Summary
The genetic contribution to common forms of osteoarthritis (OA) is well established but poorly understood. We performed a genome scan, using 302 markers for loci predisposing to distal interphalangeal joint (DIP) OA. To minimize genetic heterogeneity in our study sample, we identified siblings with a severe, radiologically defined phenotype from the nationwide registers of Finland. In the initial genome scan, linkage analysis in 27 sibships gave a pairwise LOD score (Z) >1.00 with nine of the screening markers. In the second stage, additional markers and family members were genotyped in these chromosomal regions. On 2q12-q13, IL1R1 resulted in Z=2.34 at recombination fraction (θ) 0, allowing a dominant mode of inheritance. Association analysis of markers D2S2264, IL1R1, D2S373, and D2S1789 jointly provided some evidence for a shared haplotype among the affected individuals (P value of .012). Also, multipoint nonparametric linkage analysis yielded a P value of .0001 near the locus IL1R1 and P=.0007 ∼20 cM telomeric near marker D2S1399, which, in two-point analysis, gave Z=1.48 (θ=.02). This chromosomal region on 2q harbors the interleukin 1 gene cluster and, thus, represents a good candidate region for inflammatory and autoimmune disorders. Three additional chromosomal regions—4q26-q27, 7p15-p21, and Xcen—also provided some evidence for linkage, and further analyses would be justified to clarify their potential involvement in the genetic predisposition to DIP OA.
Introduction
Osteoarthritis (OA; MIM 165720) represents a common cause of human musculoskeletal disability. This heterogeneous disorder is characterized by both degenerative and reactive changes in the joints, thought to be driven by a spectrum of environmental and genetic factors (Felson 1988; Felson et al. 1998; Hirsch et al. 1998). To study the genetic predisposition to a complex disease such as OA, accurate phenotype ascertainment is a necessity, and radiological changes are considered superior to clinical manifestations in the diagnostic definition of OA (Hart et al. 1994). The proportion of variability attributed to genetic factors in the susceptibility to radiologically defined OA depends on the sex and the joint-specific pattern of the disorder (Kaprio et al. 1996; Spector et al. 1996) and is estimated to be 39%–65% for OA of the hands and knees in female subjects (Spector et al. 1996).
The prevalence of severe hand OA in Finland is low in subjects of age <50 years (0.04% in age group 40–49 years), after which it increases rapidly (0.18% in age group 50–59 years) (Kärkkäinen 1985). There is some evidence that distal interphalangeal joint (DIP) OA, characterized by symmetry of joint involvement, female preponderance, and Heberden nodes (Stecher et al. 1941; Cooper et al. 1996), represents a specific form of OA with the strongest genetic component and, thus, is an ideal target for genetic studies aimed at dissecting the molecular background of OA (Spector et al. 1996; Felson et al. 1998; Hirsch et al. 1998).
In addition to careful dissection of the clinical phenotype, the utilization of study samples from isolated populations can reduce the complexity of polygenic disorders (Lander and Schork 1994). The population structure of Finland has proved advantageous for disease-locus mapping, because of the restricted number of ancestral founders (leading to somewhat reduced gene diversity) and subsequent genetic isolation (Mahtani et al. 1996; Kuokkanen et al. 1997). To search for DIP OA loci, we performed a genome scan in a study sample of 27 sibships, identified on the basis of nationwide study cohorts. The results provide evidence for linkage on the chromosomal region 2q12-q13 harboring a cluster of potentially relevant candidate genes for OA. Nonparametric multipoint analysis (SIMWALK 2.40) revealed evidence for two additional loci, on chromosomes 4q and 7p.
Subjects and Methods
Family Ascertainment
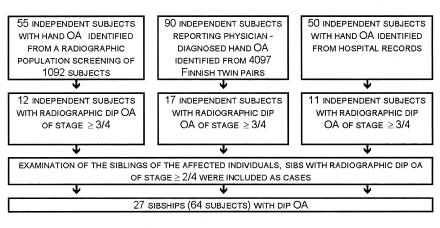
We contacted subjects who had participated in any of three studies (fig. 1), as follows: (a) 55 subjects recorded as having hand OA in the Mini-Finland Health Survey (Kärkkäinen 1985), (b) 90 twin pairs reporting physician-diagnosed hand OA in a questionnaire of the Finnish twin-cohort study in 1995 (Kujala et al., in press), or (c) 50 patients with hand OA identified by the Rheumatism Foundation Hospital in an earlier study (Vikkula et al. 1993). In all stages, the studies were approved by the institutional ethical committees, and informed consent was obtained from the participants. Volunteers responded to a questionnaire, participated in hand x-ray examination, and provided blood samples for genetic analysis. A radiologist who did not have access to the other subject information graded the osteoarthritic changes in the hand x-rays (Kellgren 1963), DIP, proximal interphalangeal joints (PIP), and metacarpophalangeal joints (MCP), and both hands separately. Subjects with rheumatoid arthritis were excluded, on the basis of clinical or radiological data. Because of the high prevalence of DIP OA in the population with of >50 years, stringent diagnostic criteria were applied. The inclusion criteria in our final study group were 3d- or 4th-degree radiographic OA in DIP joints and a positive family history of OA, with at least one sibling with DIP OA. The other siblings volunteering for the study went through the same protocol as described above, except that subjects with 2d- or 4th-degree radiographic OA in DIP joints were also included as affected individuals. The distribution of OA stages, by joint and by hand, among the subjects included in the first stage of the genome scan is given in table 1.
