Summary
The Muckle-Wells syndrome (MWS) is a hereditary inflammatory disorder characterized by acute febrile inflammatory episodes comprising abdominal pain, arthritis, and urticaria. Progressive nerve deafness develops subsequently, and, after several years, the disease is complicated by multiorgan AA-type amyloidosis (i.e., amyloidosis derived from the inflammatory serum amyloid–associated protein) (MIM 191900) with renal involvement and end-stage renal failure. The mode of inheritance is autosomal dominant, but some sporadic cases have also been described. No specific laboratory findings have been reported. The genetic basis of MWS is unknown. Using a genomewide search strategy in three families, we identified the locus responsible for MWS, at chromosome 1q44. Our results indicate that the gene is located within a 13.9-cM region between markers D1S2811 and D1S2882, with a maximum two-point LOD score of 4.66 (recombination fraction .00) at D1S2836 when full penetrance is assumed. Further identification of the specific gene that is responsible for MWS will therefore provide the first biological element for characterizing MWS, other than doing so on the basis of its variable clinical expression.
Introduction
The Muckle-Wells syndrome (MWS [MIM 191900]) was first described, in 1962, as a rare hereditary disorder with an autosomal dominant mode of inheritance (Muckle and Wells 1962). Since that time, the majority of published studies have focused on kindreds from northern Europe. The disease is characterized by acute febrile inflammatory episodes (denoted as “aguey bouts” in Derbyshire, United Kingdom, the region where the first families with MWS were described). The inflammatory episodes, which commonly manifest in childhood, include abdominal pain, joint inflammation, myalgia, urticaria, and conjunctivitis. These episodes last 24–48 h. Later in the course of the disease, nerve deafness occurs. After several years, the inflammatory condition is complicated by the development of amyloidosis of the AA type (i.e., amyloidosis derived from the inflammatory serum amyloid–associated protein), which can involve most tissues and organs but which especially involves the kidney (Muckle and Wells 1962; Muckle 1979; Linke et al. 1983; Messier et al. 1988). Twenty-six percent of patients have renal amyloidosis that is revealed by proteinuria, leads to nephrotic syndrome, and develops to end-stage renal failure, which, before the era of maintenance hemodialysis, was fatal (Muckle and Wells 1962; Lagrue et al. 1972; Prost et al. 1976; Muckle 1979; Watts et al. 1994). AA amyloidosis is not required for the diagnosis of MWS, since (1) this complication is delayed in the course of the disease, (2) it can be clinically latent (Messier et al. 1988), and (3) it apparently does not occur in every kindred.
At the time when acute clinical episodes occur, laboratory testing may show an acute-phase response with a high erythrocyte sedimentation rate, high leukocyte blood counts, and hypergammaglobulinemia. Although a circadian increase in interleukin-6 blood levels has recently been documented during urticarial flares in a patient with MWS, the cause and the pathogenesis of the disease remain unknown (Gerbig et al. 1998). No treatment has been proved to be beneficial for MWS, although our own experience with family 1 suggests that colchicine exerts a favorable effect on the intensity and repetition of aguey bouts (Messier et al. 1988). The rarity of MWS and the lack of long-term observation leave unresolved the issue of prevention of amyloidosis with protracted colchicine treatment.
The periodic occurrence of inflammatory crises and the potential threat of amyloidosis are reminiscent of characteristics of other hereditary periodic inflammatory diseases (Grateau et al. 1999). These inflammatory diseases are familial Mediterranean fever (FMF); the syndrome of hyperimmunoglobulinemia D and periodic fever (HIDS) (Drenth et al. 1994), both of which have an autosomal recessive mode of inheritance; and the group of autosomal dominant recurrent fevers (ADRF) (Mulley et al. 1998), including familial Hibernian fever (FHF), which is the best-characterized fever within this group (McDermott et al. 1997, 1998).
The gene responsible for FMF, which maps to chromosome 16p13, has been identified; it encodes a putative leukocyte transcriptional factor known as “marenostrin/pyrin” (The French FMF Consortium 1997; The International FMF Consortium 1997). More recently, mutations in the mevalonate kinase gene, which is located at 12q24, have been shown to cause HIDS (Drenth et al. 1999, Houten et al. 1999), whereas in families with FHF and in some families with ADRF the disease is caused by missense mutations in the tumor-necrosis-factor (TNF) receptor 1 (TNFR1) gene, located at chromosome 12p13 (McDermott et al. 1999). McDermott has proposed the term “TRAPS” (TNF receptor–associated periodic syndromes) to define this disease group.
