Summary
To establish the contribution of germline BRCA1 and BRCA2 mutations to familial ovarian cancer, we have analyzed both genes in DNA samples obtained from an affected individual in each of 112 families containing at least two cases of epithelial ovarian cancer. Germline mutations were found in 43% of the families; BRCA1 mutations were approximately four times more common than BRCA2 mutations. The extent of family history of ovarian and breast cancers was strongly predictive of BRCA1-mutation status. Segregation analysis suggests that a combination of chance clustering of sporadic cases and insensitivity of mutation detection may account for the remaining families; however, the contribution of other genes cannot be excluded. We discuss the implications for genetic testing and clinical management of familial ovarian cancer arising from the data presented in these studies.
Introduction
Familial ovarian cancer occurs as part of two clinically distinct syndromes. The most common of these syndromes is ovarian cancer, which is either apparently site specific or occurs in association with early-onset breast cancer (Easton et al. 1993). Predisposition to ovarian cancer also occurs as part of Lynch type II or hereditary nonpolyposis colorectal cancer (HNPCC) syndrome (Lynch et al. 1982). The results of genetic-linkage studies have suggested that, in >90% of families with multiple cases of breast and ovarian cancer, the cancers are the result of mutations in either the BRCA1 gene on chromosome 17q12-21 or the BRCA2 gene on chromosome 13q12-13 (Narod et al. 1995; Ford et al. 1998). Linkage data from a small series of families suggest that BRCA1 is also responsible for the majority of “site-specific ovarian cancer” families with three or more cases of epithelial ovarian cancer (Steichen-Gersdorf et al. 1994).
Germline mutations in the BRCA1 and BRCA2 genes have been identified in a large number of families with multiple cases of early-onset breast and/or ovarian cancer (Miki et al. 1994; Shattuck-Eidens et al. 1995; Wooster et al. 1995; Tavtigian et al. 1996; Gayther et al. 1997b). The mutation spectrum is similar in both genes: most germline mutations are predicted to result in protein truncation caused by frameshift, nonsense, or splice-site alterations, and the mutations are spread along the length of the coding region (Breast Cancer Information Core). A small number of founder mutations that are common in specific populations have been described. In particular, these include the 185delAG mutation in BRCA1 and the 6174delT mutation in BRCA2, which are common in Ashkenazi Jews (Struewing et al. 1995; Neuhausen et al. 1996a, 1996b); the 5382insC mutation in BRCA1, which is common in the eastern European population (Gayther et al. 1997a; Ramus et al. 1997); and the 999del5 mutation, which is common in the Icelandic population (Thorlacius et al. 1996). The precise functions of the BRCA1 and BRCA2 proteins are unknown; they have recently been shown to interact with each other, which suggests that they may function in the same pathway (Chen et al. 1998). Both proteins interact in vitro with RAD51, a protein that has been implicated in the repair of double-strand DNA breaks (Scully 1997; Wong et al. 1997). This finding and the results of an analysis of homozygous mutant mice suggest a possible functional role for BRCA1 and BRCA2 in DNA repair and genome integrity (Gayther and Ponder 1998; Zhang et al. 1998).
An immediate consequence of the identification of the BRCA1 and BRCA2 genes is that DNA-based predictive testing has become possible in families when the mutation is known. Since the search for the mutation is laborious, it is helpful to know the prevalence of BRCA1 and BRCA2 mutations in families with different cancer histories. In this study, we set out to obtain this information in relation to ovarian cancer. One hundred and twelve families were ascertained to have a family history in which at least two first- or second-degree relatives had been given a diagnosis of epithelial ovarian cancer. To search for disease-associated mutations, the entire BRCA1 and BRCA2 coding sequence was analyzed in one affected individual from each family. This enabled us to estimate the proportion of families with ovarian cancer that resulted from these two genes, and it also enabled us to predict whether there are likely to be other highly penetrant susceptibility genes that are responsible for a proportion of cases of familial ovarian cancer.
Material and Methods
Patient Material
One hundred and three families in which at least two first-degree relatives had epithelial ovarian cancer diagnosed at any age and nine families in which two cases of epithelial ovarian cancer were diagnosed in second-degree relatives were identified through the Familial Ovarian Cancer Register of the United Kingdom Co-ordinating Committee on Cancer Research (UKCCCR). To be included in the register, families must have at least two cases of ovarian cancer confirmed by pathology reports or by a death certificate. Borderline cases of ovarian cancer were not included in the study. Fifty-three families had ovarian cancer only, whereas the remaining 59 families had at least one case of breast cancer, in addition to ovarian cancer, diagnosed in a family member before the age of 60 years. These families included a total of 330 cases of ovarian cancer and 133 cases of breast cancer diagnosed before the age of 60 years. Confirmation of diagnosis was obtained from either pathology reports or a death certificate, for a total of 281 cases of ovarian cancer and 70 cases of breast cancer. Initial DNA analysis was performed with genomic DNA obtained from one affected individual from each family, and, when material was available, mutations were confirmed in other affected individuals from the family.
