Abstract
Nonsyndromic hearing loss is one of the most genetically heterogeneous traits known. A total of 30 autosomal dominant nonsyndromic hearing–loss loci have been mapped, and 11 genes have been isolated. In the majority of cases, autosomal dominant nonsyndromic hearing loss is postlingual and progressive, with the exception of hearing impairment in families in which the impairment is linked to DFNA3, DFNA8/12, and DFNA24, the novel locus described in this report. DFNA24 was identified in a large Swiss German kindred with a history of autosomal dominant hearing loss that dates back to the middle of the 19th century. The hearing-impaired individuals in this kindred have prelingual, nonprogressive, bilateral sensorineural hearing loss affecting mainly mid and high frequencies. The DFNA24 locus maps to 4q35-qter. A maximum multipoint LOD score of 11.6 was obtained at 208.1 cM at marker D4S1652. The 3.0-unit support interval for the map position of this locus ranges from 205.8 cM to 211.7 cM (5.9 cM).
Hearing loss is a common form of sensory impairment that affects millions of individuals worldwide (Wilson 1985). Of the hereditary forms of hearing loss, ∼70% of the cases are nonsyndromic in nature—that is, hearing loss is the only clinical feature (Morton 1991). Nonsyndromic hearing loss is one of the most genetically heterogeneous traits known (Van Camp et al. 1997). More than 60 loci have been mapped, and a total of 15 genes have been identified (Hereditary Hearing Loss Homepage). The extreme genetic heterogeneity of nonsyndromic hearing loss suggests that there are many different processes that can malfunction within the inner ear to cause hearing loss (Heller and Hudspeth 1998). Because of the extreme genetic heterogeneity of nonsyndromic hearing loss, an important strategy in mapping novel loci has been to use individual kindreds that can independently establish linkage.
Thus far, for autosomal dominant nonsyndromic hearing loss, a total of 30 loci have been mapped and 11 genes have been isolated (Hereditary Hearing Loss Homepage). In the majority of cases, autosomal dominant nonsyndromic hearing loss is postlingual and progressive, with the exception of hearing impairment in families in which the hearing loss is linked to DFNA3 (MIM 601544), DFNA8/12 (MIM 601543/601842), and DFNA24, the novel locus described here. For DFNA3, hearing impairment is due to mutations in one of two genes, GJB2 (MIM 121011) or GJB6 (MIM 604418). Mutations within the TECTA gene (MIM 602574) are responsible for hearing impairment due to DFNA8/12.
The Swiss German kindred (102) presented here has a history of bilateral prelingual nonprogressive sensorineural hearing loss that dates back to the middle of the 19th century. Kindred 102 (fig. 1) was ascertained through the Department of Otolaryngology at the University Hospital of Zurich. Informed consent was obtained from all study participants and from the parents of minor study participants. A family history was obtained through an oral interview. Audiometric and DNA samples were obtained from pedigree members in three generations.
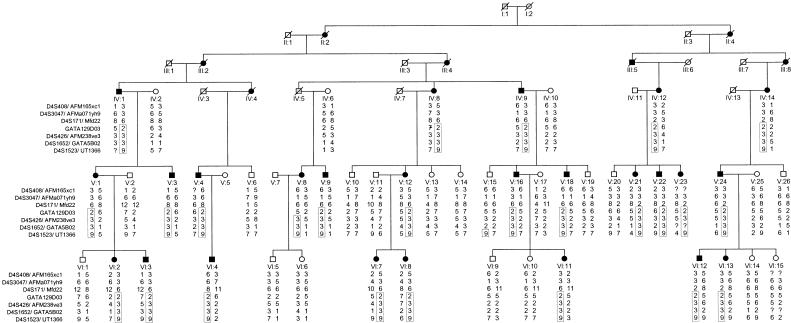
Figure 1.