In an effort to localize the MWS gene, we undertook a genomewide scan, using 276 microsatellite markers, in one English family and in two French families. This led to mapping the disease locus to the long arm of chromosome 1 at 1q44.
Patients, Material, and Methods
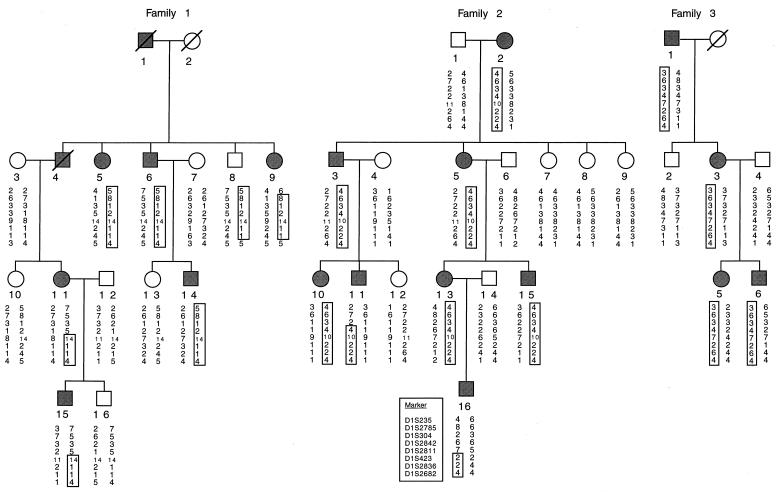
Patients
This project was approved by the ethics committees of Cochin Hospital, Paris, and Norfolk and Norwich Hospital, Norwich, United Kingdom. Three families comprising a total of 35 individuals, including 18 affected individuals, were studied. Each participant was interviewed and examined and provided written informed consent. Studies of some members of these families have been published elsewhere (Lagrue et al. 1972; Prost et al. 1976; Messier et al. 1988; Watts et al. 1994), and the pedigree structure of those individuals participating in this study is shown in figure 1. Characteristic signs of the disease, such as inflammatory crises with urticaria, conjunctivitis, arthralgias, and nerve deafness, were present in all three families. However, amyloidosis of the AA type was observed only in family 1 (Lagrue et al. 1972; Messier et al. 1988). Intermittent oligoarticular synovitis, which affected the large joints and was unresponsive to treatment with colchicine, was reported in family 3 (Watts et al. 1994).
Figure 1.
Pedigree of the three families with haplotype analysis in the region of linkage on chromosome 1. Affected individuals are shown as blackened circles (females) or blackened squares (males). Deceased individuals are denoted by diagonal slashes.
Genotyping
Peripheral blood was obtained from all individuals, and DNA was extracted, by means of the standard phenol/chloroform method, from a lymphocyte pellet. The genomewide screening was performed with the Association Française contre les Myopathies/Généthon panel of 276 fluorescent microsatellite markers at a resolution of 12–14 cM. The PCR products for each template were appropriately pooled, and an aliquot was loaded onto a 5% standard denaturating polyacrylamide gel and was run in an Applied Biosystems (ABI) 373A automated DNA sequencer. Allele sizes were determined on the basis of an internal size standard in each lane (500 Tamra; PE Biosystems ABI), and results were processed by GeneScan 2.1.1. (ABI) software. After allele assignment was done with the use of Genotyper 1.1.1. (ABI) software, each allelic profile was verified by two individuals, and the genotypes were checked for Mendelian segregation, by use of the Marksyn (Généthon) software.
Linkage Analysis
Statistical analyses were performed on the basis of an autosomal dominant disease with full penetrance. The frequency of the abnormal allele was set at .0001, in view of the rarity of the disorder. The feasibility of the linkage study was explored by use of SLINK (Ott 1989). Pairwise LOD scores were calculated, with isofrequencies of the marker alleles being assumed, by use of the MLINK subroutine of the FASTLINK 4.0 package (which also contains ILINK and LINKMAP) (Cottingham et al. 1993). Values for the maximum LOD score (Zmax) were calculated by the ILINK program. The recombination frequency was assumed to be equal for both sexes. Multipoint LOD scores were calculated by use of the LINKMAP subroutine. Haplotypes, which were constructed and assigned in such a way as to minimize the number of crossovers in each family, were analyzed to determine the minimal candidate region for the location of MWS. Homogeneity was tested by means of the HOMOG program (Ott 1991).