Figure 1.
Schematic presentation of the identification of subjects with DIP OA, who were included in the first stage of the genome scan (for details, see Subjects and Methods).
Table 1.
Distribution of OA Stages (Kellgren 1963), by Joint and by Hand, among Subjects Included in the First Stage of the Genome Scan
|
Distribution of OA in (%) |
||||||
| DIP Joints |
PIP Joints |
MCP Joints |
||||
| Stage | Right | Left | Right | Left | Right | Left |
| 0 | 0 | 0 | 0 | 0 | 29 | 28 |
| I | 0 | 0 | 35 | 30 | 39 | 43 |
| II | 25 | 26 | 21 | 28 | 19 | 21 |
| III | 21 | 23 | 16 | 18 | 8 | 5 |
| IV | 54 | 51 | 26 | 23 | 5 | 3 |
Our final study group comprised a total of 64 affected subjects (11 men and 53 women; mean age at x-ray 58.9 years), derived from 27 independent families (3 families with 4 affected sibs, 4 families with 3 affected sibs, and 20 families with affected sib pairs [ASP]). Only affected sibs were genotyped in the first stage, but samples were collected from all available parents, for phase information. In the second stage, 16 additional affected family members (5 mothers, 3 children, 4 additional siblings, and 4 first cousins) were also genotyped for denser marker maps. To obtain control-allelic frequencies and haplotypes for the IL1R1 locus and flanking markers, 16 Finnish core families were genotyped.
Genotyping
For the initial low-resolution scan, we genotyped 302 microsatellite markers, with an average spacing of ∼13 cM, from a modified Cooperative Human Linkage Center (CHLC) human-screening set (Weber version 6.0). The order and sex-averaged distances of these markers were determined on the basis of the published CHLC map (CHLC database). For the second stage, additional markers were genotyped in the regions yielding pairwise or ASP LOD scores (Z) >1.00. Markers were selected from the Genome Database, the Genetic Location Database, and the Marshfield Medical Research Foundation. Intragenic markers from relevant candidate genes were chosen if they were located on the chromosomal regions yielding positive Z values, and the marker order and the distances between the markers were determined in accordance with the above-mentioned maps. For the higher-resolution map of the chromosomal region 2q12-q24, the marker order was determined on the basis of Whitehead contigs WC2.8, WC2.9, and WC2.11 and on the contig map containing critical region of NPH1 (familial juvenile nepronophthisis), all accessed with the Mapview function from the Genome Database. The location of marker IL1R1 was determined by radiation-hybrid mapping with a G3 RH panel (Research Genetics), the Stanford Human Genome Center radiation hybrid–mapping e-mail server, and the program RHMAP (v. 2.01; M. Boehnke). Two DQ microsatellite markers (DQCAR and G51152, allelic frequencies of which were available in the Finnish population) on 6p21.3 (Lin et al. 1997) were genotyped to thoroughly cover the human leukocyte antigen (HLA) region. The fluorecently labeled PCR products were electrophoretically separated, either with an automated laser fluorecence DNA sequencer (Pharmacia Biotech) or with an ABI 377 sequencer (PE Biosystems) with GENESCAN (version 2.1) peak-calling software. The alleles were identified with ALLELE LINKS (Pharmacia Biotech) or with GENOTYPER (version 2.0; PE Biosystems).
