Summary
Dilated cardiomyopathy (DCM) is a leading cause of heart failure and the most frequent indication for heart transplantation in young patients. Probably >25% of DCM cases are of familial etiology. We report here genetic localization in a three-generation German family with 12 affected individuals with autosomal dominant familial DCM characterized by ventricular dilatation, impaired systolic function, and conduction disease. After exclusion of known DCM loci, we performed a whole-genome screen and detected linkage of DCM to chromosome 2q14-q22. Investigation of only affected individuals defines a 24-cM interval between markers D2S2224 and D2S2324; when unaffected individuals are also included, the critical region decreases to 11 cM between markers D2S2224 and D2S112, with a peak LOD score of 3.73 at recombination fraction 0 at D2S2339. The identification of an additional locus for familial autosomal dominant DCM underlines the genetic heterogeneity and may assist in the elucidation of the causes of this disease.
Introduction
Dilated cardiomyopathy (DCM) (MIM 102540, MIM 115200, MIM 212110, MIM 300069, MIM 302045, MIM 302060, MIM 510000, MIM 600884, MIM 601154, MIM 601493, MIM 601494, and MIM 602067) is a heart-muscle disease characterized by ventricular dilatation and impaired systolic function. The incidence of DCM is 7/100,000 per year, and the prevalence is 37/100,000 (Codd et al. 1989; Rakar et al. 1997). DCM is a leading cause of heart failure and the most frequent indication for heart transplantation in young patients (Dec and Fuster 1994). Heart failure is a progressive clinical syndrome, with a median survival time of 1.7 years in men and 3.2 years in women (Ho et al. 1993). Most patients die from pump failure or sudden cardiac death.
Although familial occurrence of DCM has been reported for the first time by Whitfield (1961), the frequency of familial etiology has been systematically examined only recently (Mestroni and Giacca 1997). Systematic examination of relatives showed that probably >25% of DCM cases are of familial etiology (Michels et al. 1992; Keeling et al. 1995; Grünig et al. 1998). The mode of inheritance in most families seems to be autosomal dominant (Michels et al. 1992), but autosomal recessive, X-linked, and mitochondrial transmission have also been reported (Mestroni and Giacca 1997).
To date, three genes—two X-linked genes (dystrophin and tafazzin) and cardiac actin—have been identified as causes of DCM (Muntoni et al. 1993; Bione et al. 1996; D'Adamo et al. 1997; Olson et al. 1998). Mutations in the dystrophin gene (Xp21) in males lead to early manifestation of DCM, at age of 15–21 years and to rapid progression to death, usually after <1 year, whereas affected females present with mild symptoms of heart failure during the 5th and 6th decade, with a gradual decline of cardiac function (Berko and Swift 1987; Muntoni et al. 1993; Towbin et al. 1993). The infantile X-linked DCM is caused by mutations of the tafazzin gene (G4.5 gene; Xq28) (Bione et al. 1996; D'Adamo et al. 1997). The function of tafazzin is unknown. The onset of the disease is typically during the 1st year of life, and death usually occurs during childhood. In addition, most patients may be characterized by skeletal myopathy, short stature, neutropenia, and abnormal mitochondria, also referred to as “Barth syndrome” (Neustein et al. 1979; Barth et al. 1983; D'Adamo et al. 1997). The only known gene causing autosomal dominant–transmitted DCM is the cardiac actin gene (15q14) (Olson et al. 1998). Two different missense mutations in the immobilized end of the actin filament are known to cause the disease (Olson et al. 1998). Age at onset in the two families showed a wide range, 1–41 years, and affected individuals presented with left-ventricular (LV) dilatation and impaired systolic function, without apparent conduction disease (Olson et al. 1998).
In addition, seven loci for autosomal dominant DCM have been reported on chromosomes 1p1-q1 (CMD1A), 9q13-q25 (CMD1B), 10q21-q23 (CMD1C), 1q32 (CMD1D), 3p25-p22 (CMD1E), 6q23 (CMD1F), and 2q31 (CMD1G) (Kass et al. 1994; Durand et al. 1995; Krajinovic et al. 1995; Bowles et al. 1996; Olson and Keating 1996; Messina et al. 1997; Siu et al. 1999). CMD1A and CMD1E are characterized by conduction abnormalities preceding cardiac dilatation and systolic dysfunction (Kass et al. 1994; Olson and Keating 1996). The first abnormalities are sinus and/or atrioventricular-node dysfunction, resulting in sinus bradycardia, pauses and arrest, first- to third-degree atrioventricular block (1°AVB–3°AVB), and/or atrial fibrillation. CMD1F is also associated with AVB and, in addition, with mild proximal limb-girdle muscular dystrophy, which can be recognized initially during the late 3d decade (Messina et al. 1997). CMD1C is associated with mitral-valve prolapse in all cases (Bowles et al. 1996). DCM loci at CMD1B, CMD1D, and CMD1G are not associated with either conduction disease or skeletal muscular dystrophy (Durand et al. 1995; Krajinovic et al. 1995; Siu et al. 1999). Although many candidate genes in these loci have been examined, the responsible genes are yet unknown. We describe here a novel locus for autosomal dominant DCM associated with conduction disease (CMD1H), on chromosome 2q14-q22 in a three-generation German kindred.