Results
Segregation of the disease in the families suggests the presence of a single disease gene with an autosomal dominant mode of inheritance and very high penetrance. Because MWS is reminiscent of other hereditary periodic inflammatory diseases, we first investigated linkage analyses at loci 16p13 (for FMF), 12q24 (for HIDS), and 12p13 (for FHF). Two-point LOD scores between the disease and markers (surrounding 10–18-cM regions) were strongly negative in the three families (data not shown), and no single haplotypes were shared among affected individuals. This finding excluded the genomic regions of FMF, HIDS, and FHF as being responsible for MWS. SLINK simulation showed that linkage detection by means of a genomewide search was feasible with these three families, and a semiautomated genomewide search for linkage was initiated with 276 fluorescent microsatellite markers at a resolution of 12–14 cM. Under the assumption of full penetrance, two-point LOD scores showed linkage to the subterminal region of the long arm of chromosome 1. Initially, a suggestive LOD score of 2.76 (recombination fraction [θ] = .05) was obtained for the marker at locus D1S2682, and all the Généthon-map microsatellite markers in the region were typed (to an average density of five markers per 23 cM). Linkage was obtained with a Zmax of 4.66 (θ = .00) at marker D1S2836 (table 1). The LOD scores did not change when they were calculated with penetrance values of .95, .90, and .85 (data not shown).
Table 1.
Two-Point Linkage Analysis Data for the MWS Gene and Markers Spanning a Sex-Averaged Interval of 20 cM at Chromosome 1q44
|
LOD Score at θ = |
|||||||||
| Marker | .00 | .01 | .05 | .1 | .2 | .3 | .4 | Zmax | θ |
| D1S235 | −∞ | −2.69 | –.79 | −.15 | .23 | .25 | .15 | .26 | .258 |
| D1S2785 | −∞ | −6.62 | −2.67 | −1.18 | −.04 | .29 | .26 | .31 | .336 |
| D1S304 | −∞ | −1.66 | −.36 | .1 | .37 | .36 | .22 | .43 | .236 |
| D1S2842 | −∞ | −1.05 | .82 | 1.40 | 1.53 | 1.17 | .59 | 1.59 | .167 |
| D1S2811 | −∞ | −1.00 | .32 | .78 | .98 | .80 | .41 | .99 | .190 |
| D1S423 | 1.43 | 1.43 | 1.33 | 1.19 | .86 | .61 | .19 | 1.43 | 0 |
| D1S2836 | 4.66 | 4.59 | 4.30 | 3.89 | 2.96 | 1.93 | .86 | 4.66 | 0 |
| D1S2682 | −∞ | 2.38 | 2.76 | 2.64 | 2.5 | 1.29 | .61 | 2.77 | .053 |
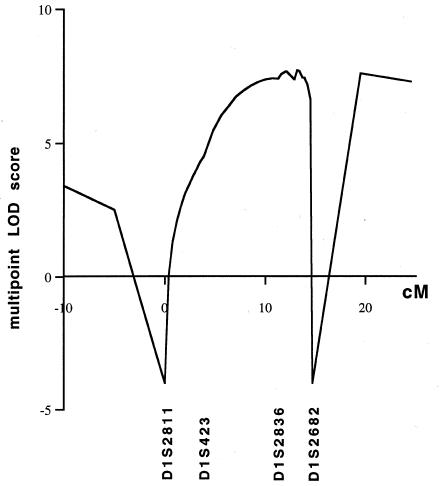
Haplotypes were constructed by parsimony (fig. 1). Several recombinants that defined the limits of the MWS region were detected. Analysis of individual 16 in family 2 showed that the centromeric limit of the MWS interval is located between markers D1S2811 and D1S2836. This limit could not be defined more precisely, since the affected mother (individual 13) was homozygous for marker D1S423. The recombinant event in family 1 (individual 9) suggested that the telomeric limit would be located between markers D1S2836 and D1S2682, but this could not be confirmed, since the haplotype of the affected father (individual 1) is unknown. No common haplotype was apparent in these three families. The HOMOG program detected no evidence to support genetic heterogeneity. A maximum log likelihood of 10.73 was obtained for α = 1, at marker D1S2836, where α is the proportion of families with linkage to chromosome 1q (table 2). Therefore, the minimal region of linkage is defined by a 13.9-cM critical interval, extending from marker D1S2811 to marker D1S2682, that must contain the MWS gene. Multipoint linkage analysis (fig. 2) confirmed the localization.