Statistical Analysis
The data were analyzed with the ANALYZE program, by FASTLINK version 2.2 (Cottingham et al. 1993), with application of both “model-based” and “model-free” tests. In the first stage, two-point affecteds-only linkage analysis was performed with MLINK (Lathrop and Lalouel 1984), allowing both dominant and recessive mode-of-inheritance models, with phenocopy penetrance of .01 and disease-allele frequencies of .1 and .0001. Allele frequencies for marker loci were estimated under the null hypothesis of no linkage, on the basis of all individuals included in the analysis, with use of the DOWNFREQ program of ANALYZE (Göring and Terwilliger 1999). Nonallelic heterogeneity among the families was assessed with HOMOG (Ott 1991, p. 223; Kuokkanen et al. 1996). ASP analysis was performed with a maximum likelihood–pseudomarker method, as implemented in the SIBPAIR program (Kuokkanen et al. 1996). In the second stage, the regions followed up were analyzed with the program POLYLOCUS (Terwilliger and Ott 1993), which adds information from neighboring markers to the two-point analysis when the primary marker being analyzed is uninformative for linkage, thus increasing the power. For regions showing suggestive evidence of linkage in the two-point linkage analysis, multipoint nonparametric analysis was performed with SIMWALK 2.40 (Sobel and Lange 1996). This program slides an imaginary trait locus across the marker map and calculates several sharing statistics at each position of the map. The “statistic B” measures the maximum number of genes among affecteds identical by descent (IBD) to any founder and tends to have greatest power against dominant disease transmission. Association analyses for a single locus and for multiple loci were performed with the DISLAMB and DISMULT programs, respectively (Terwilliger 1995). Haplotypes were constructed with GENEHUNTER (Kruglyak et al. 1996).
Results
First-Stage Genome Scan
We performed the initial genome scan in 27 sibships with DIP OA. Complete linkage data of the genome scan, done with 302 screening markers providing the average marker density of ∼13 cM, are available on our web site (Department of Human Molecular Genetics, National Public Health Institute, Finland).
In pairwise linkage analysis, Z>1.00 was obtained with seven screening-set markers, in the chromosomal regions 2q, 7p, 8q, 9p, 9q, and 12q. In addition, two markers on 4q and 10p gave Z>1.00 in ASP analysis. Three consecutive markers on Xcen yielded Z<1.00, but they were still included in the second-stage analysis because of the width of the chromosomal region showing mildly positive Z values.
Second-Stage Genome Scan
In the second stage, the promising regions were further examined by genotyping of additional markers and family members (see Subjects and Methods). The complete results of the second-stage analyses are available on our web site (Department of Human Molecular Genetics, National Public Health Institute, Finland).
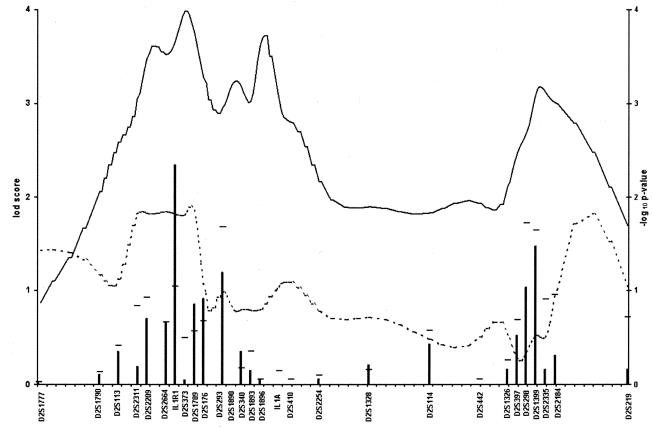
In the initial genome scan, marker D2S410 gave a two-point Z of 2.51 at recombination fraction (θ) 0 (dominant mode). D2S1399, ∼20 cM telomeric, had a two-point Z of 1.48 (θ=.02 [dominant mode]), and an ASP Z of 1.68. Additional markers genotyped in the second stage included intragenic microsatellite markers for two candidate genes in the region: 222/223 for interleukin 1 alpha (IL1A; MIM 147760) and IL1R1 (for interleukin 1 type I receptor; MIM 147810). IL1R1 gave a maximum pairwise Z of 2.34 (θ=0 [dominant mode]), and a higher resolution map of this 20-cM chromosomal region was constructed (fig. 2). The order of the markers was established on the basis of clone contigs and physical maps available for the region (see Subjects and Methods).
Figure 2.
Results of pairwise linkage analysis (dominant inheritance, disease-allele frequency of .0001) and multipoint nonparametric analysis (statistic B) of markers genotyped in a 62-cM region on 2q11-q24. The spacing between hash marks on the X-axis is 1 cM. The columns represent the pairwise dominant Zmax values. Two-point Z values obtained with information from all markers of the figure in the polylocus analysis are indicated by short horizontal lines above the corresponding columns. The continuous line represents the −log10 P values for statistic B results, measuring the maximum number of genes among affecteds IBD to any founder gene, as described in the article by Sobel and Lange (1996). NPL scores, as implemented in GENEHUNTER, are represented by the dashed line and are included for comparison. The marker order was determined on the basis of the contig maps (see Subjects and Methods).