Subjects and Methods
Phenotypic Characterization
After informed consent was obtained in accordance with the guidelines of the ethical commission of the Max-Delbrück-Center for Molecular Medicine, clinical evaluation was performed, and blood samples from a German family (fig. 1) were drawn in the Franz-Volhard-Klinik. Family members were evaluated on the basis of medical history, physical examination, two-dimensional and M-mode echocardiography, 12-lead electrocardiogram, and, in some cases, Holter electrocardiography (Holter ECG) (table 1). Five affected members had left ventriculography showing a reduced LV ejection fraction, as determined by the area-length method (Sandler and Dodge 1968). In each case, coronary-artery disease (CAD) as a cause of the decreased LV function was excluded by selective coronary angiography. LV dilatation was defined as LV end-diastolic diameter >95% for body-surface area (Henry et al. 1980). Impaired systolic function was defined as either a shortening fraction of <28% by M-mode echocardiography or decreased ejection fraction of <50% (Mestroni et al. 1999). In addition, global LV hypokinesia as determined by two-dimensional echocardiography was taken to be a sign of DCM. ECG abnormalities were defined according to established criteria.
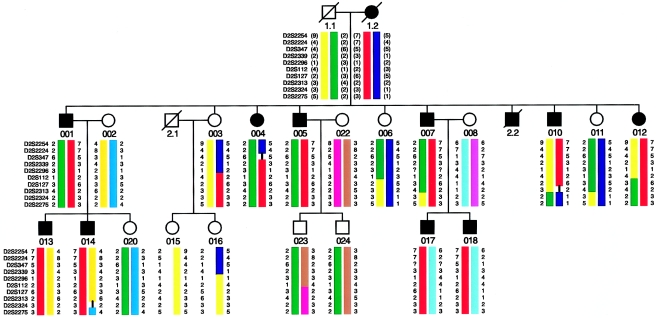
Figure 1.
Pedigree of family with autosomal dominant DCM, with most likely marker haplotypes from the linked region on chromosome 2q. Key recombination events were observed with marker D2S2224, in individual 004; with marker D2S112, in individual 003; and with marker D2S2324, in individual 010.
Table 1.
Evaluation of Clinical Data[Note]
| LV |
|||||||||||||||||
|
Interval |
Diameter(mm) |
||||||||||||||||
| Individual(Age [years]) | Height(cm) | Weight(kg) | Symptom(s)a | Rhythmb | HR(1/min) | PR(ms) | QRS(ms) | 24-h HolterECGc | PM/AICDd | End Diastolic | End Systolic | Dilatation | GlobalHypokinesiah | FSe(%) | LV-EFf and NoCAD | Total Pointsg | AffectionStatus |
| 1.1 (53i) | 167 | 45 | … | 1°AVB(+) | 1 | Absent | |||||||||||
| 1.2 (55i) | 172 | 57 | … | Presentj | 4 | Affected | |||||||||||
| 2.2 (36i) | … | … | NYHA II, palpitations | SB, 1°AVB(+) | 50 | 200 | <110 | … | DDD | 57 | … | Present | Present | … | NDk | 12 | Affected |
| 001 (50i) | 172 | 63 | NYHA III, palpitations, stroke | 1°AVB(+), intermittent 2°AVB(−), LBBB | 86 | 280 | 140 | SVT | AICD | 74 | 68 | Present | Present | 20 | 12 | 21 | Affected |
| 002 (50) | None | … | … | … | … | … | … | … | … | … | … | … | … | 0 | Unaffected | ||
| 003 (52) | 164 | 66 | None | 1°AVB(−) | 81 | 230 | 80 | … | … | 50 | 30 | Absent | Absent | 40 | … | 1 | Unaffected |
| 004 (50) | … | … | NYHA II, palpitations | AF, intermittent SB, 1°AVB(−), 3°AVB(−) | 41 | … | <110 | UVT | … | 52 | 38 | Absent | Present | 27 | … | 13 | Affected |
| 005 (46) | 168 | 63 | NYHA II–III, palpitations | 1°AVB(+), LBBB, intermittent AF | 89 | 250 | 120 | VF | AICD | 70 | 60 | Present | Present | 14 | 25 | 19 | Affected |
| 006 (42) | … | … | VBI | SB | 54 | 160 | 90 | Absent | … | 45 | 28 | Absent | Absent | 38 | … | 1 | Unaffected |