Mutation Analysis
BRCA1 and BRCA2 were both screened for germline mutations, by use of a combination of the protein-truncation test (PTT) and nonradioactive single-strand conformation analysis/heteroduplex analysis (SSCA/HA). PTT was performed, as described elsewhere (Friedman et al. 1997), for the two largest exons of BRCA2 and for the largest exon of BRCA1. For SSCA/HA, coding exons 2, 3, 5–10, and 12–24 of BRCA1 and coding exons 2–9 and 12–27 of BRCA2 were amplified from genomic DNA. The 5′ and 3′ splice boundaries of exon 11 of BRCA1 and exons 10 and 11 of BRCA2 were also amplified from genomic DNA. Nonradioactive SSCA/HA was performed as described elsewhere, and gels were visualized by silver staining (Gayther et al. 1995). Direct sequence analysis was used to characterize the nucleotide alteration associated with PTT and/or SSCA/HA variants, as described elsewhere (Gayther et al. 1995).
We also screened for a recently described 6-kb duplication near exon 13 of BRCA1 (Puget et al. 1999) in families in which no BRCA1 or BRCA2 coding mutation was identified. Duplication-specific primers (forward primer, 5′-GATTATTTCCCCCCAGGCTA-3′; reverse primer, 5′-AGATCATTAGCAAGGACCTGTG-3′) will only produce a PCR-amplification product if the duplication is present in the DNA sample. Two positive genomic DNA controls for the duplication were included in each experiment. In addition, for each sample, a second, independent PCR reaction was simultaneously performed with the use of forward primer 5′-TCACAATTCCGAGACATC-3′ and reverse primer 5′-AACGGCTACTGCACAGTTCT-3′. This second reaction amplified a product with a size similar to that of the duplication and was used to control both for the efficacy of each genomic DNA sample and for PCR amplification.
Statistical Analysis
The effects, on mutation prevalence, of the numbers of individuals affected with breast and ovarian cancer were assessed with the use of χ2 tests for trend. To evaluate the evidence for existence of a third susceptibility gene, in addition to BRCA1 and BRCA2, we performed segregation analysis using the MENDEL program (Lange et al. 1988). To allow for the ascertainment of families on the basis of multiple affected individuals, analyses were based on the conditional likelihood Lik(M,D/D), where M was the observed mutation status of the index case and D was the observed disease phenotype in the family. BRCA1 and BRCA2 were initially assumed to confer the age-specific risks for breast and ovarian cancer given in previous analyses (Ford et al. 1995). Carriers of a susceptibility allele, at a hypothetical third gene locus (OVCA) and unlinked to BRCA1 and BRCA2, had an increased relative risk (r) of ovarian cancer compared with noncarriers, but they had no increased risk for breast cancer. Dominant, recessive, and codominant models for OVCA were explored. The absolute risks by genotype were determined by the constraint that the overall incidence of the disease must agree with the incidence rates for England and Wales. We then estimated r and the frequencies p1, p2, and p3 of the susceptibility alleles of BRCA1, BRCA2, and OVCA, respectively, according to maximum likelihood. The sum p1+p2 was constrained to be no higher than .003, which is the combined frequency, given by the model of Claus et al. (1991), of breast cancer–susceptibility alleles and the highest plausible frequency for BRCA1 and BRCA2 mutations combined. We also allowed for a sensitivity parameter, γ, which was used to determine the probability of detecting a mutation, if one was present (the probability was assumed to be the same for BRCA1 and BRCA2).
We restricted consideration to those genetic models that gave familial risks of ovarian cancer that were consistent with those observed in epidemiological studies. Stratton et al. (1998) have previously estimated that, among first-degree relatives of patients with ovarian cancer, the relative risk for ovarian cancer is 2.4, with a 95% confidence interval of 1.9–3.2, on the basis of a meta-analysis of published cohort studies. We incorporated this estimate into our analysis by adding a “penalty” to the log-likelihood, to reflect deviation of risk from 2.4: P=[log(2.4)-log(λo)]2/2v, where λο is the predicted familial relative risk from the model and where v=.0177 is the variance of the log (relative risk) from the meta-analysis.
Results
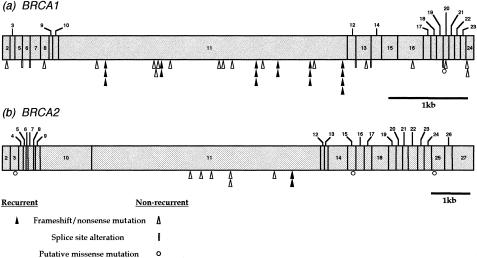
Germline mutations, which are thought to be disease associated, were detected in 48 (43%) of the 112 families. These data are listed in table 1. Twenty-nine different BRCA1 mutations were identified in 40 (36%) of the 112 families. The spectrum of BRCA1 mutations in this data set is illustrated in figure 1a. Mutations are spread quite evenly along the length of the gene. Six mutations were detected on more than one occasion; these six mutations represent 43% of all BRCA1 mutations identified in this study. Haplotype analysis does not indicate a common founder mutation in the British population (Gayther et al. 1995; Neuhausen et al. 1996b). Thirty-five (88%) of the 40 mutations are either frameshift or nonsense mutations that would be predicted to result in premature truncation of the BRCA1 protein. Four mutations occur at the boundaries between exons and introns and are expected to affect splicing. The results of analysis of reverse-transcribed RNA samples obtained from two affected individuals with splice variants (4304G→A in family OV161 and 4476+6T→C in family OV003) have previously shown that these alterations cause aberrant splicing (Gayther et al. 1995). There was no available RNA with which to test the effect of the remaining two splice-site mutations, although both are predicted to abolish highly conserved splice-site consensus sequences. Pro1749Arg, the putative missense mutation in family OV073, has not been described elsewhere and was not detected in 346 normal chromosomes. The functional significance of this missense mutation has been assessed elsewhere (Chapman and Verma 1996). The ability of the C terminus of BRCA1 to cause transcriptional activation in vitro was abolished by a GAL4-BRCA1 fusion protein containing the Pro1749Arg missense alteration, a finding that suggests this change is functionally significant.