Table 2.
HOMOG Analysis
| Hypothesisa | df | χ2 | Likelihood Ratio |
| H2 vs. H1 | 1 | .000 | 1.0000 |
| H1 vs. H0 | 1 | 21.460 | 4.571x104 |
| H2 vs. H0 | 2 | 21.460 | 4.571x104 |
H0 = no linkage; H1 = linkage with homogeneity; H2 = linkage with heterogeneity.
Figure 2.
Multipoint linkage analysis between MWS and selected markers on chromosome 1q. D1S2811 was arbitrarily assigned position 0, and the order and genetic distances of the maker loci were determined by the use of the Généthon genetic map (Dib et al. 1996).
Discussion
The results of the present study have identified, in three families, a major-gene locus for MWS, at chromosome 1q44. We first excluded linkage to the loci for FMF, HIDS, and ADRF, which are located at 16p13.3, 12q24, and 12p13, respectively. A genomewide search strategy was then adopted, and we found a suggestive LOD score of 2.76 (θ = .05) at locus D1S2682. In this region, we tested all available markers from the Généthon map and obtained a Zmax of 4.66 (θ = .00) at marker D1S2836. The genomewide scan did not support linkage to any other site. Intrafamilial recombination analyses placed the MWS locus within a 13.9-cM interval between markers D1S2811 and D1S2682. This localization was confirmed by a multipoint linkage analysis, whereas the results obtained with the HOMOG program were consistent with genetic homogeneity in the three families. However, because of the small number of families in this study, genetic heterogeneity cannot be definitively excluded for MWS.
In these three families, MWS appeared to segregate as an autosomal dominant trait, with complete penetrance and variable expression being consistent with current data on MWS. In particular, variable expression was observed in family 1, in which individual 5, a 70-year-old woman, has arthralgia only, although several of her children (who refused to participate in the study) exhibit a complete clinical phenotype. Sporadic cases of MWS have also been reported. It is unclear whether these sporadic cases are, in fact, familial cases in kindreds with incomplete penetrance, new mutations, variable clinical expression, and either underdiagnosis of mild forms of the disease or misdiagnosis of such close entities as the chronic infantile neurological cutaneous articular syndrome (Prieur et al. 1987). Given the lack of specific laboratory markers to substantiate the diagnosis of MWS, mild or atypical forms of the disease may be underdiagnosed. When genetically linked DNA markers become available, this should no longer be the case, and an accurate tool for diagnosis of MWS, even in the absence of explicit symptomatology, will then be provided.
Family 3 is English, and families 1 and 2 are, respectively, from the western and northern regions of France. Historically, both of these regions of France were occupied by the English armies for a long period during the Hundred Years War. Virtually all members of family 1 are fair haired and blue eyed. We traced the origins of this family back to the year 1600 and to a small town near Beauvais, France, where a common (village green) is still named “Le Clos aux Anglais” (Englishmen's Field). Therefore, given the rarity of MWS, it may be hypothesized that there is a founder effect. Haplotype analysis of the three families did not show any common region in the disease-bearing chromosomes. This finding suggests that the mutation is different in each family. However, we cannot exclude an ancient founder effect, which would not be detected with markers spaced only a few centimorgans from each other. Only characterization of the mutation or analysis of the polymorphisms within the gene would address this hypothesis.
In the families that we studied, some degree of clinical variability was observed with regard to urticaria, deafness, and joint involvement, which suggests the existence of modulating genes. Amyloidosis, which is a main complication of MWS, has been observed only in family 1 (Messier et al. 1988). This is not surprising, since, for the reasons given above, AA amyloidosis is not an obligatory finding in MWS. This is also the case for such other hereditary inflammatory syndromes as FMF and ADRF, in which amyloidosis affects only some individuals in a given family and never even develops in some families. Amyloidosis is also known to occur, in some individuals, as a complication of such nonhereditary chronic inflammatory disorders as rheumatoid arthritis. As in the aforementioned inflammatory disorders, the occurrence of amyloidosis in MWS probably depends on additional genetic and environmental factors.