With markers telomeric to IL1R1, a two-point Z of 1.19 (θ=.10) with marker D2S293 was obtained. Near marker D2S1399 (2q21), marker D2S298 (Z=1.03; θ=.10) also resulted in Z>1.00 (fig. 2). Polylocus two-point analysis, which extracts information from multiple markers jointly, was also conducted with all the markers on 2q11-q24 and gave higher Z values for markers D2S293 (Z=1.68), D2S298 (Z=1.72), and D2S1399 (Z=1.65), but did not increase the Z value obtained in two-point analysis with IL1R1 (see fig. 2). In these analyses, the dominant mode of inheritance systematically yielded the highest Z values in this region. In nonparametric multipoint analysis, done with SIMWALK 2.40 (Sobel and Lange 1996), statistic B gave P= .0001 near marker IL1R1 and gave P=.0007 near marker D2S1399 (fig. 2). SIMWALK 2.40 also calculates the nonparametric statistics as implemented in GENEHUNTER, and this nonparametric linkage (NPL) curve is also shown in figure 2.
We wanted to monitor for potential linkage disequilibrium at the IL1R1 locus, and this and flanking microsatellite markers were genotyped in a control-sample set of 16 Finnish core families yielding a total of 64 control haplotypes. The allelic frequencies were found to differ slightly between the independent DIP OA cases (n=37) and the control subjects (n=32). The IL1R1 94-bp allele was more prevalent in individuals affected with DIP OA (55% vs. 39%), whereas the 92-bp allele was found at a lower frequency in the affected than in the control sample (15% vs. 26%) (P=.05). A putative shared haplotype, with markers D2S2264–1 cM–IL1R1–1 cM–D2S373–1 cM–D2S1789, was identified in six DIP OA families, but in only 2 of 64 control chromosomes. Association analysis for multiple loci jointly, done with the program DISMULT (Terwilliger 1995), yielded a P value of .012 between markers D2S373 and D2S1798. Parts of this potential risk haplotype were also more prevalent in chromosomes of affected individuals than in controls (table 2).
Table 2.
The Common D2S2264–1 cM–IL1R1–1 cM–D2S373–1 cM–D2S1798 Haplotypes Observed in the DIP OA Pedigrees
| Marker | Haplotypesa | |||
| D2S2264 | 12 | 12 | ||
| IL1R1 | 10 | 10← | 10 | 10← |
| D2S373 | 10← | 10 | 10← | 10 |
| D2S1789 | 6 | 6 | ||
| No. of DIP OA Pedigreesb (n=27) |
||||
| 6 (4) | 8 (5) | 13 (8) | 18 (10) | |
| No. of Control Chromosomes (n=64) |
||||
| 2 | 2 | 7 | 8 | |
| Pc | .012 | .049 | .012 | .049 |
An arrow (←) within the corresponding haplotype indicates the map position revealing the strongest association.
The number of families in which the corresponding haplotype is shared by all affected individuals is shown in parentheses.
One randomly selected affected individual from each DIP OA family (54 affected chromosomes) and 64 control chromosomes were included in the association analysis for multiple loci jointly, done with DISMULT (Terwilliger 1995), to obtain the P value.
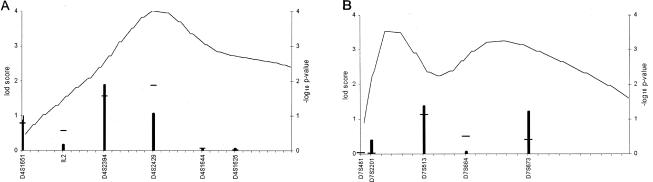
The results of two-point analysis of markers genotyped on other chromosomal regions in the second stage are presented in table 3. On 4q26-q27, marker D4S2394 resulted in Zmax=1.89 (θ=.10) with the recessive mode of inheritance and Zmax=1.72 (θ=0) when the dominant mode was applied. When combined information from markers D4S1651, IL2 (the intragenic marker for the interleukin 2 gene), D4S2394, D4S2429, and D4S1644—flanking ∼5 cM—was used, polylocus analysis gave two-point Z values of 1.89 and 1.85 with markers D4S2394 and D4S2429, respectively (fig. 3). Statistic B gave P=.0001 in the vicinity of the marker D4S2394 (fig. 3).
Table 3.