Table 1.
Germline Mutations in BRCA1 and BRCA2
|
No. of Confirmed (Unconfirmed) Cases of Cancer |
Description of Mutation |
|||||||
| Family | Ovarian | Breast | Gene | Exon orIntron | Codon | Nucleotide | Alteration | Type |
| OV010 | 3 (1) | 1 (3) | BRCA1 | Exon 2 | 23 | 185 | 185delAG | Frameshift |
| OV022 | 3 | 1 | BRCA1 | Intron 5 | — | 331 | 331+2T→C | Splice site |
| OV131 | 2 | 1 (2) | BRCA1 | Intron 6 | — | 421 | 421-2delA | Splice site |
| OV047 | 3 (2) | 2 (2) | BRCA1 | Exon 8 | 169 | 624T→G | Gln169ter | Nonsense |
| OV008 | 3 (1) | 2 (2) | BRCA1 | Exon 11 | 367 | 1220 | 1220insC | Frameshift |
| OV002 | 6 | 3 (1) | BRCA1 | Exon 11 | 392 | 1294 | 1294del40 | Frameshift |
| OV110 | 2 | 1 | BRCA1 | Exon 11 | 392 | 1294 | 1294del40 | Frameshift |
| OV150 | 4 | 2 | BRCA1 | Exon 11 | 392 | 1294 | 1294del40 | Frameshift |
| OV132 | 3 | 0 (1) | BRCA1 | Exon 11 | 607 | 1938A→T | lys607ter | Nonsense |
| OV014 | 3 (3) | 0 | BRCA1 | Exon 11 | 608 | 1942 | 1942del4 | Frameshift |
| OV069 | 2 | 0 | BRCA1 | Exon 11 | 627 | 2000 | 2000del4 | Frameshift |
| OV001 | 3 | 1 (1) | BRCA1 | Exon 11 | 652 | 2073 | 2073delA | Frameshift |
| OV020 | 3 | 1 (1) | BRCA1 | Exon 11 | 652 | 2073 | 2073delA | Frameshift |
| OV175 | 2 | 2 (1) | BRCA1 | Exon 11 | 655 | 2082 | 2082insG | Frameshift |
| OV252 | 3 | 2 (2) | BRCA1 | Exon 11 | 690 | 2187 | 2190delA | Frameshift |
| OV181 | 2 | 2 (1) | BRCA1 | Exon 11 | 871 | 2731 | 2731insT | Frameshift |
| OV133 | 2 (1) | 1 (1) | BRCA1 | Exon 11 | 885 | 2774 | 2774delCT | Frameshift |
| OV171 | 2 | 0 | BRCA1 | Exon 11 | 916 | 2867 | 2867insA | Frameshift |
| OV013 | 2 | 0 | BRCA1 | Exon 11 | 1001 | 3121 | 3121delA | Frameshift |
| OV029 | 6 | 1 (1) | BRCA1 | Exon 11 | 1001 | 3121 | 3121delA | Frameshift |
| OV160 | 2 | 0 | BRCA1 | Exon 11 | 1001 | 3121 | 3121delA | Frameshift |
| OV189 | 3 | 0 | BRCA1 | Exon 11 | 1056 | 3286 | 3286delC | Frameshift |
| OV012 | 7 (1) | 1 | BRCA1 | Exon 11 | 1112 | 3452 | 3452del4 | Frameshift |
| OV033 | 2 | 0 | BRCA1 | Exon 11 | 1112 | 3452 | 3452del4 | Frameshift |
| OV007 | 4 (3) | 0 (1) | BRCA1 | Exon 11 | 1252 | 3875 | 3875del4 | Frameshift |
| OV206 | 3 (4) | 0 | BRCA1 | Exon 11 | 1252 | 3875 | 3875del4 | Frameshift |
| OV282 | 3 | 1 | BRCA1 | Exon 11 | 1252 | 3875 | 3875del4 | Frameshift |
| OV025 | 3 (3) | 2 | BRCA1 | Exon 11 | 1289 | 3986 | 3986delAA | Frameshift |
| OV019 | 3 | 1 | BRCA1 | Exon 11 | 1355 | 4184 | 4184del4 | Frameshift |
| OV042 | 2 (2) | 1 (5) | BRCA1 | Exon 11 | 1355 | 4184 | 4184del4 | Frameshift |
| OV044 | 3 (7) | 0 (7) | BRCA1 | Exon 11 | 1355 | 4184 | 4184del4 | Frameshift |
| OV245 | 3 | 0 (1) | BRCA1 | Exon 11 | 1355 | 4184 | 4184del4 | Frameshift |
| OV161 | 6 | 1 | BRCA1 | Exon 12 | 1395 | 4304 | 4304G→A | Splice site |
| OV226 | 2 (1) | 0 (1) | BRCA1 | Exon 13 | 1443 | 4446C→T | Arg1443ter | Nonsense |
| OV003 | 2 (1) | 2 | BRCA1 | Intron 13 | — | 4476 | 4476+6T→C | Splice site |
| OV148 | 2 (1) | 2 | BRCA1 | Exon 16 | 1648 | 5061 | 5061delA | Frameshift |
| OV073 | 3 | 0 (1) | BRCA1 | Exon 20 | 1749 | 5365C→G | Pro1749Arg | Missense |
| OV139 | 2 | 0 (1) | BRCA1 | Exon 20 | 1755 | 5382 | 5382insC | Frameshift |
| OV074 | 2 | 0 (1) | BRCA1 | Exon 24 | 1835 | 5622C→T | Arg1835ter | Nonsense |
| OV224 | 2 (1) | 1 (7) | BRCA1 | Exon 24 | 1837 | 5629 | 5629delG | Frameshift |
| OV092 | 2 (1) | 2 (1) | BRCA2 | Exon 11 | 1366 | 4326 | 4326insCATC | Frameshift |
| OV072 | 3 | 0 | BRCA2 | Exon 11 | 1491 | 4701 | 4701delGAAA | Frameshift |