| 007 (44) | 176 | 107 | NYHA II–III, palpitations | 1°AVB(+), LBBB | 72 | 250 | 140 | SB, UVT | AICD | 63 | 47 | Present | Present | 25 | 20 | 20 | Affected |
| 008 (42) | 169 | 99 | None | SR | 84 | 120 | 80 | … | … | 55 | 30 | Absent | Absent | 45 | … | 0 | Unaffected |
| 010 (35) | … | … | NYHA II–III, palpitations, stroke | 1°AVB(−), LBBB | 55 | 260 | 140 | UVT | AICD | … | … | Present | Present | … | 48 | 16 | Affected |
| 011 (31) | 159 | 57 | None | 1°AVB(−) | 80 | 200 | 80 | … | … | 44 | 31 | Absent | Absent | 30 | … | 1 | Unaffected |
| 012 (32) | 159 | 58 | Palpitations | 1°AVB(+) | 67 | 227 | 90 | SVTl | … | 52 | 38 | Present | Present | 27 | … | 9 | Affected |
| 013 (28) | 170 | 62 | None | SR | 70 | 160 | 100 | SB | … | 52 | 43 | Absent | Present | 17 | … | 5 | Affected |
| 014 (23) | 171 | 54 | NYHA II | SR | 70 | 150 | 110 | SB | … | 45 | 33 | Absent | Present | 27 | … | 7 | Affected |
| 015 (30) | 169 | 62 | None | SR | 73 | 140 | 80 | … | … | 48 | 30 | Absent | Absent | 38 | … | 0 | Unaffected |
| 016 (24) | 168 | 59 | None | SR | 51 | 140 | 90 | … | … | 50 | 32 | Absent | Absent | 36 | … | 0 | Unaffected |
| 017 (18) | 189 | 81 | None | SR | 81 | 150 | 110 | … | … | 50 | 38 | Absent | Present | 24 | … | 5 | Affected |
| 018 (17) | 170 | 63 | None | ST | 115 | 120 | 90 | … | … | 50 | 37 | Absent | Present | 26 | … | 5 | Affected |
| 020 (13) | 150 | 52 | None | SR | 77 | 125 | 90 | … | … | 44 | 24 | Absent | Absent | 45 | … | 0 | Unaffected |
| 022 (35) | 157 | 55 | None | SR | 75 | 140 | 80 | … | … | NDm | … | Absent | Absent | Normal | … | 0 | Unaffected |
| 023 (18) | 175 | 64 | None | SR | 82 | 180 | 90 | … | … | 50 | 30 | Absent | Absent | 40 | … | 0 | Unaffected |
| 024 (17) | 167 | 64 | None | SR | 65 | 160 | 90 | … | … | 45 | 28 | Absent | Absent | 38 | … | 0 | Unaffected |
Note.—Only abnormal values are documented.
NYHA = symptoms on exertion, as evaluated on the basis of the New York Heart Association functional classification; palpitations are defined as irregular and fast heart beats; stroke is as diagnosed by neurologists; VBI = vertebrobasilar ischemia.
(+) and (−) = with and without medication slowing atrioventricular conduction, respectively; SB = sinus bradycardia; LBBB = left bundle-branch block; AF = atrial fibrillation; SR = sinus rhythm; ST = sinus tachycardia.
SVT = sustained ventricular tachycardia; UVT = unsustained ventricular tachycardia; VF = ventricular fibrillation; SB = sinus bradycardia; SVT = supraventricular tachyarrhythmia.
Pacemaker/automatic cardioverter defibrillator implanted. DDD = pacemaker in DDD mode.
Fractional shortening, determined by echocardiography. NA = not applicable (since, although LV-EF was significantly decreased, it could not be exactly determined).
LV dilation and ejection fraction, determined angiographically; CAD absence determined by selective coronary angiography.
Symptoms and signs scored on the basis of data in table 2.
Determined by echocardiography or angiography.
Age at death.
Biventricular dilation of all cardiac chambers, on autopsy.
LV-EF significantly decreased but unable to be exactly determined.
190/min.
Not dilated.
