Summary
The availability of robust quantitative biological markers that are correlated with qualitative psychiatric phenotypes can potentially improve the power of linkage methods to detect quantitative-trait loci influencing psychiatric disorders. We apply a variance-component method for joint multipoint linkage analysis of multivariate discrete and continuous traits to the extended pedigree data from the Collaborative Study on the Genetics of Alcoholism, in a bivariate analysis of qualitative alcoholism phenotypes and quantitative event-related potentials. Joint consideration of the DSM-IV diagnosis of alcoholism and the amplitude of the P300 component of the Cz event-related potential significantly increases the evidence for linkage of these traits to a chromosome 4 region near the class I alcohol dehydrogenase locus ADH3. A likelihood-ratio test for complete pleiotropy is significant, suggesting that the same quantitative-trait locus influences both risk of alcoholism and the amplitude of the P300 component.
Introduction
Many psychiatric disorders, such as schizophrenia or alcoholism, are typically diagnosed, at a nominal level, as a dichotomous trait—that is, on a binary or presence/absence basis. With some disorders (e.g., bipolar disorder), a more refined diagnosis may be possible on an approximately ordinal scale of severity, but the number of classes distinguished is relatively few, and the diagnostic classes themselves may not be mutually exclusive. In general, psychiatric diagnostic systems are largely classificatory rather than metrical in nature and are subject to high imprecision, large interobserver variability, low repeatability, and questions of validity and relevance (Blackwood et al. 1991; Moldin 1994). In statistical genetic analysis, the discrete nature of psychiatric phenotypes can result in increased type I error, decreased power, and reduced ability to detect, localize, and estimate the effect size of quantitative-trait loci (QTLs) that influence these traits (Xu and Atchley 1996; Duggirala et al. 1997; Wijsman and Amos 1997).
Although inherently continuous biological characters that directly quantify psychiatric disorders are unavailable, there are often correlated quantitative characters, disease precursors, or associated risk factors that can be measured easily. For example, schizophrenia is correlated with a number of altered psychophysiological paradigms, such as eye-tracking dysfunction (Holzman et al. 1973), reduced visual and auditory P300 amplitudes (Blackwood et al. 1991), and diminished inhibition of the P50 auditory-evoked response to the second of paired stimuli (Freedman et al. 1997). In individuals at risk for alcoholism, the amplitude of the visual P300 component of event-related potentials (ERPs) is significantly decreased (Begleiter et al. 1984; Polich et al. 1994; Porjesz and Begleiter 1996; Porjesz et al. 1998). These correlated biological markers, or endophenotypes (Ott 1995), can be used to identify relatives of affected individuals who, on the basis of conventional diagnostic criteria, would be considered unaffected but who nevertheless are at risk for the disease. To some extent, the correlated character may also serve to quantify the degree of the associated risk (Blackwood et al. 1991; Porjesz et al. 1998).
The availability of a quantitative biological marker that is correlated with a qualitative disease trait suggests the possibility of exploiting the joint information content of the pair of phenotypes, in the search for mediating genetic factors (McGuffin 1984; Lander 1988; Moldin 1994; Ott 1995; Blangero et al. 1997; Williams et al. 1999 [in this issue]; Czerwinski et al., in press). As a demonstration of the potential utility of considering correlated biological markers in elucidating the genetics of a psychiatric disorder, we apply the method described by Williams et al. (1999 [in this issue]) to data, on alcoholism and event-related potentials, from the Collaborative Study on the Genetics of Alcoholism (COGA).
ERPs are neuroelectrical signals in the brain that are elicited in response to stimuli such as light and sound. These signals are complex convolutions of exogenous potentials, influenced by the characteristics of the stimulus, and endogenous potentials, influenced by cognitive processes, and can be regarded as direct measures of cognitive activity when the brain is engaged in perception, memory, and attention. Other properties of ERPs appear to reflect changes due to aging and brain maturation. Although ERPs are highly variable between individuals, they are relatively stable within individuals (Porjesz and Begleiter 1985, 1996; Regan 1989). Unlike electroencephalograms (EEGs), which monitor collective neuroelectrical activity, ERPs are highly sensitive to specific brain functions and provide useful indices to the cognitive process (Porjesz and Begleiter 1996).
The P3(00) ERP component has been studied extensively in relation to alcoholism (Polich et al. 1994; Porjesz and Begleiter 1996; Porjesz et al. 1998). The P300 component is a positive-going signal that occurs ∼300 ms after presentation of a stimulus. The amplitude of the P300 signal is related to cognitive activity, such as attention and memory, and to the significance of the stimulus—that is, factors such as task relevance, unpredictability, infrequency, and motivation (Pritchard 1981; Regan 1989). The COGA experimental protocol for visual and auditory P300 paradigms has been described in detail by Alexander et al. (1994) and Cohen et al. (1994).