| OV140 | 2 | 1 | BRCA2 | Exon 11 | 1602 | 5032 | 5032insA | Frameshift |
| OV027 | 2 | 1 (1) | BRCA2 | Exon 11 | 1782 | 5573 | 5573delAA | Frameshift |
| OV085 | 2 | 0 | BRCA2 | Exon 11 | 1782 | 5573 | 5573delA | Frameshift |
| OV254 | 2 (1) | 0 (2) | BRCA2 | Exon 11 | 1970 | 6137C→A | Ser1970ter | Nonsense |
| OV016 | 3 | 4 | BRCA2 | Exon 11 | 2092 | 6503 | 6503delTT | Frameshift |
| OV162 | 3 | 1 | BRCA2 | Exon 11 | 2092 | 6503 | 6503delTT | Frameshift |
Figure 1.
Location and type of germline mutations detected in (a) the BRCA1 gene and (b) the BRCA2 gene, in families containing two or more confirmed cases of epithelial ovarian cancer.
Seventy-two families without identifiable BRCA1 mutations were analyzed for mutations throughout the BRCA2 coding sequence. Seven different mutations that were considered to be pathogenic were identified in eight (7%) of these families. The spectrum of these mutations throughout the gene is illustrated in figure 1b. In seven families, the mutation was a frameshift deletion, and, in one family, it was a nonsense mutation. One mutation, 6503delTT, was detected in two different families. Two distinct mutations seen in families OV085 and OV027 occur at nucleotide 5573, which is the beginning of a repeat of seven adenines; this finding possibly suggests this region is a mutation hotspot. Three putative missense mutations were also identified: A75P in exon 3 (family OV120), R2502H in exon 15 (family OV222), and Y3098H in exon 25 (family OV045). These mutations were not detected in 344, 326, and 310 normal chromosomes, respectively. However, because these mutations have an unclear disease association and because there is no available functional assay with which to further investigate the significance of the mutations, they have not been included in the statistical analyses.
Subsequent to our analysis of BRCA1 and BRCA2 for coding mutations, large genomic alterations involving the BRCA1 gene have been reported. These alterations would not have been detected by the PCR-based approaches to mutation screening used in our study. One of these genomic alterations, a duplication and insertion involving 6 kb around exon 13 of BRCA1, is predicted to lead to protein truncation and may be of British origin (Puget et al. 1999). A PCR-based assay has been specifically designed to detect this mutation. We were able to screen for the exon 13 duplication in 55 families that did not have a coding mutation in BRCA1 or BRCA2; the mutation was not detected in any of the families (data not shown).
Mutation Prevalence, by Family Type
Table 2 gives the proportion of families with BRCA1 and BRCA2 mutations, according to the number of cases of breast and ovarian cancer in the family. The numbers of ovarian cancers and breast cancers in the family were strongly predictive of BRCA1-mutation status (χ 2/1=18.2, P=.0001 and χ 2/1=15.29, P<.0001, respectively). BRCA2-mutation status was significantly associated with the number of cases of breast cancer in the family (χ 2/1=3.80, P=.05), but there was no significant association with the number of cases of ovarian cancer. BRCA1 mutations were more frequent than BRCA2 mutations in families with large numbers of ovarian cancers; in particular, 14 (35%) of 40 families with BRCA1 mutations had four or more ovarian cancers, whereas none of the families with BRCA2 mutations fell into this category. However, this difference did not reach formal statistical significance (χ 2/1=2.89, P=.08).