Two-Point Linkage Analysis of the Markers Outside 2Q, 4Q, and 7P, Analyzed in the Second Stage of the Genome Scan
|
Zmax |
Zmax Polylocus |
||||||
| Region | DistancebetweenMarkers (cM) | Marker or Locus | Dominant | Recessive | Dominant | Recessive | ASP |
| 8q21-q24 | D8S1018 | .00 | .00 | .19 (.22) | .00 | .00 | |
| 2 | GAAT1A4 | 1.62 (.00) | .70 (.16) | 1.39 (.00) | .56 (.18) | .75 | |
| 2 | D8S257 | .00 | .00 | .00 | .00 | .00 | |
| 9p-p22 | D9S54 | .01 (.34) | .00 | .17 (.16) | .12 (.28) | .01 | |
| 4 | D9S178 | .00 (.38) | .01 (.38) | .26 (.10) | .35 (.22) | .02 | |
| 1 | D9S2169 | .76 (.00) | 1.14 (.10) | .65 (.00) | .78 (.14) | .99 | |
| 6 | D9S921 | .00 | .00 | .00 | .00 | .00 | |
| 9q33-q34 | D9S1850 | .00 | .00 | .06 (.22) | .10 (.26) | .00 | |
| 4 | D9S915 | 1.12 (.00) | 2.23 (.00) | 1.12 (.00) | 2.23 (.00) | 1.51 | |
| 4 | DBH | .16 (.16) | .21 (.24) | .35 (.10) | .31 (.22) | .19 | |
| 1 | D9S158 | .00 | .00 | .00 | .00 | .00 | |
| 10p14-p15 | D10S552 | .00 (.42) | .03 (.32) | .00 | .08 (.28) | .08 | |
| 4 | D10S189 | .78 (.12) | 1.05 (.12) | .58 (.16) | .52 (.18) | 1.20 | |
| 8 | D10S1412 | .00 | .02 (.36) | .00 | .01 (.38) | .00 | |
| 12q24 | D12S367 | .00 | .00 | .00 | .03 (.36) | .00 | |
| 3 | GATA32F05 | .23 (.24) | .00 | .06 (.34) | .00 | .18 | |
| 10 | D12S1045 | .00 | .00 | .00 | .00 | .00 | |
| Xcen | DXS6810 | .00 | .00 | .00 | |||
| 1 | MAOA | .00 | .04 (.26) | .04 | |||
| 4 | DXS1003 | .65 (.04) | .92 (.02) | .34 | |||
| 3 | DXS1039 | .93 (.00) | 1.72 (.00) | .84 | |||
| 6 | AR | .67 (.06) | .99 (.06) | .28 | |||
| 2 | DXS7132 | .64 (.10) | .47 (.14) | .55 | |||
| 8 | DXS6800 | .00 | .00 | .31 | |||
Disease-allele frequencies producing Zmax values under dominant mode of inheritance were .0001, for the markers on 8q and 10p, and .1, for the markers on 9p, 9q, and Xcen. When recessive mode of inheritance was allowed, disease-allele frequency was set at .1 for all the chromosomal regions. Information from the neighboring markers (shown in this table) was extracted in two-point analyses with POLYLOCUS.
Figure 3.
Results of pairwise and multipoint nonparametric analysis (statistic B) of markers in chromosomal regions 4q26-q27 (A) and 7p15-p21 (B). The spacing between hash marks on the X-axis is 1 cM. The columns represent the pairwise dominant Zmax values. Two-point Z values obtained with information from all markers of the figure in the polylocus analysis are indicated by short horizontal lines above the corresponding columns. The line represents the −log10 P values for statistic B results, measuring the maximum number of genes among affecteds IBD to any founder gene (Sobel and Lange 1996).
On 7p15-p21, the screening-set marker D7S513 yielded Zmax=1.29 (θ=0) for the recessive and Zmax=1.38 (θ=0) for the dominant mode, and marker D7S673, 10-cM centromeric, gave Z=1.22 (θ=0) (at disease-allele frequency of .0001). When information was extracted from markers D7S481, D7S2201, D7S513, D7S664, and D7S613—with average spacing of ∼5 cM—the combined Zmax obtained in the polylocus analysis was 1.11 with marker D7S513. In multipoint allele-sharing analysis (SIMWALK 2.40), P values of .001 were obtained near markers D7S664 and D7S673 (fig. 3).
On 9q33-q34, the screening-set marker D9S915 resulted in Zmax=2.23 (θ=0 [recessive mode]). Flanking markers D9S1850 and DBH gave Zmax=0 and Zmax=0.22 (θ=.21 [recessive mode]), respectively, and the polylocus Z value obtained with these markers was 2.23 with D9S915. Multipoint allele-sharing analysis with SIMWALK 2.40 (statistic B) resulted in P=.03, near marker D9S158.