MWS can be viewed as a composite syndrome germane to such recurrent inflammatory syndromes as FMF, HIDS, and ADRF, as well as to familial urticaria and hereditary deafness. Many cytokine-network derangements have been described in FMF and HIDS. Some of the main cytokine or cytokine-receptor genes have been considered as candidate genes for these diseases. However, since the genes responsible for both of these syndromes do not encode this class of molecules, cytokine dysregulation appears to be a secondary event in these inflammatory diseases. Conversely, it has been shown that a gene responsible for at least one form of ADRF encodes TNFR1 (McDermott et al. 1999); the mutations of this gene lead to unmatched binding of soluble forms of TNFR1 to TNF. An early study of circulating cytokines and cytokine receptors did not find any significant modifications in either the soluble form or the TNFR1 concentration during the inflammatory episode (Mc Dermott and Powell 1997). However, McDermott et al. (1999) recently reported that p55 levels were significantly depressed in carriers of the TNFR1 mutations, compared with such levels in normal individuals, and that, although they do rise during an inflammatory episode, the increase is much less than that seen during an inflammatory response in individuals without such mutations. Thus, although dysregulation of the circadian rhythm of interleukin-6 has recently been shown in one family with MWS (Gerbig et al. 1998), one must be cautious in approaching the cause of the disease via cytokines.
The MWS localization that we have characterized also constitutes a candidate region for such diverse familial forms of urticaria as cold hypersensitivity (MIM 120100), aquagenic urticaria (MIM 191850), mast-cell disease (MIM 154800), dermodistortive urticaria (MIM 125630), and localized heat urticaria (MIM 191950), for which none of the responsible genes has yet been cloned or even localized.
The most intriguing clinical feature of MWS is nerve deafness, which best distinguishes MWS from other inflammatory disorders. To the best of our knowledge, in only one case (Muckle and Wells 1962) was the inner ear of a patient with MWS studied at autopsy. Dissection excluded inner-ear amyloidosis, and deafness was attributed to absence of both Corti's organ and the vestibular sensory epithelium, in addition to cochlear nerve atrophy and basilar membrane ossification. However, we are not aware of any data regarding a gene that underlies syndromic or nonsyndromic deafness and that would be located within the interval containing the MWS gene (Kalatzis and Petit 1998). In addition, by use of the GeneMap'98 database, no obvious candidate gene was identified in the disease interval
Our localization of the MWS gene on the long arm of chromosome 1 opens the way for important developments. First, mapping of the gene will provide a biological marker in a disease with various clinical expressions. Second, cloning—presumably, positional cloning—of the gene may lead to elucidation of the links between urticaria, the inflammatory response leading to AA amyloidosis, and deafness.
Acknowledgments
We are grateful to the study participants for their kind cooperation in this research effort. The research was performed with the financial support of Contrat de Recherche Clinique CRC 950162, 1995, from the Délégation à la Recherche Clinique, Assistance Publique-Hôpitaux de Paris. J.P.H.D. is a recipient of both a grant from The Niels Stensen Foundation and Netherlands Organisation of Scientific Research travel grant F92-189. The authors express their gratitude to Dr. Marielle Berlioz-Thibal for invaluable help in tracing the genealogy and geographical origin of family 1 back to 1600.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genemap'98, http://www.ncbi.nlm.nih.gov/genemap98 (for identification of candidate genes)
- Généthon, http://www.genethon.fr (for microsatellite markers)
- National Institutes of Health FTP Site, ftp://fastlink.nih.gov/pub/fastlink (for FASTLINK software)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for MWS [191900], cold hypersensitivity [120100], aquagenic urticaria [191850], mast-cell disease [154800], dermodistortive urticaria [125630], and localized heat urticaria [191950])