The clinical findings were rated by a point assignment, without knowledge of genotype (table 2). Affection status was based on major and minor criteria. Major criteria were LV dilatation, reduced systolic function, documented ventricular tachycardias and fibrillation, implantation of automatic cardioverter defibrillators, 2°AVB or 3°AVB, or implantation of pacemakers. The greatest accuracy of diagnosis is achieved by LV angiography and exclusion of CAD and by autopsy. Therefore, these criteria were assigned the highest-scoring points. Minor criteria were supraventricular tachyarrhythmias, 1°AVB, increased QRS duration, left bundle-branch blocks, symptoms or signs of reduced myocardial performance, such as dyspnoe on exertion, and signs of tachyarrhythmias sensed as palpitations. The only other family study in which DCM was evaluated by a scoring system did not differentiate between major and minor criteria and rated non–DCM-specific cardiovascular events such as stroke at age <40 years as having the highest-scoring points (Olson and Keating 1996). Recently, Mestroni et al. (1999) have suggested guidelines for the study of familial DCM. The authors stated that the complete criteria required for the diagnosis of DCM appeared to be too restrictive in familial DCM. We therefore used slightly different major and minor criteria, all of which are suggestive of DCM associated with conduction abnormalities, in order to obtain a higher sensitivity to detect DCM in the absence of knowledge of the genotype.
Table 2.
Quantitative Phenotype Scoring[Note]
| Criterion | Points |
| Major: | |
| Dilated left ventricle with reduced ejection fraction and exclusion of CAD, by angiography | 4 |
| Dilation of all cardiac chambers, on autopsy | 4 |
| LV dilation >95% (corrected for body-surface area) | 2 |
| Fractional shortening <28% | 2 |
| Global LV hypokinesia | 2 |
| 2°AVB or 3°AVB | 2 |
| Ventricular tachycardia | 2 |
| Pacemaker/automatic cardiac defibrillator implantation | 2 |
| Minor: | |
| New York Heart Association classification >2 | 1 |
| Palpitations | 1 |
| Stroke | 1 |
| Sinus bradycardia (heart rate <60/min) | 1 |
| 1°AVB, PR>200 ms | 1 |
| Unexplained supraventricular tachyarrhythmia | 1 |
| Unexplained atrial fibrillation | 1 |
| Increased QRS duration (QRS>110 ms) | 1 |
| Left bundle-branch block (QRS>120 ms) | 1 |
Note.—Points were summed up for each individual and then the status of the latter was classified as unaffected (0 or 1 point), uncertain (2 or 3 points), or affected (⩾4 points).
Genotyping
DNA was isolated by standard techniques. Microsatellite markers were from the MDC-Généthon microsatellite mapping panel, with an average spacing of 11 cM (M. Jung, unpublished data). Loci were chosen from the final Généthon linkage map (Dib et al. 1996). Markers were amplified by simplex PCR on a MJ Tetrad cycler, were separated on an ABI 377 DNA sequencer, and were analyzed by GENESCAN version 2.2 and GENOTYPER version 2.0 software (ABI). All genotypes were checked for Mendelian segregation, by LINKRUN (T. F. Wienker, personal communication). No DNA was available from individuals 2.1 and 2.2.
Linkage Analysis
Power calculations, prior to genome scan, by the program [FAST]SLINK (Weeks et al. 1990) revealed a maximum two-point LOD score of 4.14 and an expected LOD score of 2.24, at recombination fraction (θ) .05. Two-point LOD scores were calculated by the LINKAGE-program package (Lathrop and Lalouel 1984), and, for each chromosome, the exclusion map was displayed by LODVIEW, an Microsoft Excel application (Hildebrandt et al. 1993). Multipoint analyses were performed by VITESSE (O'Connell and Weeks 1995). The most likely haplotypes were constructed manually.
Since manifestation of DCM is age dependent, penetrance values were modified as follows: 60% for unaffected individuals ⩽20 years of age (individuals 020, 023, and 024), 90% for those ⩽30 years of age (individuals 015 and 016), those ⩽45 years of age (individuals 006 and 011), and 99% in those ⩾45 years of age (individual 003). Asymptomatic individuals <30 years of age who were scored clinically as affected (individuals 013, 014, 017, and 018) were assigned a penetrance value of 90%, whereas all other affecteds were scored as fully penetrant.
Results
A total of 25 members of a family with autosomal dominant inheritance of DCM (fig. 1) were examined in detail (table 1). On the basis of a scoring system shown in table 2, patients with >3 points were classified as affected. Of the 12 individuals who were diagnosed as affected, 5 had invasive angiography to exclude other causes of ventricular dilatation and impaired LV systolic function; in all 5 patients, CAD was excluded by selective coronary angiography, and the diagnosis was confirmed in 1 patient by autopsy. A total of 8 family members were clinically unaffected. There were no clinical data available from the deceased individual, 2.1.