On Xcen, several markers in a 30-cM region showed slightly positive ASP Z values, and polylocus analysis for markers in this region resulted in the following Z values: 0.92 with DXS1003, 1.72 with DXS1039, and 0.99 with AR (an intragenic marker for the androgen-receptor gene).
No increased evidence of linkage emerged in the pairwise linkage analyses with markers in the chromosomal regions 8q21-q24, 9p-p22, 10p14-p15, and 12q24 (table 3).
Discussion
OA is a common feature in many monogenic connective-tissue syndromes, and mutations have been identified in genes encoding proteins influencing cartilage structure and metabolism (Vikkula and Olsen 1996). Unlike these syndromic forms of OA, the genetic factors predisposing to common OA remain largely unknown. Most investigations have focused on OA in large, weight-bearing joints, and positive evidence for linkage or association has been reported for candidate genes or loci on 12q12-q14 (COL2A1, by Knowlton et al. [1990], and VDR, by Uitterlinden et al. [1997]), 8q (Baldwin et al. 1995), and 1p (MATN1 by Meulenbelt et al. 1997). Recently, a two-stage genome scan in British sib pairs detected a putative female-specific susceptibility locus for hip and knee OA, on chromosome 11q (Chapman et al. 1999). However, findings in one data set have usually not been replicated in other study samples, consistent with the expectations in studies of complex multifactorial diseases. Similarly, association of DIP OA with HLA haplotypes has been suggested by some (Pattrick et al. 1989) but not by all of the studies (Ercilla et al. 1977; Benavides et al. 1985). In our genome scan for DIP OA loci, neither linkage nor association could be detected in the HLA region (see the Subjects and Methods section and the Department of Human Molecular Genetics, National Public Health Institute, Finland web site). Suggestive evidence of linkage was found, however, with markers on 2q and 4q, providing support for the potential involvement of gene(s) in these chromosomal regions in the predisposition to DIP OA.
The inheritance pattern of DIP OA could be consistent with the presence of a single major genetic component modified by additional genes and environmental factors (Felson et al. 1998). Linkage analysis has proved useful in the mapping of complex traits in rare multiplex families (Mahtani et al. 1996; Kuokkanen et al.1997), but this method has also been criticized (Curtis and Sham 1995; Kruglyak 1997). However, in pedigrees rising from genetically homogeneous populations—such as the Finns—with potentially only few ancestors, linkage analyses of complex traits could be especially powerful if the phenotype is properly dissected. We performed the initial analyses by using robust two-point linkage analyses, which are less sensitive to map and genotyping errors than are multipoint methods (Risch and Giuffra 1992), and applied polylocus linkage analyses in the second stage to increase the informativeness of a given genomic region. In our data set, the highest pairwise Z values were obtained with the assumption of a dominant mode of inheritance and the absence of phenocopies, most probably a result of the selection strategy of our study sample, which emphasized a very severe and familial form of the disease (H. Göring and J. Terwilliger, unpublished data).
Multipoint nonparametric statistics for allele sharing among affecteds were calculated with SIMWALK 2.40 (Sobel et Lange 1996; see Subjects and Methods). In multipoint analysis, it is more important to use “model-free” methods because of the high false-negative rates associated with multipoint “model-based” linkage analysis done under a wrong model. The statistic B of SIMWALK 2.40, measuring the maximum number of genes among affecteds IBD to any one founder gene, showed significant evidence for linkage on 2q12-q21. In our data set, this analysis had greater power than the NPL statistics of GENEHUNTER (Kruglyak et al. 1996), as is shown in figure 2. This statistic provided additional support for linkage of chromosomal regions 4q26-q27 and 7p15-p21 as well, but did not increase the evidence for linkage obtained in pairwise linkage analyses of chromosomal region 9q33-q34.
Interestingly, two loci on 2q13-q32 and 2q33-q35 revealed association with finger-joint OA, in a study of British sib pairs (Wright et al. 1996). These regions are located at some distance from our most informative markers; however, if all the problems associated with the distance approximations in the linkage analyses of complex diseases are taken into consideration, this could still suggest that the loci on 2q have a more general impact on DIP OA in Finns and also in other populations. The chromosomal region 2q12-q14 providing evidence for linkage in this genome scan harbors the interleukin 1 (IL1) gene cluster (genes coding for IL1A on 2q13, IL1B on 2q13, IL1R1 and IL1R2 on 2q12-q22, and IL1RN on 2q14.2), a group of relevant candidate genes for rheumatoid diseases (McDowell et al. 1995). The degree of linkage disequilibrium in the IL1 gene cluster has been studied recently (Cox et al. 1998), allowing for the rational design of further experiments in this region. Since DIP OA often aggregates with OA in large, weight-bearing joints—particularly knee OA (Spector et al. 1996)—this potential predisposing locus could also be involved in other forms of common arthritis.