Pedigree drawing of the Swiss German kindred 102, segregating DFNA24. Blackened symbols represent individuals with hearing impairment due to DFNA24. Grayed symbols represent individuals whose hearing status is unknown. Unblackened symbols represent unaffected individuals. Haplotypes for the most closely linked STRPs are shown below each symbol. Question marks (?) indicate unknown alleles. Haplotypes segregating with the disease allele appear in rectangles. All hearing-impaired individuals have the same disease haplotype for markers GATA129D03, D4S426, D4S1652, and D4S1523, except individual V.8, who shares this haplotype only for markers D4S426, D4S1652, and D4S1523. Unaffected individual V.15 shares the same allele as hearing-impaired family members at marker D4S1523, the most telomeric marker.
Blood samples were collected from 47 pedigree members, and a buccal swab was collected from one family member aged <6 years. Of the study subjects from whom DNA samples were obtained, 26 individuals were hearing impaired because of the DFNA24 locus, 16 had normal hearing, and 7 were the spouses of hearing-impaired pedigree members.
Auditory thresholds were determined by standard pure-tone audiometry with air conduction at 125–8,000 Hz and bone conduction at 250–4,000 Hz. In addition, where possible, previous audiologic tests were collected. Audiograms for the 26 hearing-impaired family members displayed mild to profound hearing loss, mainly in the frequencies between 0.5 KHz and 4 KHz. The hearing-impaired family members all had sloping audiograms affecting mainly the mid to high frequencies. Hearing-impaired individuals with severe to profound hearing loss in the high frequencies also had moderate hearing loss in the mid and low frequencies. The hearing loss appears to be nonprogressive, with no correlation between severity of hearing loss and current age. No evidence of acquired risk factors predisposing to hearing loss was observed for the hearing-impaired family members.
On the basis of medical history, there was no evidence that the hearing loss in this kindred is part of a syndrome. It is of interest to note that facioscapulohumeral (FSH) muscular dystrophy (FSHMD1A [MIM 158900]), which maps to the same region as DFNA24, is associated with early-onset sensorineural hearing loss. The family members of kindred 102 do not present with muscle weakness, ruling out FSHMD1A as a diagnosis.
DNA was extracted from 10 ml of blood by means of standard techniques (Miller et al. 1988). A genome scan was performed on 46 DNA samples at the National Heart, Lung, and Blood Institute (NHLBI [NIH]) mammalian genotyping service, using the Marshfield Screening set 9 (Yuan et al. 1997). A total of 386 fluorescently labeled simple tandem-repeat (STRP) markers with an average heterozygosity of 0.76 were genotyped. These markers are spaced ∼10 cM apart and are located on the 22 autosomes and the X and Y chromosomes.
Linkage analysis was performed under a fully penetrant autosomal dominant mode of inheritance with no phenocopies. A disease frequency of .001 was used in the analysis. The marker allele frequencies were estimated from the data by means of both observed and reconstructed genotypes of founders within the pedigrees. We performed two-point linkage analysis on all autosomal markers from the genome scan, using MLINK of the FASTLINK computer package (Cottingham et al. 1993). A maximum two-point LOD score of 4.9 with marker D4S1652 was obtained. D4S1652 was the most telomeric chromosome 4 marker included in the genome scan. No other marker gave a LOD score >3. Flanking distal markers to D4S1652, D4S408, and D4S241 produced maximum LOD scores of 2.03 and 0.71, respectively. Multipoint linkage analysis with LINKMAP of the FASTLINK computer package ruled out that the DFNA24 locus lies between D4S408 and D4S241 (data not shown).