The severity of the disease showed great variability in affected members. Disease manifestation in this family began during adolescence, with sinus- or atrioventricular-node dysfunction. Clinical manifestations were sinus bradycardia, sinus tachycardia, supraventricular tachyarrythmias, and/or 1°AVB–3°AVB. Then intraventricular conduction was delayed, resulting in either prolonged QRS duration or left bundle-branch block. In the early stages, LV systolic function was only mildly decreased, but ventricular dilatation could not yet be observed. Later, malignant tachyarrhythmias were recorded, leading to implantation of automatic cardioverter defibrillators in four of the five living affected individuals >35 years of age. Both moderate dilatation of the left atria and ventricles and severe systolic dysfunction occurred as late signs. The end stage of disease manifested clinically as overt congestive heart failure.
The known loci on chromosome 1p1-q1, 1q32, 3p25-p22, 6q23, 9q13-q22, and 10q21-q23 and the actin gene locus on chromosome 15q14 were excluded by linkage analysis. Then a genomewide screen was initiated, and a total of 310 microsatellite markers were typed. Initially, two regions, on chromosomes 9q and 2q, showed, by two-point analysis, evidence for linkage to DCM. Haplotype analyses clearly excluded the region on chromosome 9 (data not shown). In contrast, the family was concordant for a region on 2q14-q22. Recombination events in only the affected individuals defined a 24-cM critical region delineated by recombination events, in individual 004, by marker D2S2224, and in individual 010, by marker D2S2324 (figs. 1 and 2). A further recombination event was observed in unaffected individual 003, which reduces the critical region to an 11-cM segment between markers D2S2224 and D2S112. This individual was 52 years of age when last seen and had only 1°AVB but otherwise was healthy. Using the scoring system devised prior to genotyping she was diagnosed as unaffected. When disease penetrance was assumed to be 60%–100%, dependent on the age of the individuals, two-point analyses gave a peak LOD score, for D2S2339, of 3.73 at θ=0 (table 3).
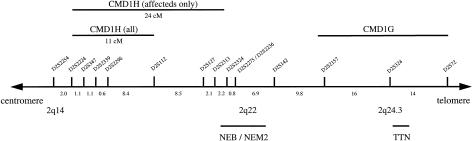
Figure 2.
Part of genetic map of human chromosome 2q. The region including the DCM gene is marked. Genetic distances between markers are given (in cM). Microsatellite loci were obtained from the final Généthon linkage map (Dib et al. 1996). The locations of nebulin (NEB), the disease gene of autosomal recessive nemaline myopathy 2 (NEM2), titin (TTN), and the adjacent CMD1G locus are shown.
Table 3.
Pairwise LOD Scores between DCM and Markers on Chromosome 2[Note]
|
LOD Score at θ = |
|||||||||
| Marker (Position [cM]a) | .0 | .01 | .05 | .1 | .2 | .3 | .4 | Maximum LOD Score | Maximum θ |
| D2S2254 132.6 | −5.57 | 1.67 | 2.13 | 2.11 | 1.72 | 1.11 | .40 | 2.15 | .07 |
| D2S2224 134.6 | −5.69 | 1.08 | 1.57 | 1.60 | 1.31 | .82 | .27 | 1.61 | .08 |
| D2S347 135.7 | 1.05 | 1.03 | .95 | .84 | .61 | .35 | .11 | 1.08 | .00 |
| D2S2339 136.8 | 3.73 | 3.67 | 3.41 | 3.06 | 2.32 | 1.48 | .55 | 3.73 | .00 |
| D2S2296 137.4 | 3.26 | 3.20 | 2.95 | 2.63 | 1.95 | 1.20 | .40 | 3.26 | .00 |
| D2S112 145.8 | 1.09 | 1.76 | 2.15 | 2.12 | 1.72 | 1.12 | .40 | 2.16 | .07 |
| D2S127 154.3 | 1.73 | 1.97 | 2.20 | 2.14 | 1.73 | 1.12 | .41 | 2.20 | .06 |
| D2S2313 156.4 | 1.36 | 1.34 | 1.23 | 1.10 | .82 | .50 | .19 | 1.36 | .00 |
| D2S151 156.4 | .71 | .70 | .63 | .55 | .38 | .23 | .09 | .71 | .00 |
| D2S2324 158.6 | −5.69 | −.80 | .32 | .66 | .73 | .52 | .21 | .75 | .19 |
| D2S2275 159.4 | −5.21 | −.23 | .87 | 1.16 | 1.12 | .77 | .29 | 1.20 | 0.14 |
a
On the genetic map, according to Dib et al. (1996).
Discussion
We have identified a three-generation family with autosomal dominant DCM and conduction-system disease. In a genomewide scan for linkage to DCM, we were able to exclude >90% of the autosomal genome. Two regions showed evidence for linkage. After haplotype analysis, the region on chromosome 9 could be excluded (data not shown). In contrast, the region on chromosome 2q could be confirmed by both detailed analysis of numerous markers (table 3) and extensive haplotyping (fig. 1). Linked markers have been assigned to the long arm of chromosome 2, in the region 2q14-q22.
