Summary
Over the past 4 years, a number of investigators have reported findings suggestive of linkage to schizophrenia, with markers on chromosomes 13q32 and 8p21, with one recent study by Blouin et al. reporting significant linkage to these regions. As part of an ongoing genome scan, we evaluated microsatellite markers spanning chromosomes 8 and 13, for linkage to schizophrenia, in 21 extended Canadian families. Families were analyzed under autosomal dominant and recessive models, with broad and narrow definitions of schizophrenia. All models produced positive LOD scores with markers on 13q, with higher scores under the recessive models. The maximum three-point LOD scores were obtained under the recessive-broad model: 3.92 at recombination fraction (θ) .1 with D13S793, under homogeneity, and 4.42 with α=.65 and θ=0 with D13S793, under heterogeneity. Positive LOD scores were also obtained, under all models, for markers on 8p. Although a maximum two-point LOD score of 3.49 was obtained under the dominant-narrow model with D8S136 at θ=0.1, multipoint analysis with closely flanking markers reduced the maximum LOD score in this region to 2.13. These results provide independent significant evidence of linkage of a schizophrenia-susceptibility locus to markers on 13q32 and support the presence of a second susceptibility locus on 8p21.
Introduction
Schizophrenia is a serious neuropsychiatric illness affecting ∼1% of the general population. Family, twin, and adoption studies have demonstrated that schizophrenia is predominantly genetically determined and has high heritability (McGuffin et al. 1994). No gene for schizophrenia has yet been identified, and no significant linkage finding has yet been reproduced in an independent sample (Moldin 1997). Two regions that have recently yielded reports of significant linkage to schizophrenia are chromosomes 13q32 and 8p21 (Blouin et al. 1998). Several groups had previously reported positive, but not significant, findings of linkage to markers on chromosomes 13q (Lin et al. 1995; Pulver et al. 1996; Lin et al. 1997; Straub et al. 1997; Shaw et al. 1998) and 8p (Moises et al. 1995; Pulver et al. 1995; Kendler et al. 1996; Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6, and 8 1996; Kaufmann et al. 1998; Shaw et al. 1998; Wildenauer and Schwab 1998).
Table 1 summarizes positive linkage findings for chromosomes 13q and 8p. The chromosome 13 linkage results cluster within a region of ∼12 cM in 13q32, except for the findings of Shaw et al. (1998), which are located within two regions, ∼15 cM centromeric on 13q31 (the findings presented in Table 1) and ∼50 cM centromeric on 13q12–q13. For all of the chromosome 13 studies, results were obtained by use of narrow definitions of affected phenotype: schizophrenia only or schizophrenia and schizoaffective disorder. Studies that also evaluated evidence for linkage under broader phenotypic models found, in all cases, that a narrowly defined phenotype produced the strongest evidence for linkage to 13q (Lin et al. 1997; Straub et al. 1997; Shaw et al. 1998). Results were not as consistent with respect to recessive and dominant models of inheritance; some studies favored a dominant model, and others a recessive model (table 1). Maximum LOD scores under both inheritance models were not routinely reported, however, making intrastudy comparisons difficult. Interestingly, in the case of two studies using the same set of 54 families but different parametric-linkage methods and marker sets, Pulver et al. (1996) found greater support for linkage to 13q32 under the dominant model of inheritance, whereas Blouin et al. (1998) found greater support under the recessive model of inheritance.
Table 1.
Previous Significant and Suggestive Linkage Findings of Schizophrenia to Chromosomes 13q and 8p[Note]
|
Parametric Analyses |
||||
| Model-FreeAnalyses | Dominant | Recessive | No. of Families | Reference |
| Chromosome 13q: | ||||
| P=.00002a | hLOD=1.84a | hLOD=3.19a | 54b | Blouin et al. (1998) |
| P=.0002a | hLOD=3.24 | hLOD=2.53 | 54b | Pulver et al. (1996) |
| LOD=2.58a | hLOD=1.54a | … | 21 | Lin et al. (1997) |
| P=.02a | … | hLOD=1.36a | 275 | Straub et al. (1997) |
| P=.03a | hLOD=1.25 | … | 70 | Shaw et al. (1998) |
| Chromosome 8p: | ||||
| P=.0001a | hLOD=4.54a | hLOD=2.01a | 54b | Blouin et al. (1998) |
| P=.00004 | LOD=2.35 | LOD=2.20 | 57b | Pulver et al. (1995) |
| P=.006a | hLOD=2.34a | hLOD=2.52a | 265 | Kendler et al. (1996) |
| P=.005a | hLOD=.99 | hLOD=2.22 | 463c | SLCG (1996) |
| P=.07a | … | LOD=1.99 | 70 | Shaw et al. (1998) |
| P=.013a | … | … | 30 | Kaufmann et al. (1998) |
| P=.04 | … | … | 5 | Moises et al. (1995) |
Note.—P=nominal P values; LOD=maximum homogeneity LOD score; and hLOD=maximum heterogeneity LOD score.