Table 2.
Observed and Predicted Number of Mutations, by Family Type
|
No. (%) of Families with Cancer |
||||
| Confirmed Casesb |
All Casesc |
|||
| Family History of Cancer and Mutation Statusa | Observed | Predictedd | Observed | Predictedd |
| At least two cases of ovarian cancer and at least two cases of breast cancer: | ||||
| BRCA1 | 10 (56) | 11.40 | 18 (56) | 20.13 |
| BRCA2 | 1 (5) | 1.18 | 3 (13) | 1.73 |
| None | 7 (39) | 5.42 | 11 (31) | 10.14 |
| At least three cases of ovarian cancer and no more than one case of breast cancer: | ||||
| BRCA1 | 17 (63) | 16.37 | 12 (52) | 11.10 |
| BRCA2 | 2 (7) | .68 | 2 (13) | .90 |
| None | 8 (30) | 9.75 | 9 (35) | 11.00 |
| Two cases of ovarian cancer and one case of breast cancer: | ||||
| BRCA1 | 5 (29) | 6.89 | 5 (26) | 6.86 |
| BRCA2 | 3 (18) | .77 | 2 (11) | .66 |
| None | 9 (53) | 9.34 | 12 (63) | 11.49 |
| Two cases of ovarian cancer and no cases of breast cancer: | ||||
| BRCA1 | 8 (16) | 8.33 | 5 (13) | 4.91 |
| BRCA2 | 2 (4) | 1.42 | 1 (3) | .96 |
| None | 40 (80) | 40.25 | 32 (84) | 32.13 |
| Totals: | ||||
| BRCA1 | 40 (36) | 43.00 | 40 (36) | 43.00 |
| BRCA2 | 8 (7) | 4.25 | 8 (7) | 4.25 |
| None | 64 (57) | 64.76 | 58 (57) | 64.76 |
Ovarian cancer diagnosed at any age, breast cancer diagnosed at age ⩽60 years.
χ2=4.61 (P=.47) for comparison of observed and predicted numbers.
χ2=4.32 (P=.50) for comparison of observed and predicted numbers.
For the best-fitting model (model 4a) in table 3.
As shown in table 2, mutations were found in 66% of families with either three or more reported cases of ovarian cancer or four or more reported cases of breast or ovarian cancer; however, they were found in only 20% of families with two cases of ovarian cancer alone. If families are classified only on the basis of confirmed cases, then these frequencies are altered to 71% and 22%, respectively. In the families with a BRCA1 mutation, the average patient age at diagnosis of ovarian cancer (49.9 years) was significantly lower than that in the families without the BRCA1 mutation (53.9 years) (P=.003). In the families with the BRCA2 mutation, the average age at diagnosis of ovarian cancer (55.0 years) did not differ significantly from that in either the families with the BRCA1 mutation or the families with no mutation.
Model Fitting
To evaluate the evidence for further ovarian cancer–susceptibility genes, in addition to BRCA1 and BRCA2, we performed a segregation analysis. We tested several models that allowed for a third gene (OVCA), which was unlinked to BRCA1 or BRCA2 and which had two alleles; in these models, carriers of the susceptibility allele had an increased risk for ovarian cancer, compared with noncarriers, but they had no increased risk for breast cancer. Models were constrained for plausible BRCA-gene frequencies and by the requirement that the predicted overall incidence of breast and ovarian cancer must agree with known incidence. The frequencies of BRCA1 and BRCA2 were estimated in all models. We did not assume any values for these parameters. The only assumption was that their combined frequency should not be >.003. To assess the goodness of fit of different genetic models to the data, we computed, for each model, the predicted probability of observing a BRCA1 or BRCA2 mutation in families, in different categories of family history. We then compared these predictions with what we actually observed.
Table 3summarizes the estimated parameters for some of these models. In all but one of the models (model 4), the relative risk conferred by the OVCA susceptibility allele converged to 1 (i.e., there was no effect). This was true whether OVCA was assumed to be dominant, recessive, or codominant. In the case of model 4, the likelihood differed only slightly from that of the corresponding model showing no effect of OVCA (model 4a). We also fitted models in which the penetrance estimates for BRCA1 and BRCA2 were reduced so that the incidence rates for both breast and ovarian cancer were 50% of the Breast Cancer Linkage Consortium (BCLC) estimates. However, these models also converged to a relative risk of 1 for OVCA (data not shown). Models in which the risk for breast cancer conferred by BRCA1 was given by the BCLC values (models 1 and 2) had a poor fit, with the observed number of BRCA1 and BRCA2 mutations being greater than the predicted number of such mutations. The models with the reduced incidence rates, in which BRCA1 and BRCA2 were at 50% of the BCLC rates, showed a better fit than did the models in which the actual BCLC rates were used; however, they did not show a better fit than did the average heterogeneity rates. Better fits were obtained in the models in which the average of the two penetrance estimates given by Easton et al. (1995) were assumed. In each case, models assuming 90% sensitivity for mutation detection showed better fit than did models assuming 64% sensitivity. Table 2 shows that, when model 4a was used, the predicted mutation frequencies were close to those observed.