Clinical symptoms manifest only at a late stage of DCM. Therefore, linkage mapping is difficult in this late-onset disease. Sensitive diagnostic procedures are needed to identify minimal cardiac dysfunction, especially in younger individuals. On the basis of a scoring system similar to that suggested by Mestroni et al. (1999), individuals in this family were classified as either affected or unaffected. Severe arryhthmias were rated as a major criterion, since they represent a special feature of the DCM family described by us. Three other DCM families, with linkage to 1p1-q1, 3p25-p22, and 6q23, showed a combination of DCM and sinus- and/or atrioventricular-node disease (Kass et al. 1994; Olson and Keating 1996; Messina et al. 1997) similar to that seen in the family described here. The bradyarrhyhthmias required implantation of pacemakers in most affected individuals of the family with CMD1A and in some patients of the family with CMD1F (Kass et al. 1994; Messina et al. 1997). Only one individual in the family that we studied received a pacemaker, because of symptomatic AVB. In contrast to the other three families with DCM and conduction disease, 5 of the 12 affected individuals of the family that we studied had documented ventricular tachycardias, and 4 of them received an automatic cardioverter defibrillator. Clinical overt congestive heart failure occurred only at an older age (table 1).
Using a conservative approach, which considered affecteds only, we were able to identify, on chromosome 2, a critical region of 24 cM defined by recombination events in individuals 004 and 010 (fig. 1). When we consider the unaffected individuals as well, the region can be reduced to an 11-cM interval, because of a recombination event in individual 003. This clinically unaffected 52-year-old woman shows only 1°AVB. Her affected siblings had a much earlier manifestation of the disease. We therefore scored her as unaffected.
Recently, Siu et al. (1999) have reported mapping of a DCM locus with early onset and a rapidly progressive course of the disease and apparently reduced penetrance. They have identified linkage to markers on chromosome 2q31, distal to the region described in the present study, and have proposed titin, a giant sarcomeric protein, as a candidate gene. However, sequence analysis of the cardiac-specific N2-B domain of titin failed to detect disease-causing mutations. The clinical manifestation in the family that we studied is markedly different from that in the family described by Siu et al. (1999). Affected members in the family described here are characterized by early onset of conduction disease, followed by malignant ventricular arrhythmias and mild ventricular dilatation and overt heart failure at later stages. In contrast, affected individuals described by Siu et al. (1999) are characterized by early onset of ventricular dilatation and a rapid progressive deterioration without conduction abnormalities. The different clinical manifestation and course of the disease in both families, considered together with the different, although adjacent genetic localization (fig. 2), argues for the existence of two DCM loci on chromosome 2. Only identification of the disease-causing mutations will prove this likely assumption.
Disrupted force transmission from the sarcomere to the extracellular matrix has been suggested as a cause of DCM (Olson et al. 1998). Structural proteins such as dystrophin, dystrophin-associated glycoproteins such as sarcoglycans, muscle LIM, and sarcomeric proteins responsible for force transmission may therefore be involved in the etiology of DCM. Mutations in dystrophin cause X-linked DCM in humans (Muntoni et al. 1993; Milasin et al. 1996), and mutations in ∂-sarcoglycan lead to DCM in hamsters (Sakamoto et al. 1997). Muscle LIM knockout in mice also causes DCM (Arber et al. 1997). The giant sarcomeric protein nebulin, which is a component of the cytoskeletal matrix, is located within the 24-cM region defined by the affecteds-only model (fig. 2) (Wallgren-Pettersson et al. 1995). Recently, mutations in this gene have been identified as the cause of autosomal recessive nemaline myopathy 2, a syndrome with an increased number of muscle fibers with central nuclei and rods (Arts et al. 1978; Wallgren-Pettersson et al. 1988; Pelin et al. 1999). This disorder is characterized by infantile onset of slowly progressive or nonprogressive weakness of facial and proximal-limb skeletal muscles (Wallgren-Pettersson et al. 1988; Goebel 1998). Nebulin is apparently not expressed in cardiac tissue (Schiaffino and Reggiani 1996); therefore, although nebulin is located within the 24-cM critical region, it seems an unlikely candidate gene for DCM in this family. In addition, it clearly would be excluded from the 11-cM region defined by the recombination event in unaffected individual 003. Titin, the candidate proposed by Siu et al. (1999) for the disease in the family with CMD1G, maps outside the critical region defined in the family described by us. No other obvious candidate gene coding for cytoskeletal proteins has been mapped to this region.