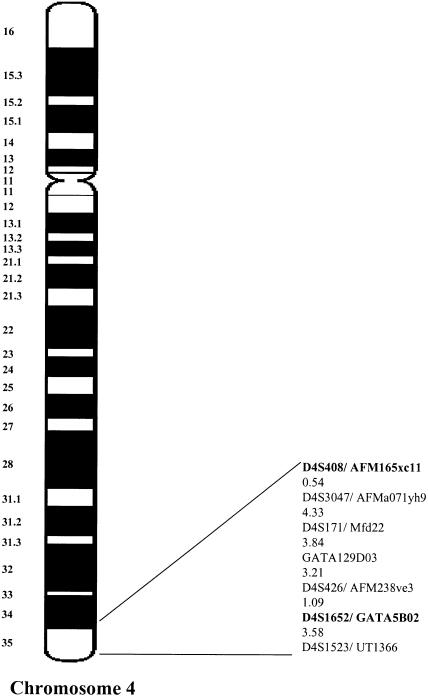
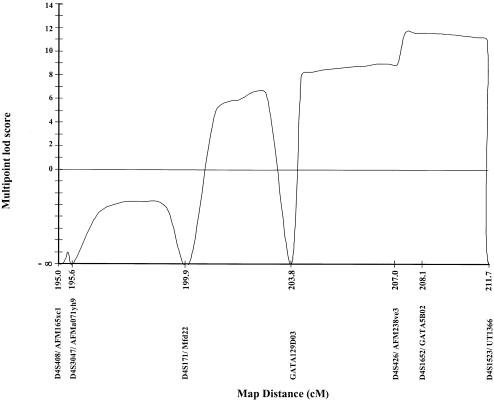
For fine mapping, five additional markers were selected from the Marshfield map (Broman et al. 1998); four of these markers—D4S3047, D4S171, GATA129D03, D4S426—lie between D4S408 and D4S1652, and one additional marker—D4S1523—is telomeric to D4S1652 (fig. 2). No other markers were available from this region that would allow for finer mapping of the DFNA24 locus. Forty-eight DNA samples were genotyped for these markers: the DNA samples included in the genome scan, plus two additional DNA samples, sample V.23, which was ascertained after the initial genome scan was carried out, and sample VI.15, for which only a limited amount of DNA was available, since a buccal swab had been collected. The data were reanalyzed by means of two-point and multipoint linkage analyses. The results of the two-point linkage analysis are displayed in table 1. A maximum two-point LOD score of 8.57 was obtained at recombination fraction (θ) .01 for marker D4S1523, which is the most telomeric marker on chromosome 4. A graph of the results of the multipoint analysis is displayed in figure 3. A maximum multipoint LOD score of 11.6 occurred at marker D4S1652 at 208.1 cM, with a 3.0-unit support interval from 205.08 cM to 211.7 cM (5.9 cM).
Figure 2.
Genetic map location of linked markers on chromosome 4, delineating the DFNA24 locus. Map distances from the Marshfield map are given in centimorgans. Bold markers were genotyped for the genome scan. The disease interval, determined on the basis of recombination events, is flanked by markers GATA129D03 and UT1366/D4S1523 (7.88 cM).
Table 1.
Two-Point LOD Score Results between the DFNA24 Locus and Chromosome 4 Markers
|
LOD Score at θ = |
|||||||
| Marker | .0 | .01 | .05 | .10 | .20 | .30 | .40 |
| D4S408 /AFM165xc1 | −∞ | −2.66 | .35 | 1.25 | 1.50 | 1.12 | .55 |
| D4S3047 /AFMa071yh9 | −∞ | −1.39 | 1.58 | 2.40 | 2.45 | 1.79 | .88 |
| D4S171 /Mfd22 | −∞ | .60 | 2.80 | 3.29 | 3.02 | 2.18 | 1.11 |
| GATA129D03 | −∞ | 6.02 | 6.21 | 5.83 | 4.62 | 3.16 | 1.55 |
| D4S426 /AFM238ve3 | 6.63 | 6.52 | 6.04 | 5.42 | 4.11 | 2.73 | 1.34 |
| D4S1652 /GATA5B02 | 4.86 | 4.75 | 4.34 | 3.81 | 2.73 | 1.67 | .74 |
| D4S1523 /UT1366 | −∞ | 8.57 | 8.52 | 7.85 | 6.11 | 4.11 | 1.98 |
Figure 3.
