. Deletion of residue E233 would result in a phase shift in the 1,4 periodicity and would perturb an otherwise highly conserved property among IF proteins. Furthermore, although sequence drift in the central rod domain of IF proteins is substantial, there are occasional sites and small motifs where conservation of specific residues is quite strong. Mutations in these highly conserved sites are the genetic basis for several inherited human disorders in skin and cornea (Fuchs 1994; Fuchs et al. 1994; Irvine et al. 1997). The deleted residue E233 is conserved in phakinins from cow, human, mouse, and chicken. In addition, as shown in figure 2, alignment of phakinin with 24 other human IF proteins shows only one exception (a conservative E→D substitution in hk20) to absolute conservation of this residue, suggesting that it is critical to IF proteins.
Figure 2.
Domain structure of human phakinin (hphak) (top). “Coil” domains predicted to mediate assembly of coiled-coil dimers are noted. Shown below are the amino-acid sequences of residues 188–237 of hphak and of homologous regions from 26 other IF proteins. Residues 1 and 4 of the alpha helical heptad repeats of hphak are starred. Glutamic acid 233 of hphak is underlined and appears in boldface type. Residues in the other IF proteins identical to those at homologous positions of hphak are indicated by dashes. Abbreviations for other IF proteins are as follows: mphak, murine phakinin; bphak, bovine phakinin; cphak, chicken phakinin; hk1–hk20, human keratins type 1–type 20; hvim, human vimentin (type 3); hdesmin, human desmin (type 3); hperiph, human peripherin; hgfap, human glial fibrillary acidic protein; hnfl, human neurofilament light chain; and hnfm, human neurofilament medium chain.
The actual mechanism by which a mutation in BFSP2 gives rise to cataracts is unknown, but it could be clarified by studies of a mouse model. Such a model may already exist. The murine autosomal-dominant cataract mutation Coc maps to a region of mouse chromosome 16 that is syntenic with human chromosome 3q21-q24 (Sidjanin et al. 1997); hence, the murine homologue of BFSP2 may be the site of the mutation responsible for Coc. Characterization of the molecular defect in the Coc mouse, as well as creation of a transgenic mouse model that expresses the ΔE233 mutation in the murine homologue of BFSP2, may elucidate the role of this protein in maintaining lens transparency.
Mutations that have previously been reported to cause ADCC are found in genes that encode crystallins (Litt et al. 1997, 1998; Kannabiran et al. 1998; Heon et al. 1999; Stephan et al. 1999), gap-junction proteins (Shiels et al. 1998; Mackay et al. 1999), or a homeobox protein (Semina et al. 1998). The BFSP2 mutation reported here is the first example of an ADCC mutation in a cytoskeletal structural protein. As this article was being prepared, we learned that another group had found a nonconservative R278W mutation in exon 4 of BFSP2 associated with juvenile-onset cataract (Conley et al. 2000).
Acknowledgments
This work was funded by National Institutes of Health grants EY11710 (to M.L.) and EY08747 (to P.F.). R.G.W. was supported in part by unrestricted funds from Research to Prevent Blindness. We thank the Vollum Institute sequencing core facility for DNA sequencing. We thank an anonymous reviewer of our manuscript (Kramer et al., in press) for the suggestion to screen BFSP2 for mutations.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BFSP2 [MIM 603212])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for cDNA [U48224] and genomic DNA [AF195044 and AF195849])
References
- Conley YP, Erturk D, Keverline A, Mah TS, Keravala A, Barnes LR, Bruchis A, et al (2000) A juvenile-onset, progressive cataract locus on chromosome 3q21-q22 is associated with a missense mutation in the beaded filament structural protein-2 (BFSP2). Am J Hum Genet 66:1426–1431 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PG (1990) Methods for the circumvention of problems associated with the study of the ocular lens cytoskeleton. Curr Eye Res 9:1083–1097 [DOI] [PubMed] [Google Scholar]
- FitzGerald PG, Casselman JC (1991) Immunologic conservation of the fiber cell beaded filament. Curr Eye Res 10:471–478 [DOI] [PubMed] [Google Scholar]
- FitzGerald PG, Gottlieb W (1989) The 115-kd fiber cell specific protein is a component of the lens cytoskeleton. Curr Eye Res 8:801–811 [DOI] [PubMed] [Google Scholar]
- Francis PJ, Berry V, Moore AT, Bhattacharya S (1999) Lens biology, development and human cataractogenesis. Trends Genet 15:191–196 [DOI] [PubMed] [Google Scholar]