In summary, we have identified a novel DCM locus associated with conduction-system disease, on chromosome 2q14-q22. This confirms that DCM is genetically heterogeneous. Identification of the responsible gene would improve our understanding of both DCM and conduction-system disorders in general. Ultimately, new diagnostic and, it is hoped, therapeutic strategies, such as genetic reconstitution of the disease gene (Greelish et al. 1999), may evolve from this knowledge.
Acknowledgments
We thank the family for their cooperation in this study. The Mikrosatellitenzentrum is supported by a grant-in-aid from the German ministry of research to A.R. and T.F.W. The clinical study was supported, in part, by the Dr.-Karl-Wilder-Stiftung, Bonn. We also thank Franz Rüschendorf, for his contribution of haplotyping and multipoint analysis, and Elke Szczech, for technical assistance during the clinical studies.
Electronic-Database Information
The accession numbers and URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for DCM [MIM 102540, MIM 115200, MIM 212110, MIM 300069, MIM 302045, MIM 302060, MIM 510000, MIM 600884, MIM 601154, MIM 601493, MIM 601494, and MIM 602067])
References
- Arber S, Hunter JJ, Ross J, Hongo M, Sansig G, Borg J, Perriard JC, et al (1997) MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88:393–403 [DOI] [PubMed]
- Arts WF, Bethlem J, Dingemans KP, Eriksson AW (1978) Investigations on the inheritance of nemaline myopathy. Arch Neurol 35:72–77 [DOI] [PubMed]
- Barth PG, Scholte HR, Berden JA, Van der Klei Van Moorsel JM, Luyt Houwen IE, Van't Veer Korthof ET, Van der Harten JJ, et al (1983) An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 62:327–355 [DOI] [PubMed]
- Berko BA, Swift M (1987) X-linked dilated cardiomyopathy. N Engl J Med 316:1186–1191 [DOI] [PubMed]
- Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D (1996) A novel X-linked gene, G4.5 is responsible for Barth syndrome. Nat Genet 12:385–389 [DOI] [PubMed]
- Bowles KR, Gajarski R, Porter P, Goytia V, Bachinski L, Roberts R, Pignatelli R, et al (1996) Gene mapping of familial autosomal dominant dilated cardiomyopathy to chromosome 10q21-23. J Clin Invest 98:1355–1360 [DOI] [PMC free article] [PubMed]
- Codd MB, Sugrue DD, Gersh BJ, Melton LJ (1989) Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy: a population-based study in Olmsted County, Minnesota, 1975–1984. Circulation 80:564–572 [DOI] [PubMed]
- D'Adamo P, Fassone L, Gedeon A, Janssen EAM, Bione S, Bolhuis PA, Barth PG, et al (1997) The X-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet 61:862–867 [DOI] [PMC free article] [PubMed]
- Dec WG, Fuster V (1994) Idiopathic dilated cardiomyopathy. N Engl J Med 331:1564–1575 [DOI] [PubMed]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Durand JB, Bachinski LL, Bieling LC, Czernuszewicz GZ, Abchee AB, Yu QT, Tapscott T, et al (1995) Localization of a gene responsible for familial dilated cardiomyopathy to chromosome 1q32. Circulation 92:3387–3389 [DOI] [PubMed]
- Goebel HH (1998) Congenital myopathies with inclusion bodies: a brief review. Neuromuscul Disord 8:162–168 [DOI] [PubMed]
- Greelish JP, Su LT, Lankford EB, Burkman JM, Chen H, Konig SK, Mercier IM, et al (1999) Stable restoration of the sarcoglycan complex in dystrophic muscle perfused with histamine and a recombinant adeno-associated viral vector. Nat Med 5:439–443 [DOI] [PubMed]
- Grünig E, Tasman JA, Kücherer H, Franz W, Kübler W, Katus HA (1998) Frequency and phenotypes of familial dilated cardiomyopathy. J Am Coll Cardiol 31:186–194 [DOI] [PubMed]
- Henry WL, Gardin JM, Ware JH (1980) Echocardiographic measurements in normal subjects from infancy to old age. Circulation 62:1054–1061 [DOI] [PubMed]
- Hildebrandt F, Pohlmann A, Omran H (1993) LODVIEW: a computer program for the graphical evaluation of lod score results in exclusion mapping of human disease genes. Comput Biomed Res 26:592–599 [DOI] [PubMed]
- Ho KKL, Anderson KM, Kannel WB, Grossmann W, Levy D (1993) Survival after the onset of congestive heart failure in Framing heart study subjects. Circulation 88:107–115 [DOI] [PubMed]
- Kass S, MacRae C, Graber HL, Sparks EA, McNamara D, Boudoulas H, Basson CT, et al (1994) A gene defect that causes conduction system disease and dilated cardiomyopathy maps to chromosome 1p1-1q1. Nat Genet 7:546–551 [DOI] [PubMed]