Results from the multipoint linkage analysis. A maximum multipoint LOD score of 11.6 occurred at marker D4S1652 at 208.1 cM. The 7.9-cM interval to which DFNA24 maps is flanked by recombination events at markers GATA129D03 and UT1366/D4S1523.
SIMWALK2 (Weeks et al. 1995) was used to generate the haplotypes with the highest likelihood (fig. 1). The interval to which DFNA24 maps is flanked by recombination events at marker GATA129D03 and UT1366/D4S1523. The interval between these two markers is 7.9 cM.
In the region 4q35-qter, there are two known expressed sequence tags (ESTs) that are expressed in the cochlea, SHGC-33691 and SHGC-35919 (Human Cochlear cDNA Library and EST Database). Genes and ESTs expressed in the cochlea are potential candidates for DFNA24, because all known mutations leading to deafness in humans occur in genes that are expressed in the specialized cell types of the cochlea.
The EST SHGC-35919 belongs to the unigene cluster Hs.16506. The ESTs within this cluster are widely expressed (i.e., colon, heart, kidney, lung, and stomach). The EST SHGC-33691 (unigene Hs.9817) is within the ArgBP2 gene (Wang et al. 1997). ArgBP2 is widely expressed in human tissue and is associated with the actin cytoskeleton. ArgBP2 is a potential candidate gene for DFNA24, because another widely expressed actin-associated protein, the human homologue of Drosophila melanogaster diaphanous (DIAPH1 [MIM 602121]), is responsible for nonsyndromic hearing loss due to DFNA1 (MIM 124900).
Relatively few genes have been identified within the interval where DFNA24 is localized. Current knowledge of genes other than ArgBP2 within this region does not suggest they are potential candidates for DFNA24. These genes include adenine nucleotide translocator 1 (ANT1 [MIM 103220]), coagulation factor XI (F11 [MIM 264900]), homologue of Drosophila FAT tumor suppressor (FAT [MIM 600976], interferon regulatory factor 2 (IRF2 [MIM 147576]), melatonin receptor 1A (MTNR1A [MIM 600665]), and toll-like receptor 3 (TLR3 [MIM 603029]).
It is also of interest to note that FSH muscular dystrophy (MIM 158900) maps to this region. A number of FSH muscular dystrophy cases with early-onset bilateral high-frequency sensorineural hearing loss have been reported (Small 1968; Taylor et al. 1982; Padberg et al. 1992). In some cases, the hearing loss was progressive and, over time, also involved lower frequencies (Voit et al. 1986). These findings make the FSH muscular dystrophy region gene 1 (FRG1 [MIM 601278]), once identified, a potential candidate for DFNA24.
In several cases, different mutations within the same gene are responsible for both syndromic and nonsyndromic forms of hearing loss. For example, the myosin VIIA gene (MYO7A [MIM 276903]) is responsible for Usher syndrome type 1B (USH1B [MIM 276903]), DFNB2 (MIM 600060), and DFNA11 (MIM 601317). The pendrin gene (PDS [MIM 274600]) is responsible for Pendred syndrome (MIM 274600) and DFNB14 (MIM 603678). Mutations in the COL11A2 gene (MIM 120290) cause otospondylomegaepiphyseal dysplasia (OSMED [MIM 215150]), Weissenbacher-Zwymuller syndrome (WZS [MIM 277610]), Stickler syndrome nonocular type (STL2 [MIM 184840]), and DFNA13 (MIM 601868). Linkage of the DFNA24 locus to 4q35-qter is an essential first step in gene identification that will provide insight into the pathophysiology of this disorder.