Multipoint analysis result.
The same 54 families were studied by Pulver et al. (1996) and Blouin et al. (1998); 46 of these families overlapped with those studied by Pulver et al. (1995).
No known overlap with samples from other studies referenced.
The chromosome 8p linkage results shown in table 1 cluster within a region of ∼18 cM, centered around 8p21, except in one study, in which positive results extended over a 50-cM region (Kaufmann et al. 1998). A recent meeting report summarized additional evidence in support of an 8p21 schizophrenia locus (Wildenauer and Schwab 1998), including a LOD score of 2.16, found by Gurling and colleagues, under a dominant model of inheritance, using a set of 23 families from Iceland and the United Kingdom, and an increase in the maximum LOD score to 3.16, in the sample previously reported by Kendler et al. (1996). Most of the chromosome 8p studies reported analyses using a single, narrow definition of schizophrenia (Moises et al. 1995; Pulver et al. 1995; Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6, and 8 1996; Blouin et al. 1998; Kaufmann et al. 1998). Of two studies that considered broader phenotypic definitions, one reported maximum LOD scores under a very broad diagnostic definition, with significant decreases in scores under narrower definitions (Kendler et al. 1996), and another found the strongest evidence for linkage on chromosome 8p using a very narrow definition of disease (Shaw et al. 1998). As with the chromosome 13 findings, positive linkage results on 8p were seen under both dominant and recessive models of inheritance (table 1). Among studies reporting LOD scores for both dominant and recessive models, two studies, using overlapping samples, reported greater support for linkage under the dominant model (Pulver et al. 1995; Blouin et al. 1998). In contrast, two other studies reported higher maximum LOD scores under recessive models of inheritance (Kendler et al. 1996; Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6, and 8 1996).
Despite the differences in the samples used, the range of diagnostic and genetic models tested, and the variety of analytical approaches taken, the multiple findings of positive linkage support the possibility of schizophrenia-susceptibility genes located in relatively small regions of chromosomes 8p and 13q. Given the history of unreplicated findings in the field of psychiatric genetics (Moldin 1997), however, other significant linkage findings in studies using an independent sample of families would help establish these chromosomal regions as more definitively linked to schizophrenia.
As part of an ongoing genome scan, the present study evaluated the evidence for linkage of schizophrenia to chromosomes 8 and 13 in a sample of 21 extended Canadian families. Parametric-linkage analyses were conducted under both autosomal dominant and recessive models, using two diagnostic classifications, narrow and broad. We found significant linkage of schizophrenia to markers on 13q32, providing independent replication of significant linkage results for this chromosomal region.
Subjects and Methods
Subjects
Canadian families of Celtic (n=20) or German (n=1) descent were recruited for the linkage study if schizophrenic illness appeared to be segregating in a unilineal autosomal dominant manner (Bassett et al. 1993; Bassett and Honer 1994). Twenty-one moderately large families (n=285 subjects) were assessed, and 276 subjects had DNA samples available for the current study. All subjects were enrolled in this study after informed consent was obtained, and all procedures conformed to human-subjects protocols approved by the University of Toronto and Rutgers University. The average family size was 22 individuals, with an average of 13 individuals per family participating. Most families consisted of two or three generations with only first- and second-degree relatives participating, although some large complex families contained relatives as distant as ninth degree. Direct interviews—the Structured Clinical Interview for DSM-III-R (SCID-I) for major disorders and the SCID-II for personality disorders— along with collateral information and medical records were used to make consensus diagnoses on the basis of DSM III-R criteria. Further details of the diagnostic and ascertainment methods have been described elsewhere (Bassett et al. 1993; Bassett and Honer 1994). The diagnostic classifications narrow and broad were used, with 71 and 107 affected individuals in each category, respectively. Individuals were considered affected under the narrow diagnostic classification if they were diagnosed with schizophrenia (n=54) or chronic schizoaffective disorder (n=17). Individuals were considered affected under the broad diagnostic classification if they had been diagnosed with one of those disorders or with a nonaffective psychotic disorder (n=13), schizotypal personality disorder (n=17), or paranoid personality disorder (n=6). Diagnoses were available on all 285 subjects, and all individuals not categorized as affected under a given diagnostic scheme were classified as unaffected.