Twin and family studies have demonstrated that P300 characteristics are highly heritable (h2≈.49–.60) (Polich and Burns 1987; Rogers and Deary 1991; O'Connor et al. 1994; van Beijsterveldt et al. 1996; Katsanis et al. 1997; Almasy et al. 1999). Studies have also found that alcoholics and persons at elevated risk for alcoholism (e.g., sons of alcoholic fathers) exhibit reduced P300 amplitudes (Begleiter et al. 1984; O'Connor et al. 1987; Polich et al. 1994). The amplitude reduction is also observed among unaffected and alcohol-naive relatives of alcoholic probands and persists even in long-term–abstinent alcoholics (Porjesz and Begleiter 1985, 1996). The reduction in the P300 component in alcoholics and high-risk males is correlated strongly with number of alcohol-dependent individuals in a family, rather than with alcohol intake (Pfefferbaum et al. 1991). These findings suggest that P300 abnormalities are not functional consequences of alcoholism but, instead, that they antecede alcohol use and dependence and reflect physiological processes that mediate liability to alcoholism and other complex, multifactorial psychiatric disorders (Begleiter et al. 1984; Polich et al. 1994; Porjesz and Begleiter 1996).
Data
The families used in this study come from COGA, a large, multicenter, and multidisciplinary research effort initiated in 1989 to investigate the genetic factors that influence susceptibility to alcohol dependence and abuse (Begleiter et al. 1995). One of the many strengths of the COGA project is the collection of quantitative neuropsychological and neurophysiological data to complement the qualitative assessments related to alcoholism and associated behavioral characteristics.
Probands were systematically recruited from inpatient and outpatient alcohol-treatment facilities. Subjects who were affected under both DSM-III-R “dependence” and Feighner “definite” criteria and who had a qualifying family structure (either a sibship of at least three individuals, if parents were available for study, or a larger sibship, if parents were unavailable) were considered to be probands and their families were considered to be stage I families. Probands were not accepted into the study if they were intravenous drug users, were HIV positive, suffered a terminal illness unrelated to alcohol dependence, or were otherwise unable to participate.
All probands, their spouses, and first-degree relatives who met admission criteria were interviewed by means of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al. 1994), to assess adult lifetime psychiatric status. The SSAGA enables alcoholism dependence, abuse, and harmful use to be diagnosed by means of Feighner criteria (Feighner et al. 1972), DSM-III, DSM-III-R, and DSM-IV (American Psychiatric Association 1980, 1987, 1994), and ICD-10 (World Health Organization 1992) criteria. These diagnostic systems are broadly similar in their criteria for identification of substance dependence, but they differ considerably in their criteria for substance abuse or harmful use (Schuckit et al. 1994). The ICD-10 system focuses on psychological and physical harm occurring in the context of substance use, and it deliberately excludes social, interpersonal, and legal problems, whereas the DSM-III-R and DSM-IV systems are broader and include psychosocial items. The DSM-III-R and DSM-IV systems differ with regard to whether certain symptoms are indicative of dependence or abuse/harmful use (Schuckit et al. 1994).
The COGA criterion for alcoholism requires an individual to meet both the DSM-III-R criteria for alcohol dependence and the Feighner criteria for alcoholism at the “definite” level. Subjects who consumed alcohol but who had no symptoms of alcohol dependence, alcohol abuse, or harmful use by any diagnostic system were considered to be “unaffected.” Subjects who had never been exposed to alcohol were labeled “unknown,” as were subjects who had one or more symptoms of alcohol dependence or abuse but who nevertheless did not meet the full COGA criterion for alcohol dependence.
If the proband and at least two first-degree relatives were affected under the COGA diagnosis (parents were included in the pool of first-degree relatives) and if at least one parent was unaffected (to avoid bilineal matings), then the family was considered to be a stage II family. The pedigree was extended, and family members were interviewed by means of the SSAGA. Blood samples were drawn for biochemical analyses and extraction of DNA, and a battery of neurophysiological and neuropsychological tests were administered. Stage II families were redesignated as stage III when they became available for genotyping and after the pedigree had been reviewed for completeness. Stage IV families are those that were actually selected for genotyping.
For this study, 105 multigenerational stage IV pedigrees comprising 1,219 individuals were available for genetic linkage analysis. The limiting factor on the use of these data is the availability of phenotype information; in the data examined here, there are 11 full or partially phenotyped pedigrees with one generation, 55 pedigrees with two generations, 32 with three generations, and 7 with four generations. The COGA genetic map used for these analyses comprises 263 markers, with an average intermarker distance of 13.2 cM.