- The Lab of Statistical Genetics, Rockefeller University, ftp://linkage.rockefeller.edu/software/ (for SLINK and HOMOG software)
References
- Cottingham RW Jr, Idury RW, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed]
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Drenth JPH, Cuisset L, Grateau G, Vasseur C, van de Velde-Visser SD, de Jong JG, Beckmann JS, et al (1999) Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome: International Hyper-IgD Study Group. Nat Genet 22:178–181 [DOI] [PubMed]
- Drenth JPH, Haagsma CJ, van der Meer JW (1994) Hyperimmunoglobulinemia D and periodic fever syndrome: the clinical spectrum in a series of 50 patients: International Hyper-IgD Study Group. Medicine (Baltimore) 73:133–144 [PubMed]
- French FMF Consortium, The (1997) A candidate gene for familial Mediterranean fever. Nat Genet 17:25–31 [DOI] [PubMed]
- Gerbig AW, Dahinden CA, Mullis P, Hunziker T (1998) Circadian elevation of IL-6 levels in Muckle-Wells syndrome: a disorder of the neuro-immune axis? QJM 91:489–492 [DOI] [PubMed]
- Grateau G, Drenth JPH, Delpech M (1999) Hereditary fevers. Curr Opin Rheumatol 11:75–78 [DOI] [PubMed]
- Houten SM, Kuis W, Duran M, de Koning TJ, van Royen-Kerkhof A, Romeijn GJ, Frenkel J, et al (1999) Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nat Genet 22:175–177 [DOI] [PubMed]
- International FMF Consortium, The (1997) Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90:797–807 [DOI] [PubMed]
- Kalatzis V, Petit C (1998) The fundamental and medical impacts of recent progress in research on hereditary hearing loss. Hum Mol Genet 7:1589–1597 [DOI] [PubMed]
- Lagrue G, Vernant JP, Revuz J, Touraine R, Weil B (1972) Syndrome de Muckle et Wells: cinquième observation familiale. Nouv Presse Med 1:2223–2226 [PubMed]
- Linke RP, Heilmann KL, Nathrath WB, Eulitz M (1983) Identification of amyloid A protein in a sporadic Muckle-Wells syndrome N-terminal amino acid sequence after isolation from formalin-fixed tissue. Lab Invest 48:698–704 [PubMed]
- McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, et al (1999) Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 97:133–144 [DOI] [PubMed]
- McDermott MF, Ogunkolade BW, McDermott EM, Jones LC, Wan Y, Quane KA, McCarthy J, et al (1998) Linkage of familial Hibernian fever to chromosome 12p13. Am J Hum Genet 62:1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott MF, Powell RJ (1997) Circulating cytokine concentrations in familial Hibernian fever. In: Sohar E, Gafni J, Pras M (eds) Familial Mediterranean fever. Freund Publishing, London and Tel Aviv, pp 189–192 [Google Scholar]
- McDermott EM, Smillie DM, Powell RJ (1997) Clinical spectrum of familial Hibernian fever: a 14-year follow-up study of the index case and extended family. Mayo Clin Proc 72:806–817 [DOI] [PubMed]
- Messier G, Meyrier A, Rainfray M, Coste T, Callard P (1988) Overt or occult renal amyloidosis in the Muckle-Wells syndrome. Kidney Int 34:566 [Google Scholar]
- Muckle TJ (1979) The `Muckle-Wells' syndrome. Br J Dermatol 100:87–92 [DOI] [PubMed]
- Muckle TJ, Wells M (1962) Urticaria, deafnesss and amyloidosis: a new heredo-familial syndrome. QJM 31:235–248 [PubMed] [Google Scholar]
- Mulley J, Saar K, Hewitt G, Ruschendorf F, Phillips H, Colley A, Sillence D, et al (1998) Gene localization for an autosomal dominant familial periodic fever to 12p13. Am J Hum Genet 62:884–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed]
- ——— (1991) Analysis of human genetic linkage. Johns Hopkins University Press, Baltimore [Google Scholar]
- Prieur AM, Griscelli C, Lampert F, Truckenbrodt H, Guggenheim MA, Lovell DJ, Pelkonnen P, et al (1987) A chronic, infantile, neurological, cutaneous, and articular (CINCA) syndrome: a specific entity analysed in 30 patients. Scand J Rheumatol Suppl 66:57–68 [DOI] [PubMed]
- Prost A, Barriere H, Legent F, Cottin S, Wallez B (1976) Rhumatisme intermittent révélateur d'un syndrome familial arthrites-éruption urticarienne-surdité: syndrome de Muckle et Wells sans amylose rénale. Rev Rhum Mal Ostéoartic 43:201–208 [PubMed]
- Watts RA, Nicholls A, Scott DG (1994) The arthropathy of the Muckle-Wells syndrome. Br J Rheumatol 33:1184–1187 [DOI] [PubMed]