Genotyping
DNA was extracted from blood samples or lymphoblastoid cell lines by use of the GenePure system (Gentra Systems). DNA from each subject was genotyped by use of 11 chromosome 13 markers (table 2) and 19 chromosome 8 markers (table 3) from the Weber Screening Set, version 6.0, spanning chromosomes 8 and 13 at an average spacing of 10 cM. Two additional markers from chromosome 8, D8S560 and D8S298, were also genotyped. Genotyping was conducted in our laboratory and the laboratories of the Center for Inherited Disease Research (CIDR; Johns Hopkins University, Baltimore). Approximately 45% of all genotypes were generated by both laboratories. In our laboratory, genotypes were generated by PCR amplification incorporating radiolabeled dCTP. Sample handling, PCR amplification, gel electrophoresis, and genotype-interpretation procedures have all been described elsewhere (Brzustowicz et al. 1997). Genotype generation by CIDR used automated fluorescent microsatellite analysis (further details are available at the CIDR website). The error rate for the CIDR genotypes, calculated from 4,384 genotypes generated for 12 blind duplicate pairs, was 0.08% per genotype. Seventeen genotypes (0.2% of the total used in this study), although concordantly scored on repeat genotyping within and/or between labs, nonetheless were not consistent with Mendelian inheritance. These cases were apparently due to microsatellite instability, but since the source of the original allele could not always be determined, we consistently deleted these genotypes from our analyses.
Table 2.
Maximum Pairwise LOD Scores for Schizophrenia and Chromosome 13 Markers[Note]
|
Model |
||||||||||
| Recessive-Narrow |
Recessive-Broad |
Dominant-Narrow |
Dominant-Broad |
|||||||
| Locus | MarkerPositiona | Hetero-zygosityb | LOD (hLODc) | θ (αc) | LOD (hLOD) | θ (α) | LOD (hLOD) | θ (α) | LOD (hLOD) | θ (α) |
| D13S787 | 0 | .80 | 0 (0) | .4 (.75) | 0 | .5 | .53 | .3 | .28 | .4 |
| D13S1493 | 17 | .75 | 0 (0) | .5 (.05) | 0 (.07) | .4 (.20) | .76 | .2 | 0 (.11) | .5 (.10) |
| D13S894 | 24 | .68 | 0 | .5 | .02 (.03) | .4 (.50) | .09 | .4 | 0 | .5 |
| D13S325 | 30 | .79 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D13S788 | 37 | .85 | 0 | .5 | 0 | .5 | 0 (.01) | .4 (.65) | 0 | .5 |
| D13S800 | 47 | .74 | .65 (.65) | .2 (.95) | .06 | .4 | .74 (.96) | .2 (.30) | .21 (.46) | .3 (.25) |
| D13S317 | 56 | .78 | .49 | .3 | 0 | .5 | 1.03 (1.11) | .2 (.80) | 0 | .5 |
| D13S793 | 68 | .77 | 2.09 (2.10) | .1 (.95) | 1.71 (1.93) | .1 (.55) | 1.11 (1.52) | .2 (.40) | .07 (.08) | .4 (.35) |
| D13S779 | 75 | .65 | .77 | .2 | 1.78 (2.20) | .2 (.50) | .82 (1.09) | .2 (.45) | 0 (.75) | .5 (.20) |
| D13S796 | 86 | .81 | .31 (.83) | .3 (.25) | 1.60 (1.84) | .2 (.50) | .45 | .3 | .08 (.74) | .4 (.10) |
| D13S285 | 103 | .80 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
Note.—LOD=maximum homogeneity LOD score; hLOD=maximum heterogeneity LOD score.
In Kosambi centimorgans.
Determined from 30 unrelated family members.
For HOMOG analyses with α<1.
Table 3.