Table 3.
Parameters of Genetic Models Fitted to the Ovarian-Cancer Data Set
|
Frequency of |
||||||||
| Model | BRCA1 | BRCA2 | OVCA | Relative Risk of OVCA | Mutation-Detection Sensitivity | Assumed Rate | Predicted Familial Riska | Log Likelihood |
| 1 | .0011 | .0019 | 0 | 1 | .64 | Homogeneityb | 3.15 | −114.1 |
| 2 | .0010 | .0020 | 0 | 1 | .90 | Homogeneityb | 2.96 | −111.2 |
| 3 | .0017 | .0013 | 0 | 1 | .64 | Average heterogeneityc | 2.00 | −95.4 |
| 4 | .0014 | .0016 | .41 | 16,940 | .90 | Average heterogeneityc | 2.05 | −93.9 |
| 4a | .0014 | .0016 | 0 | 1 | .90 | Average heterogeneityc | 1.89 | −94.2 |
Discussion
We identified germline mutations of BRCA1 or BRCA2 in slightly less than half (43%) of 112 families with at least two confirmed cases of epithelial ovarian cancer in close relatives. The ratio of BRCA1 mutations to BRCA2 mutations was 5:1. Our data confirm previous data suggesting that BRCA1 mutations are more commonly associated with inherited susceptibility to ovarian cancer than are BRCA2 mutations. However, they also demonstrate that BRCA2 mutations are responsible for a significant proportion of families in which only cases of ovarian cancer have been observed.
No BRCA1 or BRCA2 mutation was found in ∼30% of families with an extensive family history of cancer (families containing either three or more cases of ovarian cancer or two or more cases of both breast and ovarian cancer). Some mutations undoubtedly would have been missed by the methods of mutation detection used in the present study; PTT would have missed missense mutations, and, although SSCA/HA would have been likely to detect all small insertions and deletions, it may have missed some single–base-pair substitutions. Because only the coding sequence of both genes was analyzed, mutations occurring in the regulatory regions, which affect transcription, would not have been detected. Evidence of regulatory mutation was sought by looking for a reduction to homozygosity, in cDNA, for polymorphisms heterozygous in genomic DNA, in the 23 families for which RNA samples were available; however, none was found. Because the methods used in this study were PCR based, large genomic deletions and rearrangements in BRCA1, similar to those that recently have been reported elsewhere (Petrij Bosch et al. 1997; Puget et al. 1997; Swensen et al. 1997), may have been missed. In the present study, for those families in which no BRCA1 or BRCA2 mutation was reported, there was insufficient available material with which to perform Southern-blot analysis to look for similar deletions and rearrangements. However, using a mutation-specific PCR assay, we were able to screen for a recently described duplication in BRCA1 (Puget et al. 1999); this mutation was not detected in any of the families without BRCA1 and BRCA2 mutations. Finally, in the majority of families, a blood sample was available from only one affected individual; therefore, it is possible that, in some families, mutation analysis will have been performed on an individual who was a phenocopy.
That a substantial proportion of families had no observed mutation raises the possibility that other genes (or nongenetic familial factors) may be important determinants of familial risk. In the families with either three or more ovarian cancers or four or more early-onset breast or ovarian cancers, those cancers are unlikely to be caused by chance. The failure to detect mutations in ∼30% of these families is, in fact, consistent with the findings of the BCLC studies (Ford et al. 1998). In those studies, combined linkage and mutation analysis indicated that >95% of all families with multiple cases of breast and ovarian cancer are accounted for by BRCA1 and BRCA2 mutations, if a mutation-detection sensitivity of 64% is assumed. We can therefore conclude, with some confidence, that, if there are additional genes, they are of minor importance, compared with BRCA1 and BRCA2, in families with breast and ovarian cancer. However, because there are wide confidence limits on the aforementioned estimates, the possibility of there being one or more additional highly penetrant genes cannot be completely excluded. To search for evidence for such genes, specifically in relation to ovarian cancer, we have so far analyzed, both by linkage and by loss of heterozygosity at the BRCA1 and BRCA2 loci in tumors, 11 families in which there were at least three cases of ovarian cancer and no more than one case of breast cancer diagnosed at age <60 years and in which no BRCA1 or BRCA2 mutation was detected. The best estimate based on these data (S. Ramus, unpublished data) is that the mutation is in either BRCA1 or BRCA2 in ∼50% of such apparently mutation-negative families. Although no single family showed strong evidence against linkage to BRCA1 and BRCA2, it remains possible that some of the disease in these families is the result of a mutation in other genes. One possibility would be the mismatch-repair genes, such as MLH1 and MSH2 (Leach et al. 1993; Papadopoulos et al. 1994). Our preliminary results (P. Harrington, unpublished data) indicate that the replication error–positive (RER+) phenotype expected in such cases is less frequent in tumors from familial cases without mutation than it is in sporadic ovarian cancers. This finding argues against there being a major contribution of mutations in mismatch-repair genes in these families with ovarian cancer, a conclusion that must be confirmed by direct mutation analysis of germline DNA.