- Keeling PJ, Gang Y, Smith G, Seo H, Bent SE, Caforio ALP, McKenna WJ (1995) Familial dilated cardiomyopathy in the United Kingdom. Br Heart J 73:417–421 [DOI] [PMC free article] [PubMed]
- Krajinovic M, Pinamonti B, Sinagra G, Vatta M, Severini GM, Milasin J, Falaschi A, et al (1995) Linkage of familial dilated cardiomyopathy to chromosome 9. Am J Hum Genet 57:846–852 [PMC free article] [PubMed]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed]
- Messina DN, Speer MC, Pericak-Vance MA, McNally EM (1997) Linkage of familial dilated cardiomyopathy with conduction defect and muscular dystrophy to chromosome 6q23. Am J Hum Genet 61:909–917 [DOI] [PMC free article] [PubMed]
- Mestroni L, Giacca M (1997) Molecular genetics of dilated cardiomyopathy. Curr Opin Cardiol 12:303–309 [DOI] [PubMed]
- Mestroni L, Maisch B, McKenna WJ, Schwartz K, Charron P, Rocco C, Tesson F, et al (1999) Guidelines for the study of familial dilated cardiomyopathies. Eur Heart J 20:93–102 [DOI] [PubMed]
- Michels VV, Moll PP, Miller FA, Tajik J, Chu JS, Driscoll DJ, Burnett JC, et al (1992) The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med 326:77–82 [DOI] [PubMed]
- Milasin J, Muntoni F, Severin GM, Bartoloni L, Vatta M, Krajinovic M, Mateddu A, et al (1996) A point mutation in the 5′ splice site of the dystrophin gene first intron responsible for X-linked dilated cardiomyopathy. Hum Mol Genet 5:73–79 [DOI] [PubMed]
- Muntoni F, Cau M, Ganau A, Congiu R, Arvedi G, Mateddu A, Marrosu MG, et al (1993) Deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med 329:921–925 [DOI] [PubMed]
- Neustein HB, Lurie PR, Dahms B, Takahashi M (1979) An X-linked recessive cardiomyopathy with abnormal mitochondria. Pediatrics 64:24–29 [PubMed]
- O'Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed]
- Olson TM, Keating MT (1996) Mapping a cardiomyopathy locus to chromosome 3p22-p25. J Clin Invest 97:528–532 [DOI] [PMC free article] [PubMed]
- Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT (1998) Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 280:750–752 [DOI] [PubMed]
- Pelin K, Hilpela P, Donner K, Sewry C, Akkari PA, Wilton SD, Wattanasirichaigoon D, et al (1999) Mutations in the nebulin gene associated with autosomal recessive nemaline myopathy. Proc Natl Acad Sci USA 96:2305–2310 [DOI] [PMC free article] [PubMed]
- Rakar S, Sinagra G, Di Lenarda A, Poletti A, Bussani R, Silvestri F, Camerini F (1997) Epidemiology of dilated cardiomyopathy: a prospective post-mortem study of 5252 necropsies. Eur Heart J 18:117–123 [DOI] [PubMed]
- Sakamoto A, Ono K, Abe M, Jasmin G, Eki T, Murakami Y, Masaki T, et al (1997) Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, delta-sarcoglycan, in hamster: an animal model of disrupted dystrophin-associated glycoprotein complex. Proc Natl Acad Sci USA 94:13873–13878 [DOI] [PMC free article] [PubMed]
- Sandler H, Dodge HT (1968) The use of single plane angiocardiograms for the calculation of left ventricular volume in man. Am Heart J 75:325–334 [DOI] [PubMed]
- Schiaffino S, Reggiani C (1996) Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 76:371–423 [DOI] [PubMed]
- Siu BL, Niimura H, Osborne JA, Fatkin D, MacRae C, Solomon S, Benson DW, et al (1999) Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation 99:1022–1026 [DOI] [PubMed]
- Towbin JA, Hejtmancik JF, Brink P, Gelb B, Zhu XM, Chamberlain JS, McCabe ERB, et al (1993) X-linked dilated cardiomyopathy: molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation 87:1854–1865 [DOI] [PubMed]
- Wallgren-Pettersson C, Avela K, Marchand S, Kolehmainen J, Tahvanainen E, Hansen FJ, Muntoni F, et al (1995) A gene for autosomal recessive nemaline myopathy assigned to chromosome 2q by linkage analysis. Neuromuscul Disord 5:441–443 [DOI] [PubMed]
- Wallgren-Pettersson C, Rapola J, Donner M (1988) Pathology of congenital nemaline myopathy: a follow-up study. J Neurol Sci 83:243–257 [DOI] [PubMed]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis . Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Whitfield AG (1961) Familial cardiomyopathy. Q J Med 3:119–134 [PubMed] [Google Scholar]