Maximum Pairwise LOD Scores for Schizophrenia and Chromosome 8 Markers[Note]
|
Model |
||||||||||
| Recessive-Narrow |
Recessive-Broad |
Dominant-Narrow |
Dominant-Broad |
|||||||
| Locus | MarkerPositiona | Hetero-zygosityb | LOD (hLODc) | θ(αc) | LOD (hLOD) | θ (α) | LOD (hLOD) | θ(α) | LOD (hLOD) | θ(α) |
| D8S264 | 0 | .85 | .73 (.74) | .2 (.90) | .26 | .3 | .35 | .3 | .30 (.38) | .3 (.70) |
| D8S1469 | 15 | .82 | .03 | .4 | 0 | .5 | 0 | .5 | .14 | .4 |
| D8S1130 | 21 | .71 | .57 (.61) | .2 (.80) | .11 (.12) | .3 (.90) | .52 (.53) | .2 (.85) | .32 (.54) | .3 (.35) |
| D8S1106 | 25 | .69 | .72 | .2 | 0 | .5 | 0 (.18) | .5 (.15) | .67 (.88) | .3 (.45) |
| D8S1145 | 36 | .75 | .81 (.81) | .2 (.95) | .39 (.53) | .3 (.30) | .50 | .2 | .35 (.38) | .3 (.75) |
| D8S560d | 43 | .80 | 1.33 | .2 | .37 | .3 | .82 | .2 | .38 (.40) | .2 (.35) |
| D8S136 | 43 | .80 | 3.11 | .1 | .56 | .3 | 3.49 | .1 | 1.29 (1.31) | .2 (.90) |
| D8S298d | 44 | .72 | .90 | .2 | .98 | .2 | 1.43 (1.47) | .1 (.5) | .87 (1.21) | .3 (.55) |
| D8S1477 | 59 | .81 | .40 (.66) | .3 (.30) | 0 | .5 | .26 (.52) | .3 (.30) | .20 (.20) | .4 (.75) |
| D8S1110 | 66 | .78 | .26 (.27) | .3 (.15) | 0 | .5 | .70 | .3 | .17 | .4 |
| D8S1113 | 77 | .78 | .06 | .4 | .08 | .4 | 0 (0) | .5 (.20) | 0 | .5 |
| D8S1136 | 81 | .70 | .09 (.09) | .3 (.95) | 0 (0) | .5 (.50) | .21 | .3 | 0 | .5 |
| D8S2324 | 93 | .79 | .08 | .3 | .25 | .3 | .02 | .4 | 0 (0) | .5 (.25) |
| D8S1119 | 100 | .83 | 0 | .5 | 0 | .5 | .10 | .4 | .06 | .4 |
| C8S14.2 | 109 | .66 | .44 (.46) | .2 (.75) | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1132 | 118 | .87 | .05 (.11) | .3 (.40) | 0 | .5 | .30 | .3 | 0 | .5 |
| D8S592 | 124 | .67 | .05 (.05) | .4 (.60) | .15 (.21) | .3 (.15) | .15 | .3 | .08 | .4 |
| D8S1179 | 134 | .78 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1128 | 138 | .75 | 0 | .5 | .06 | .4 | .32 | .3 | .05 (.05) | .4 (.95) |
| D8S256 | 147 | .76 | .08 (.10) | .3 (.25) | 0 (.06) | .5 (.10) | 0 | .5 | 0 | .5 |
| D8S373 | 163 | .73 | .07 | .4 | .33 | .3 | 0 | .5 | 0 | .5 |
Note.—LOD=maximum homogeneity LOD score; hLOD=maximum heterogeneity LOD score.
In Kosambi centimorgans.
Determined from 30 unrelated family members.
For HOMOG analyses with α<1.
Not part of the Weber screening set.
Linkage Analysis
Linkage analyses were conducted with the FASTLINK version 4.0P programs (Lathrop and Lalouel 1984; Lathrop et al. 1984, 1986; Cottingham et al. 1993; Schaffer et al. 1994). Parametric-linkage analyses were conducted because they are more powerful than nonparametric methods (Durner et al. 1999) and are robust methods for detecting linkage despite errors or simplifications in the analyzing model, as long as both a dominant and a recessive model are used (Vieland et al. 1992, 1993; Hodge et al. 1997; Greenberg et al. 1998; Durner et al. 1999). To minimize multiple tests, we selected four genetic models to analyze data from the genome scan—one dominant and one recessive for each of the narrow and broad diagnostic classifications. Because of difficulties in determining the appropriate correction for multiple tests when highly correlated diagnostic classifications are used (Lander and Kruglyak 1995), LOD scores are reported with no correction applied. Under the narrow diagnostic classification, the dominant model was schizophrenia susceptibility–allele frequency (pA) of .0045, with penetrance of disease (f) of .75, .50, and .001 for disease homozygotes (AA), heterozygotes (Aa), and normal homozygotes (aa), respectively; the recessive model was pA=.065, f(AA)=.50, f(Aa)=.0015, and f(aa)=.0015. Under the broad diagnostic classification, the dominant model was pA=.007, f(AA)=.90, f(Aa)=.80, and f(aa)=.009; and the recessive model was pA=.10, f(AA)=.60, f(Aa)=.01, and f(aa)=.01. Marker-allele frequencies for the analysis were estimated with a set of 30 unrelated subjects from these families and were comparable to those listed in the Centre d'Etude du Polymorphisme Humain (CEPH) and Cooperative Human Linkage Center databases. Two-point linkage analyses were conducted by use of MLINK at recombination fraction (θ) 0, .01, .05, .1, .2, .3, and .4. Multipoint analyses were conducted with LINKMAP. Three-point analyses were conducted with adjacent pairs of screening-set markers and the disease locus, with intermarker distances derived from the Marshfield Map positions listed in tables 2 and 3. Additional markers flanking D8S136 were used in four-point analyses with the following map order and recombination fractions: D8S560-.005-D8S136-.0001-D8S298. Heterogeneity testing was conducted with the HOMOG program (Ott 1986, 1991). The large size and complexity of the pedigrees in this sample (Bassett and Honer 1994) made multipoint linkage analysis by the GENEHUNTER program (Kruglyak et al. 1996) infeasible because of loss of data.