A high proportion of families with only two cases of ovarian cancer and either one or no cases of breast cancer had no detectable BRCA1 or BRCA2 mutation. Even if one-third of the BRCA1 and BRCA2 mutations that were present had been missed, this would have accounted for only approximately one-third of such families. The relative contributions of chance, nongenetic familial factors, and genes to the remaining approximately two-thirds of families are unclear. If, for the mother or daughter of an affected individual, the relative risk of ovarian cancer is 2.4 (Stratton et al. 1998), then the expected proportion of affected relative pairs caused by chance would be 40%. However, this estimate of relative risk encompasses all affected pairs, including those that are part of a more extensive family history of cases; therefore, the proportion of chance occurrences will be higher for families with only two affected members.
To further explore the evidence for there being other ovarian cancer–susceptibility genes in these smaller families as well as in the multiple-case families, we performed a segregation analysis of the entire family set. We tested several models that allowed for the possibility of a third ovarian cancer–specific gene in addition to BRCA1 and BRCA2. These analyses gave no support for the existence of a further ovarian cancer–specific locus. We also looked for suggestions, both from the pathology and from familial patterns of associated cancer, that the families without mutations might include a distinct group. A review of the pathology has shown no difference in the histological subtype of ovarian cancers in familial cases with and without a BRCA1 or BRCA2 mutation, nor has it shown such a difference between familial nonmutation cases and a consecutive hospital-based series (Pharoah et al. 1999). The relatives in our 39 families, in which there are only two cases of ovarian cancer and no detectable BRCA1 or BRCA2 mutation, do not appear to show, at sites other than the ovary and breast, patterns of cancer that are distinct from those seen in the families with known mutations, although the nonuniform ascertainment and small numbers preclude a formal comparison.
What are the implications of our data for genetic testing? They indicate that testing for predisposing mutations is likely to be justified in any family in which two or more close relatives have epithelial ovarian cancer and that testing should be directed first at the BRCA1 gene and, if this is negative, at the BRCA2 gene, possibly starting with (but not being restricted to) the “ovarian cancer–cluster region” (Gayther et al. 1997b). Different germline BRCA1 and BRCA2 mutations are probably associated with different risks for breast and ovarian cancer (Easton 1997). We have previously reported that mutations found in the 3′ end of the BRCA1 gene and in a central region of the BRCA2 gene appear to be responsible for a lower risk of ovarian cancer, relative to the risk for breast cancer, compared with mutations elsewhere in the gene (Gayther et al. 1995, 1997b). Both this variation in risk and the fact that these risks may also vary according to family history and other risk factors suggest that some caution should be used when mutation data are used to provide precise risk estimates for clinical management.
In families in which there are only two cases of ovarian cancer and in which no mutation can be found, the risk to unaffected women remains unclear. If many of these families are, in fact, a chance association of cases, the risks may be quite low, and measures such as prophylactic oophorectomy may be inappropriate. Unlike the familial risks of breast cancer, familial risks of ovarian cancer do not seem to be strongly age dependent; therefore, the ages at which the affected individuals were diagnosed may not provide a reliable guide to familial risk. Prospective data from the follow-up of unaffected women at risk in such families will be needed to answer this question.
Acknowledgments
The UKCCCR Familial Ovarian Cancer Register is overseen by a steering committee including the following members: D. T. Bishop, W. Collins, D. F. Easton, G. Fraser, I. T. Jacobs, D. Lowe, J. Mackay, B. A. J. Ponder, J. Shepherd, and C. M. Steel. We thank Joanna Dearden, Paul Pharoah, Carol Pye, and Amy Storfer-Isser for maintaining the register and database. We thank Sylvie Mazoyer for providing positive genomic DNA control samples for the BRCA1 exon 13 duplication. Laboratory and statistical analyses for this study were supported by the CRC. B.A.J.P. is a Gibb Fellow of the CRC.