- Fuchs E (1994) Intermediate filaments and disease:mutations that cripple strength. J Cell Biol 125:511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Coulombe P, Cheng J, Chan YM, Hutton E, Syder A, Degenstein L, et al (1994) Genetic bases of epidermolysis bullosa simplex and epidermolytic hyperkeratosis. J Invest Dermatol 103(Suppl):25S–30S [DOI] [PubMed] [Google Scholar]
- Gounari F, Merdes A, Quinlan R, Hess J, FitzGerald P, Ouzounis C, Georgatos S (1993) Bovine filensin possesses primary and secondary structure similarity to intermediate filament proteins. J Cell Biol 121:847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, Lubsen N, Munier FL (1999) The gamma-crystallins and human cataracts: a puzzle made clearer. Am J Hum Genet 65:1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Cassleman J, FitzGerald P (1993) cDNA analysis of the 49-kDa lens fiber cell cytoskeletal protein: a new, lens-specific member of the intermediate filament family. Curr Eye Res 12:77–88 [DOI] [PubMed] [Google Scholar]
- Hess JF, Casselman JT, FitzGerald PG (1995) Chromosomal locations of the genes for the beaded filament proteins CP115 and CP47. Curr Eye Res 14:11–18 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Gene structure and cDNA sequence identify the beaded filament protein CP49 as a highly divergent type I intermediate filament protein. J Biol Chem 271:6729–6735 [DOI] [PubMed] [Google Scholar]
- Hess J, Casselman J, Kong A, FitzGerald P (1998) Primary sequence, secondary structure, gene structure, and assembly properties suggest that the lens-specific cytoskeletal protein filensin represents a novel class of intermediate filament protein. Exp Eye Res 66:625–644 [DOI] [PubMed] [Google Scholar]
- Ionides A, Francis P, Berry V, Mackay D, Bhattacharya S, Shiels A, Moore A (1999) Clinical and genetic heterogeneity in autosomal dominant cataract. Br J Ophthalmol 83:802–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland M, Maisel H (1984) A cytoskeletal protein unique to lens fiber cell differentiation. Exp Eye Res 38: 637–645 [DOI] [PubMed] [Google Scholar]
- ——— (1989 ) A family of lens fiber cell specific proteins. Lens Eye Toxic Res 6:623–638 [PubMed] [Google Scholar]
- Irvine AD, Corden LD, Swensson O, Swensson B, Moore JE, Fraze DG, Smith FJ, et al (1997) Mutations in cornea-specific keratin K3 or K12 genes cause Meesmann's corneal dystrophy. Nat Genet 16:184–187 [DOI] [PubMed] [Google Scholar]
- Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF (1998) Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaA3/A1-crystallin gene. Mol Vis 23:21 [PubMed] [Google Scholar]
- Kogan SC, Doherty M, Gitschier J (1987) An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences: application to hemophilia A. New Engl J Med 317:985–990 [DOI] [PubMed] [Google Scholar]
- Kramer PL, LaMorticella D, Schilling K, Billingslea A, Weleber R, Litt M. A new locus for autosomal dominant congenital cataracts maps to chromosome 3. Invest Ophthalmol Vis Sci (in press) [PubMed] [Google Scholar]
- Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH (1997) Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta crystallin gene CRYBB2. Hum Mol Genet 6:665–668 [DOI] [PubMed] [Google Scholar]
- Litt, M, Kramer, P, LaMorticella, DM, Murphey, W, Lovrien, EW, Weleber, RG (1998) Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet 7:471–474 [DOI] [PubMed] [Google Scholar]
- Mackay D, Ionides A, Kibar Z, Rouleau G, Berry V, Moore A, Shiels A, et al (1999) Connexin 46 mutations in autosomal dominant congenital cataract. Am J Hum Genet 64:1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D, Steinart P (1992) Intermediate filament structure. Curr Opin Cell Biol 4:94–98 [DOI] [PubMed] [Google Scholar]
- Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WLM, Reiter RS, et al (1998) A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet 19:167–170 [DOI] [PubMed] [Google Scholar]
- Shiels A, MacKay D, Ionides A, Berry V, Moore A, Bhattacharya S (1998) A missense mutation in the human connexin 50 gene (GJA8) underlies autosomal dominant “zonular pulverulent” cataract, on chromosome 1q. Am J Hum Genet 62:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjanin DJ, Grimes PA, PretschW, Neuhauser-Klaus A, Favor J (1997) Mapping of the autosomal dominant cataract mutation (Coc) on mouse chromosome 16. Invest Ophthal Vis Sci 38:2502–2507 [PubMed] [Google Scholar]
- Stephan DA, Gillers E, Verveen D, Freas Lutz D, Wistow G, et al (1999) Progressive juvenile-onset punctate cataracts caused by mutation of the γ-D-crystallin gene. Proc Nat Acad Sci USA 96:1008–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]