Results
Under all models, two-point linkage analysis produced positive LOD scores with markers on 13q, but with significantly higher LOD scores under the recessive models (table 2). The maximum two-point LOD score of 2.09 under homogeneity was obtained with the recessive model using the narrow diagnostic classification, at θ=.10 with D13S793. The maximum two-point LOD score under heterogeneity was 2.20 at θ=.10 with D13S779 and α=.5, obtained under the recessive model using the broad diagnostic classification. A pattern of suggestive positive LOD scores spanning several adjacent markers on chromosome 13q (D13S793, D13S779, and D13S796) was observed (table 2). Inspection of the linkage results by family revealed that the same subset of pedigrees was contributing to the positive linkage findings across this entire region.
Under all models, two-point linkage analysis produced positive LOD scores with markers on 8p but with significantly higher LOD scores under the models using the narrow diagnostic classification (table 3). The maximum two-point LOD score of 3.49 was obtained with the dominant model using the narrow diagnostic classification, at θ=.1 with D8S136, and was the same under homogeneity and heterogeneity. Positive but substantially lower LOD scores were obtained at markers on either side of D8S136 (table 3).
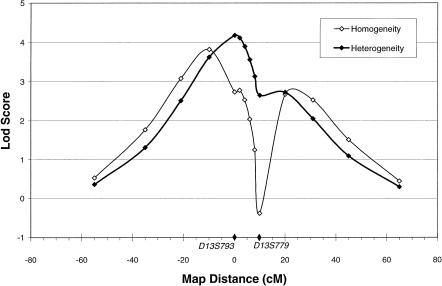
Parametric multipoint analyses of complex disorders must be used with caution, since incorrect analysis models can lead to distortions of the estimated map position of the disease locus and can exclude a true linked locus from the region between close flanking markers (Risch and Guiffra 1992). However, multipoint analyses are useful for combatting the practical limitations caused by inevitable uninformative marker typings, which can either inflate or deflate the LOD score. With large, complex pedigrees, analyzing more than two highly polymorphic marker loci at a time can be computationally prohibitive when large regions of the genome are scanned for linkage. We therefore elected to conduct three-point analyses between adjacent marker loci and the disease locus. For chromosome 13, this produced, across all analysis models under homogeneity, a maximum LOD score of 3.92 at θ=.1 on the centromeric side of D13S793, under the recessive model using the broad diagnostic classification and the marker pair D13S793 and D13S779. In none of the three-point analyses under homogeneity was the maximum LOD score located in the interval between the two marker loci, an anticipated effect of multipoint analysis under an incorrect model. Since one possible error in the model was the assumption of homogeneity, three-point results were also evaluated with HOMOG, for evidence of heterogeneity. Although the hypothesis of heterogeneity was not statistically supported, favored by a likelihood ratio of only 3:1 (χ2=2.3), the location of the maximum LOD score under heterogeneity did shift to the position of one of the marker loci. The maximum three-point LOD score under the assumption of heterogeneity was 4.42, with α=.65 at a θ=0 with D13S793, again under the recessive model using the broad diagnostic classification and the marker pair D13S793 and D13S779 (fig. 1).
Figure 1.
Three-point analysis of schizophrenia, with D13S793, and D13S779 under the recessive-broad genetic model, under homogeneity and heterogeneity. D13S793 is at map position 0. α=.65 for the heterogeneity LOD score.
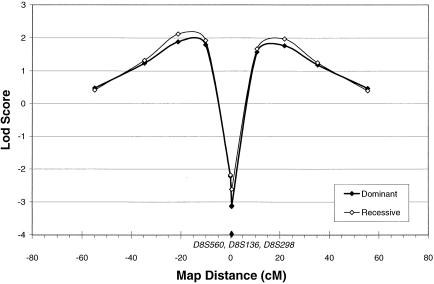
For chromosome 8, the maximum three-point LOD score for the screening-set markers, under both homogeneity and heterogeneity, was 3.51 at θ=.1 on the telomeric side of D8S136, under the dominant model using the narrow diagnostic classification and the markers D8S136 and D8S1477. This result was not significantly different from that obtained during the two-point analysis, and the screening-set markers flanking D8S136 (D8S1145 and D8S1477) did not demonstrate supportive positive LOD scores (table 3). Since these findings could indicate a LOD score inflated by uninformativeness of marker D8S136, two closely flanking markers (D8S560 and D8S298) were genotyped. The maximum two-point LOD score under both homogeneity and heterogeneity with D8S560 was 1.33 at θ=.2, under the recessive narrow model (table 3). The maximum two-point LOD score under homogeneity with D8S298 was 1.43 at θ=.1, under the dominant-narrow model, increasing to 1.47 at θ=.01 and α=.5, under heterogeneity (table 3). D8S560, D8S136, and D8S298 are very tightly linked, spanning only 0.5 cM and with no evidence of recombination observed in this set of families. There was, therefore, no concern that using all three markers in four-point analyses would falsely exclude linkage from the region between these markers. The maximum four-point LOD score under homogeneity across all analysis models was 2.13 at θ=.2 from D8S560, under the recessive model with the narrow diagnostic classification (fig. 2). Results were nearly identical when the dominant-narrow model was used (fig. 2). There was no evidence for heterogeneity in the four-point analyses.