Acknowledgments
We thank Elli Kempas, Mari Sipilä, Anne Jokiaho, and Maija Parkkonen for excellent technical assistance; Tero Hiekkalinna for his help in biocomputing; and the Academy of Finland, the University of Helsinki, and the Biocentrum of Helsinki University for financial support. Drs. Kenneth Lange and Eric Sobel are especially appreciated for their comments concerning statistical analyses. J.D.T. was supported by a Hitchings-Elion fellowship from the Burroughs-Wellcome Fund.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CHLC database, http://lpg.nci.nih.gov/CHLC/ (for marker information)
- Department of Human Molecular Genetics, National Public Health Institute, Finland, http://www.ktl.fi/molbio/ (for appendices)
- Genetic Location Database, The http://cedar.genetics.soton.ac.uk/public_html/index.html (for markers)
- Genome Database, The http://gdbwww.gdb.org (for markers)
- Marshfield Medical Research Foundation, http://marshmed.org/genetics/ (for markers)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for OA [MIM 165720], IL1A [MIM 147760], and IL1R1 [MIM 147810])
- Stanford Human Genome Center, http://www-shgc.stanford.edu/ (for marker mapping)
References
- Baldwin CT, Farrer LA, Adair R, Dharmavaram R, Jimenez S, Anderson L (1995) Linkage of early-onset osteoarthritis and chondrocalsinosis to human chromosome 8q. Am J Hum Genet 56:692–697 [PMC free article] [PubMed]
- Benavides G, Cerantes A, Silva B, Kantona G, Lardizabal J (1985) HLA and Heberden's nodes in Mexican mestizos. Clin Rheumatol 4:97–98 [DOI] [PubMed]
- Chapman K, Mustafa Z, Irven C, Carr AJ, Clipsham K, Smith A, Chitnavis J, et al (1999) Osteoarthritis-susceptibility locus on chromosome 11q, detected by linkage. Am J Hum Genet 65:167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C, Egger P, Coggon D, Hart DJ, Masud T, Cicuttini F, Doyle DV, et al (1996) Generalized osteoarthritis in women: pattern of joint involvement and approaches to definition for epidemiological studies. J Rheumatol 23:1938–1942 [PubMed]
- Cottingham RW, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed]
- Cox A, Camp NJ, Nicklin MJH, di Giovine FS, Duff GW (1998) An analysis of linkage disequilibrium in the interleukin-1 gene cluster, using a novel grouping method for multiallelic markers. Am J Hum Genet 62:1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Sham PC (1995) Model-free linkage analysis using likelihoods. Am J Hum Genet 57:703–716 [PMC free article] [PubMed]
- Ercilla MG, Brancos MA, Breysse Y, Alonso G, Vives J, Castillo R, Querol JR (1977) HLA antigens in Forestier's disease, ankylosing spondylitis, and polyarthrosis of the hands. J Rheumatol 4:89–93 [PubMed]
- Felson DT (1988) Epidemiology of hip and knee osteoarthritis. Epidemiol Rev 10:1–28 [DOI] [PubMed]
- Felson DT, Couropmitree NN, Chaisson CE, Hannan MT, Zhang Y, McAlindon TE, LaValley M, et al (1998) Evidence for a Mendelian gene in a segregation analysis of generalized radiographic osteoarthritis: the Framingham Study. Arthritis Rheum 41:1064–1071 [DOI] [PubMed]
- Göring H, Terwilliger J (1999) Linkage analysis in the presence of errors. III. Marker loci and their map as nuisance parameters. Am J Hum Genet 65:000–000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart D, Spector T, Egger P, Coggon D, Cooper C (1994) Defining osteoarthritis of the hand for epidemiological studies: the Chingford Study. Ann Rheum Dis 53:220–223 [DOI] [PMC free article] [PubMed]
- Hirsch R, Lethbridge-Cejku M, Hanson R, Scott WW Jr, Reichle R, Plato CC, Tobin JD, et al (1998) Familial aggregation of osteoarthritis: data from the Baltimore Longitudinal Study on Aging. Arthritis Rheum 41:1227–1232 [DOI] [PubMed]
- Kaprio J, Kujala UM, Peltonen L, Koskenvuo M (1996) Genetic liability to osteoarthritis may be greater in women than men. Br Med J 313:232 [DOI] [PMC free article] [PubMed]
- Kärkkäinen A (1985) Osteoarthrosis of the hand in the Finnish population aged 30 years and over. Publications of the Social Insurance Institution, Finland, ML:52, Turku, pp 1–110 [Google Scholar]