Acknowledgments
The authors would especially like to thank the family members for participating in this study. We would also like to thank Dr. Elizabeth Pugh (Center for Inherited Disease Research) for her help in binning the allele data. This work was supported by the NHLBI mammalian genotyping service, NIH–National Institute of Deafness and other Communication Disorders grant DC03594, the American Hearing Research Foundation, the Starr Center for Human Genetics (to S.M.L.), and NIH grant HG00008.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://www.marshmed.org/genetics/ (for the order and genetic distances of STRP loci on 4q)
- Hereditary Hearing Loss Homepage, http://hgins.uia.ac.be/dnalab/hhh/ (for current number of hearing loss loci and genes)
- Human Cochlear cDNA Library and EST Database, http://hearing.bwh.harvard.edu/cochlearcdnalibrary.htm (for ESTs SHGC-33691 and SHGC-35919)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for ANT1 [MIM 103220], COL11A2 [MIM 120290], DFNA1 [MIM 124900], DFNA3 [MIM 601544], DFNA8 [MIM 601543], DFNA11 [MIM 601317], DFNA12 MIM 601842], DFNA13 [MIM 601868], DFNB2 [MIM 600060], DFNB14 [MIM 603678], DIAPH1 [MIM 602121], F11 [MIM 264900], FAT [MIM 600976], FRG1 [MIM 601278], FSHMD1A [MIM 158900], GJB2 [MIM 121011], GJB6 [MIM 604418], IRF2 [MIM 147576]), MTNR1A [MIM 600665], MYO7A [MIM 276903], OSMED [MIM 215150], PDS [MIM 274600], Pendred syndrome [MIM 274600], STL2 [MIM 184840], TECTA [MIM 602574], TLR3 [MIM 603029], USH1B [MIM 276903], and WZS [MIM 277610]
References
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed]
- Heller S, Hudspeth AJ (1998) Two deaf mice, two deaf mice: murine candidate genes pinpoint the genetic bases of nonsyndromic hearing loss in humans. Nat Med 4:560–561 [DOI] [PubMed]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed]
- Morton NE (1991) Genetic epidemiology of hearing impairment. Ann N Y Acad Sci 630:16–31 [DOI] [PubMed] [Google Scholar]
- Padberg GW, Brouwer OF, de Keizer RJW, Gruter AM, Wijmenga C, Grote JJ, Frants RR (1992) Retinal vascular disease and sensorineural deafness are part of facioscapulohumeral muscular dystrophy. Am J Hum Genet Suppl 51: A104 [Google Scholar]
- Small RG (1968) Coats' disease and muscular dystrophy. Trans Am Acad Ophthalmol Otolaryngol 72:225–231 [PubMed] [Google Scholar]
- Taylor DA, Carroll JE, Smith ME, Johnson MO, Johnston GP, Brooke MH (1982) Facioscapulohumeral dystrophy associated with hearing loss and Coats syndrome. Ann Neurol 12:395–398 [DOI] [PubMed]
- Van Camp G, Willems PJ, Smith RJH (1997) Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet 60:758–764 [PMC free article] [PubMed]
- Voit T, Lamprecht A, Lenard HG, Goebel HH (1986) Hearing loss in facioscapulohumeral dystrophy. Eur J Pediatr 145:280–285 [DOI] [PubMed]
- Wang B, Golemis EA, Kruh GD (1997) ArgBP2, a multiple Src homology 3 domain-containing, Arg/Abl-interacting protein, is phosphorylated in v-Abl-transformed cells and localized in stress fibers and cardiocyte Z-disks. J Biol Chem 272:17542–17550 [DOI] [PubMed]
- Weeks DE, Sobel E, O'Connell JR, Lange K (1995) Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet 56:1506–1507 [PMC free article] [PubMed]
- Wilson J (1985) Deafness in developing countries. Approaches to a global program of prevention. Arch Otolaryngol 111:2–9 [DOI] [PubMed]
- Yuan B, Vaske D, Weber JL, Beck J, Sheffield VC (1997) Improved set of short-tandem-repeat polymorphisms for screening the human genome. Am J Hum Genet 60:459–460 [PMC free article] [PubMed]