Figure 2.
Four-point analysis of schizophrenia, with D8S560, D8S136, and D8S298, under homogeneity and the dominant-narrow and recessive-narrow genetic models. D8S560 is at map position 0. Because of their close proximity, the positions of the three markers are indicated by a single point, on the X-axis, that spans map position 0–0.5 cM. Also because of proximity, the plotted points for D8S136 and D8S298 overlap and appear as a single point on the LOD-score plot.
Discussion
To date, individual linkage studies of schizophrenia have produced mixed results, often characterized by suggestive but not significant results and by lack of replication across studies (Moldin 1997). This study produced evidence of a significant linkage on 13q32, as well as of a suggestive linkage on 8p21, <5 cM from the strongest previously reported linkages on both these chromosomes (Blouin et al. 1998). Furthermore, the results independently establish linkage of schizophrenia to markers on chromosome 13q32, satisfying significance standards appropriate for a genomewide scan of a complex disorder.
In this study, we have tested two modes of inheritance and two closely related diagnostic classifications. Lander and Kruglyak (1995) have suggested a LOD score of 3.3 as the significance threshold for linkage studies of complex traits, noting that the appropriate correction for testing multiple, closely related models is not readily known. Morton (1998) has argued that a LOD of 3 provides convincing evidence of linkage even for complex traits, as long as the assumptions of the test are not violated. Hodge et al. (1997) have demonstrated through simulation studies that a 0.3-LOD-unit correction for testing over two dominance models is conservative. Risch's (1991) proposal to subtract log10(t) for t tests from the obtained LOD score is a conservative correction for analysis over multiple diagnostic or inheritance models and may be overly conservative, given the highly correlated nature of the two affected phenotypes in the current study. Nonetheless, applying a log10(t) correction for the four tested models to the maximum LOD score of 3.92 for chromosome 13 markers obtained under the hypothesis of homogeneity would still produce a significant corrected LOD score of 3.3.
The autosomal recessive mode of inheritance was favored in our analysis of 13q, as it was in the studies by Straub et al. (1997) and Blouin et al. (1998). This support for an autosomal recessive mode of inheritance was an unexpected result, since one of our ascertainment criteria was a pattern of expression suggestive of autosomal dominant transmission (Bassett and Honer 1994). However, common recessive disease alleles in a population can produce pedigree patterns that appear to be autosomal dominant, underscoring the need for conducting parametric analyses under both dominant and recessive models for disorders with unknown modes of inheritance (Vieland et al. 1992, 1993; Durner et al. 1999).
Although the maximum LOD score for chromosome 13 was obtained by use of the broad definition of affection, prior reports of suggestive and significant chromosome 13 linkages have been obtained by use of a narrow disease definition (Pulver et al. 1996; Lin et al. 1997; Straub et al. 1997; Blouin et al. 1998; Shaw et al. 1998). Although some of these other studies did not perform an analysis under a broad definition of disease (Pulver et al. 1996; Blouin et al. 1998), those that did use multiple diagnostic classifications reported that the largest maximum LOD scores occurred under the narrow definition of disease (Lin et al. 1997; Straub et al. 1997; Shaw et al. 1998). However, since these studies did not also report the scores obtained under the broader diagnostic definitions, it is impossible to gauge the magnitude of the effect of diagnostic classification on maximum LOD score. The maximum homogeneity multipoint LOD score under the recessive narrow model was 3.08, at the same location as the recessive-broad multipoint maximum of 3.92. These scores are significantly higher than the maximum homogeneity multipoint LOD scores obtained under the dominant model: 1.38 and 0.28 with the narrow and broad definitions of disease, respectively.