Electronic-Database Information
The URL for data in this article is as follows:
- Breast Cancer Information Core Database, http://www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic/
References
- Chapman MS, Verma IM (1996) Transcriptional activation by BRCA1. Nature 382:678–679 [DOI] [PubMed]
- Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH (1998) The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci USA 95:5287–5292 [DOI] [PMC free article] [PubMed]
- Claus EB, Risch N, Thompson WD (1991) Genetic analysis of breast cancer in the Cancer and Steroid Hormone Study. Am J Hum Genet 48:232–242 [PMC free article] [PubMed]
- Easton D (1997) Breast cancer genes—what are the real risks? Nat Genet 16:210–211 [DOI] [PubMed]
- Easton DF, Bishop DT, Ford D, Crockford GP (1993) Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am J Hum Genet 52:678–701 [PMC free article] [PubMed]
- Ford D, Easton DF, Peto J (1995) Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am J Hum Genet 57:1457–1462 [PMC free article] [PubMed]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, et al (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet 62:676–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LS, Gayther SA, Kurosaki T, Gordon D, Noble B, Casey G, Ponder BA, et al (1997) Mutation analysis of BRCA1 and BRCA2 in a male breast cancer population. Am J Hum Genet 60:313–319 [PMC free article] [PubMed]
- Gayther SA, Harrington P, Russell P, Kharkevich G, Garkavtseva RF, Ponder BA (1997a) Frequently occurring germ-line mutations of the BRCA1 gene in ovarian cancer families from Russia. Am J Hum Genet 60:1239–1242 [PMC free article] [PubMed]
- Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA, Stratton MR, et al (1997b) Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet 15:103–105 [DOI] [PubMed]
- Gayther SA, Ponder BA (1998) Clues to the function of the tumour susceptibility gene BRCA2. Dis Markers 14:1–8 [DOI] [PMC free article] [PubMed]
- Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, Seal S, et al (1995) Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat Genet 11:428–433 [DOI] [PubMed]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 [DOI] [PubMed]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, et al (1993) Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75:1215–1225 [DOI] [PubMed]
- Lynch HT, Albano WA, Lynch JF, Lynch PM, Campbell A (1982) Surveillance and management of patients at high genetic risk for ovarian carcinoma. Obstet Gynecol 59:589–596 [PubMed]
- Miki Y, Swensen J, Schattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, et al (1994) A strong candidate for the breast and ovarian-cancer susceptibility gene BRCA1. Science 266:66–71 [DOI] [PubMed] [Google Scholar]
- Narod SA, Ford D, Devilee P, Barkardottir RB, Lynch HT, Smith SA, Ponder BA, et al (1995) An evaluation of genetic heterogeneity in 145 breast-ovarian cancer families. Am J Hum Genet 56:254–264 [PMC free article] [PubMed]
- Neuhausen S, Gilewski T, Norton L, Tran T, McGuire P, Swensen J, Hampel H, et al (1996a) Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet 13:126–128 [DOI] [PubMed]
- Neuhausen SL, Mazoyer S, Friedman L, Stratton M, Offit K, Caligo A, Tomlinson G, et al (1996b) Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet 58:271–280 [PMC free article] [PubMed]
- Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, et al (1994) Mutation of a mutL homolog in hereditary colon cancer. Science 263:1625–1629 [DOI] [PubMed]
- Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, et al (1997) BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 17:341–345 [DOI] [PubMed]
- Pharoah PD, Easton DF, Stockton DL, Gayther SA, Ponder BA (1999) Survival in familial, BRCA1 and BRCA2 associated epithelial ovarian cancer. Cancer Res 59:868–871 [PubMed]
- Puget N, Sinilnikova OM, Stoppa-Lyonnet D, Audoynaud C, Pages S, Lynch HT, Goldgar D, et al (1999) An Alu-mediated 6-kb duplication in the BRCA1 gene: a new founder mutation? Am J Hum Genet 64:300–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puget N, Torchard D, Serova-Sinilnikova OM, Lynch HT, Feunteun J, Lenoir GM, Mazoyer S (1997) A 1-kb Alu-mediated germ-line deletion removing BRCA1 exon 17. Cancer Res 57:828–831 [PubMed]
- Ramus SJ, Kote-Jarai Z, Friedman LS, van der Looij M, Gayther SA, Csokay B, Ponder BA, et al (1997) Analysis of BRCA1 and BRCA2 mutations in Hungarian families with breast or breast-ovarian cancer. Am J Hum Genet 60:1242–1246 [PMC free article] [PubMed]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteum J, Ashley T, et al (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 8:265–275 [DOI] [PubMed] [Google Scholar]
- Shattuck-Eidens D, McClure M, Simard J, Labrie F, Narod S, Couch F, Hoskins K, et al (1995) A collaborative survey of 80 mutations in the BRCA1 breast and ovarian cancer susceptibility gene: implications for presymptomatic testing and screening. JAMA 273:535–541 [PubMed]
- Steichen-Gersdorf E, Gallion HH, Ford D, Girodet C, Easton D, DiCioccio RA, Evans G, et al (1994) Familial site-specific ovarian cancer is linked to BRCA1 on 17q12-21. Am J Hum Genet 55:870–875 [PMC free article] [PubMed]
- Stratton JF, Pharoah PD, Smith SK, Easton D, Ponder BA (1998) A systematic review and meta-analysis of family history and risk of ovarian cancer. Br J Obstet Gynaecol 105:493–499 [DOI] [PubMed]
- Struewing JP, Abeliovich D, Peretz T, Avishai N, Kaback MM, Collins FS, Brody LC (1995) The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat Genet 11:198–200 [DOI] [PubMed]
- Swensen J, Hoffman M, Skolnick MH, Neuhausen SL (1997) Identification of a 14 kb deletion involving the promoter region of BRCA1 in a breast cancer family. Hum Mol Genet 6:1513–1517 [DOI] [PubMed]
- Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, et al (1996) The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 12:333–337 [DOI] [PubMed]
- Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson JG, Tavtigian SV, Tulinius H, et al (1996) A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet 13:117–119 [DOI] [PubMed]
- Wong AKC, Pero R, Ormonde PA, Tavtigian SV, Bartel PL (1997) RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene BRCA2. J Biol Chem 272:31941–31944 [DOI] [PubMed]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, et al (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378:789–792 [DOI] [PubMed]
- Zhang H, Tombline G, Weber BL (1998) BRCA1, BRCA2, and DNA damage response: collision or collusion? Cell 92:433–436 [DOI] [PubMed]