- Kellgren JH (1963) Atlas of standard radiographs. In: Jeffrey MR, Ball J (eds) The epidemiology of chronic rheumatism. Blackwell Scientific Publications, Oxford, pp 1–9 [Google Scholar]
- Knowlton RG, Katzenstein PL, Moskowitz RW, Weaver EJ, Malemud CJ, Pathria MN, Jimenez SA, et al (1990) Genetic linkage of a polymorphism in the type II procollagen gene (Col 2A1) to primary osteoarthritis associated with a mild chondrodysplasia. Proc Natl Acad Sci USA 87:6565–6568 [DOI] [PubMed]
- Kruglyak L (1997) Nonparametric linkage tests are model-free. Am J Hum Genet 61:254–255 [DOI] [PMC free article] [PubMed]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed]
- Kujala UM, Leppävuori J, Kaprio J, Kinnunen J, Peltonen L, Koskenvuo M Joint-specific twin and familial aggregation of recalled physician diagnosed osteoarthritis. Twin Res (in press) [DOI] [PubMed] [Google Scholar]
- Kuokkanen S, Gschwend M, Rioux JD, Daly MJ, Terwilliger JD, Tienari PJ, Wikstrom J, et al (1997) Genomewide scan of multiple sclerosis in Finnish multiplex families. Am J Hum Genet 61:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S, Sundvall M, Terwilliger JD, Tienari PJ, Wikstrom J, Holmdahl R, Pettersson U, et al (1996) A putative vulnerability locus to multiple sclerosis maps to 5p14-p12 in a region syntenic to the murine Eae2. Nat Genet 13:477–480 [DOI] [PubMed]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed]
- Lin L, Lin J, Kimura A, Carrington M, Mignot E (1997) DQ microsatellite association studies in three ethnic groups. Tissue antigens 50:507–520 [DOI] [PubMed]
- Mahtani MM, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, et al (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94 [DOI] [PubMed]
- McDowell TL, Symons JA, Ploski R, Forre O, Duff GW (1995) A genetic association between juvenile rheumatoid arthritis and a novel interleukin-1A polymorphism. Arthritis Rheum 38:221–228 [DOI] [PubMed]
- Meulenbelt I, Bijkerk C, de Wildt SC, Miedema HS, Valkenburg HA, Breedveld FC, Pols HA, et al (1997) Investigation of the association of the CRTM and CRTL1 genes with radiographically evident osteoarthritis in subjects from the Rotterdam Study. Arthritis Rheum 40:1760–1765 [DOI] [PubMed]
- Ott J (1991) Analysis of human genetic linkage, rev ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Pattrick M, Manhire A, Ward AM, Doherty M (1989) HLA-A, B antigens and alpha1-antitrypsin phenotypes in nodal and generalized osteoarthritis and erosive osteoarthritis. Ann Rheum Dis 48:470–475 [DOI] [PMC free article] [PubMed]
- Risch N, Giuffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed]
- Spector TD, Cicuttini F, Baker J, Loughlin J, Hart DJ (1996) Genetic influences on osteoarthritis in women: a twin study. Br Med J 312:940–943 [DOI] [PMC free article] [PubMed]
- Stecher RM (1941) Heberden's nodes: heredity in hypertrophic arthritis of the finger joints. Am J Med Sci 201:801 [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed]
- Terwilliger JD, Ott J (1993) A novel polylocus method for linkage analysis using the LOD-score or affected sib pair method. Genet Epidemiol 10:477–482 [DOI] [PubMed]
- Uitterlinden AG, Burger H, Huang Q, Odding E, Duijn CM, Hofman A, Birkenhager JC, et al (1997) Vitamin D receptor genotype is associated with radiographic osteoarthritis at the knee. J Clin Invest 100:259–263 [DOI] [PMC free article] [PubMed]
- Vikkula M, Nissilä M, Hirvensalo E, Nuotio P, Palotie A, Aho K, Peltonen L (1993) Multiallelic polymorphism of the cartilage collagen gene: no association with osteoarthrosis. Ann Rheum Dis 52:762–764 [DOI] [PMC free article] [PubMed]
- Vikkula M, Olsen BR (1996) Unravelling the molecular genetics of osteoarthrosis. Ann Med 28:301–304 [DOI] [PubMed]
- Wright GD, Hughes AE, Regan M, Doherty M (1996) Association of two loci on chromosome 2q with nodal osteoarthritis. Ann Rheum Dis 55:317–319 [DOI] [PMC free article] [PubMed]