The maximum LOD scores on 8p were similar under both the dominant- and recessive-narrow models (table 3 and fig. 2), consistent with the results of Pulver et al. (1995) and Kendler et al. (1996). However, there was a striking decrease in LOD scores under the broad models. Most of the prior reports of linkage on 8p considered only a narrow definition of affection (Moises et al. 1995; Pulver et al. 1995; Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6, and 8 1996; Blouin et al. 1998; Kaufmann et al. 1998). Shaw et al. (1998) considered several diagnostic classifications and obtained the strongest evidence for linkage under a very narrow definition of disease. This is opposite to the effect described by Kendler et al. (1996), who reported diminished support for linkage under a narrow diagnostic classification. Better definition of the aspects of the schizophrenia phenotype linked to this locus may help to clarify these results and to define a more appropriate genetic model for future linkage studies.
Recent simulation studies have suggested that, as long as linkage analyses are conducted under both a recessive and dominant model, parametric approaches can have significant power to detect linkage of simple or complex disorders, with little loss of power even when the analyzing model is constructed with an arbitrary disease penetrance (Hodge et al. 1997; Greenberg et al. 1998; Durner et al. 1999). Given this, it is not surprising that despite the slightly different genetic models used by different investigators for their parametric-linkage studies, the same small regions of chromosomes 8 and 13 have been implicated in schizophrenia susceptibility, by a number of groups. It is also clear, however, that not all samples have produced suggestive or significant evidence of linkage to these regions (Coon et al. 1994; Kunugi et al. 1996; Barden and Morissette 1998; Faraone et al. 1998; Levinson et al. 1998; Wildenauer and Schwab 1998), possibly because of factors such as genetic heterogeneity between samples, differences in diagnostic approaches, or sampling effects. The results presented here, as well as the results presented by Blouin et al. (1998), demonstrate that significant linkage results can be obtained in studies of complex disorders when moderate-sized samples (<500 subjects) and parametric-linkage approaches are used. Furthermore, these significant, replicable linkage results suggest that major-gene effects may be present in some samples of familial schizophrenia.
Acknowledgments
We would like to thank the participating families, whose contributions have made these studies possible; Drs. Robert Forsythe and Pamela Forsythe, for years of support; and Manzer Kahn, Jared Hayter, Laura Scutt, and Stephanie Roberts, for technical and administrative assistance. This research was supported by grants from the Medical Research Council of Canada (to A.S.B., L.M.B., and W.G.H.), the EJLB Foundation Scholar Research Programme (to L.M.B.), National Institute of Mental Health (grant K08 MH01392, to L.M.B.), the Ontario Mental Health Foundation (to A.S.B.), the Bill Jeffries Schizophrenia Endowment Fund (to A.S.B.), and the Ian Douglas Bebensee Foundation (to A.S.B.). W.G.H. is supported by a Vancouver Hospital Scientist Award. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through contract N01-HG-65403 from the National Institutes of Health to The Johns Hopkins University. Special thanks to Kim Doheny and Elizabeth Pugh of CIDR for their assistance with this project.
Electronic-Database Information
URLs for data in this article are as follows:
- Center for Inherited Disease Research, http://www.cidr.jhmi.edu/
- CEPH Genotype Database, http://www.cephb.fr/cephdb/
- Cooperative Human Linkage Center, http://www.chlc.org/
References
- Barden N, Morissette J (1998) Chromosome 13 Workshop. Psychiatr Genet 8:93–96 [DOI] [PubMed]
- Bassett AS, Collins EJ, Nuttall SE, Honer WG (1993) Positive and negative symptoms in families with schizophrenia. Schizophr Res 11:9–19 [DOI] [PMC free article] [PubMed]
- Bassett AS, Honer WG (1994) Evidence for anticipation in schizophrenia. Am J Hum Genet 54:864-870 [PMC free article] [PubMed]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 [DOI] [PubMed]
- Brzustowicz LM, Honer WG, Chow EWC, Hogan J, Hodgkinson K, Bassett AS (1997) Use of a quantitative trait to map a locus associated with severity of positive symptoms in familial schizophrenia to chromosome 6p. Am J Hum Genet 61:1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon H, Jensen S, Holik J, Hoff M, Myles-Worsley M, Reimherr F, Wender P, et al (1994) Genomic scan for genes predisposing to schizophrenia. Am J Med Genet 54:59–71 [DOI] [PubMed]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for increased power of LOD scores over nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, et al (1998) Genome scan of European-American schizophrenia pedigrees: results of the NIMH genetics initiative and millenium consortium. Am J Med Genet 81:290–295 [PubMed]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870-879 [DOI] [PMC free article] [PubMed]
- Hodge SE, Abreu PC, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed]
- Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, et al (1998) NIMH genetics initiative millenium schizophrenia consortium: linkage analysis of African-American pedigrees. Am J Med Genet 81:282–289 [PubMed]
- Kendler KS, MacLean CJ, O'Neill A, Burke J, Murphy B, Duke F, Shinkwin R, et al (1996) Evidence for a schizophrenia vulnerability locus on chromosome 8p in the Irish study of high-density schizophrenia families. Am J Psychiatry 153:1534–1540 [DOI] [PubMed]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed]
- Kunugi H, Curtis D, Vallada HP, Nanko S, Powell JF, Murray RM, McGuffin P, et al (1996) A linkage study of schizophrenia with DNA markers from chromosome 8p21-p22 in 25 multiplex families. Schizophr Res 22:61–68 [DOI] [PubMed]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed]
- Lathrop GM, Lalouel J-M (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed]
- Lathrop GM, Lalouel J-M, Julier C, Ott J (1984) Strategies for multilocus analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed]
- Lathrop GM, Lalouel J-M, White RL (1986) Construction of human genetic linkage maps: likelihood calculations for multilocus analysis. Genet Epidemiol 3:39–52 [DOI] [PubMed]
- Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Kirby A, et al (1998) Genome scan of schizophrenia. Am J Psychiatry 155:741–50 [DOI] [PubMed]
- Lin MW, Curtis D, Williams N, Arranz M, Nanko S, Collier D, McGuffin P, et al (1995) Suggestive evidence for linkage of schizophrenia to markers on chromosome 13q14.1-q32. Psychiatr Genet 5:117–126 [DOI] [PubMed]
- Lin MW, Sham P, Hwu HG, Collier D, Murray R, Powell JF (1997) Suggestive evidence for linkage of schizophrenia to markers on chromosome 13 in Caucasian but not Oriental populations. Hum Genet 99:417–420 [DOI] [PubMed]
- McGuffin P, Asherson P, Owen M, Farmer A (1994) The strength of the genetic effect: is there room for an environmental influence in the aetiology of schizophrenia? Br J Psychiatry 164:593–599 [DOI] [PubMed]
- Moises HW, Yang L, Kristbjarnarson H, Wiese C, Byerley W, Macciardi F, Arolt V, et al (1995) An international two-stage genome-wide search for schizophrenia susceptibility genes. Nat Genet 11:321–324 [DOI] [PubMed]
- Moldin SO (1997) The maddening hunt for madness genes. Nat Genet 17:127–129 [DOI] [PubMed]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62:690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1986) Linkage probability and its approximate confidence interval under possible heterogeneity. Genet Epidemiol Suppl 1:251–257 [DOI] [PubMed]
- ——— (1991) Analysis of human genetic linkage, rev. ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Pulver AE, Lasseter VK, Kasch L, Wolyniec P, Nestadt G, Blouin JL, Kimberland M, et al (1995) Schizophrenia: a genome scan targets chromosomes 3p and 8p as potential sites of susceptibility genes. Am J Med Genet 60:252–260 [DOI] [PubMed]
- Pulver AE, Wolyniec PS, Housman D, Kazazian HH, Antonarakis G, Nestadt G, Lasseter VK, et al (1996) The Johns Hopkins University Collaborative Schizophrenia Study: an epidemiologic-genetic approach to test the heterogeneity hypothesis and identify schizophrenia susceptibility genes. Cold Spring Harb Symp Quant Biol 61:797–814 [PubMed]
- Risch N (1991) A note on multiple testing procedures in linkage analysis. Am J Hum Genet 48:1058–1064 [PMC free article] [PubMed]
- Risch N, Giuffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed]
- Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed]
- Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6, and 8 (1996) Additional support for schizophrenia linkage on chromosomes 6, and 8: a multicenter study. Am J Med Genet 67:580–594 [DOI] [PubMed]
- Shaw SH, Kelly M, Smith AB, Shields G, Hopkins PJ, Loftus J, Laval SH, et al (1998) A genome-wide search for schizophrenia susceptibility genes. Am J Med Genet 81:364–376 [DOI] [PubMed]
- Straub RE, MacLean CJ, O'Neill FA, Walsh D, Kendler KS (1997) Genome scan for schizophrenia genes: a detailed progress report in an Irish cohort. Am J Med Genet Neuropsychiatr Genet 74:558 [Google Scholar]
- Vieland VJ, Greenberg DA, Hodge SE (1993) Adequacy of single-locus approximations for linkage analyses of oligogenic traits: extension to multigenerational pedigree structures. Hum Hered 43:329–336 [DOI] [PubMed]
- Vieland VJ, Hodge SE, Greenberg DA (1992) Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol 9:45–59 [DOI] [PubMed]
- Wildenauer DB, Schwab SG (1998) Chromosome 8 Workshop. Psychiatr Genet 8:57–87 [DOI] [PubMed]