Methods
Multipoint genome linkage scans were conducted at 2-cM intervals, by means of the analysis package solar (sequential oligogenic linkage analysis routines) (Almasy and Blangero 1998) and the joint qualitative-quantitative linkage method described by Williams et al. (1999 [in this issue]). Regions surrounding a signal peak were examined further with a limited multipoint scan at 1-cM resolution. As covariates in all analyses, we used age at SSAGA and sex of the individual. Age effects for males and females were modeled separately.
Given the extent to which the COGA data are ascertained, we attempted in our analyses to correct for the nonrandom sampling. The COGA ascertainment scheme comprises an initial single ascertainment event to obtain a stage I family, followed by a multiplex ascertainment event (the proband and at least two first-degree relatives must be affected) to obtain stage II families and, ultimately, stage IV families. The complexity of this ascertainment makes an exact correction difficult to specify (Comuzzie and Williams, in press), but some adjustment is essential in order to recover unbiased estimates of genetic effects and to maximize the power to detect linkage.
An approximate correction for ascertainment bias was made by conditioning the likelihood for each pedigree on the joint event represented by the observed trait values of the focal proband and two randomly selected affected first-degree relatives (Hopper and Mathews 1982; Boehnke and Greenberg 1984; Boehnke et al. 1986; Beaty and Liang 1987). We have therefore corrected for ascertainment as if the COGA stage IV pedigrees had been multiplex ascertained (Thompson 1993) on a focal proband and two secondary probands. Comuzzie and Williams (in press) have shown, for the COGA data, that this correction may be conservative and may increase the rate of type II errors.
Univariate genome scans were performed initially with the COGA, DSM-IV, and ICD-10 diagnoses of alcoholism, to identify regions for further investigation using multitrait linkage analysis. For these analyses, data were available for 661 (COGA), 593 (DSM-IV), and 488 (ICD-10) individuals, in 105 stage IV pedigrees.
The results of univariate genome scans for the visual P300 component of Cz, O2, and T8 ERPs have been reported elsewhere (Begleiter et al. 1998). These analyses involved 604 individuals from 100 stage IV COGA pedigrees and were not corrected for ascertainment bias. Subsequent reanalysis of these data to include the ascertainment correction described above confirmed the original findings (results not shown).
Bivariate genome scans were performed for the COGA, DSM-IV, and ICD-10 diagnoses, with the P300 component of the Cz, O2, and Pz ERPs. For these analyses, data on the P300 components for 607 individuals in 103 stage IV pedigrees were added to the univariate data on alcoholism. ERPs at the Cz and O2 electrodes were chosen for study because comparative univariate genome scans for these ERPs have been reported (Begleiter et al. 1998). The Pz electrode was included because P300 responses have their maximum amplitude at this location and because previous studies of reduced P300 amplitude in abstinent alcoholics and in high-risk individuals have focused on this electrode (Porjesz and Begleiter 1996; Porjesz et al. 1998).
Results
Univariate Genome Scan of Alcoholism Diagnosis
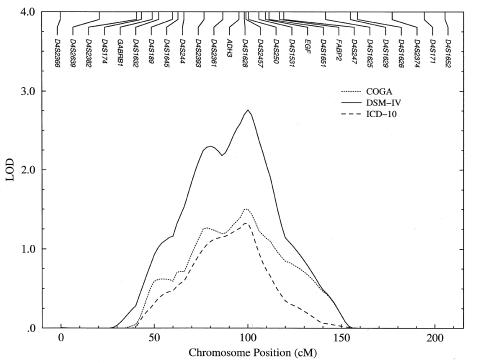
Results for the univariate genome scan are summarized in table 1 and figure 1. Linkage analysis with each alcoholism diagnosis reveals the presence of a signal in the ADH region of chromosome 4. The location of the peak is estimated consistently at 99–100 cM, near the ADH3 locus at 95.2 cM on the COGA map. The strongest evidence for linkage (LOD=2.76) is obtained with the DSM-IV diagnosis, although the COGA and ICD-10 diagnoses each give a peak LOD score >1.0. The relative strength of the linkage signal on chromosome 4 is evident irrespective of the diagnostic system—no other signals in the univariate analysis of these diagnoses had a LOD score >2.0, and only one additional LOD score was >1.0.
Table 1.
Maximum Multipoint LOD Scores on Chromosome 4, in Univariate Linkage Analysis of Three Alcoholism Diagnoses
| Diagnosis | Position(cM) | LOD Score | λ | Pa |
| COGA | 99 | 1.51 | 6.95 | .0042 |
| DSM-IV | 100 | 2.76 | 12.71 | .0002 |
| ICD-10 | 99 | 1.33 | 6.13 | .0066 |
Under the null hypothesis, the likelihood-ratio statistic λ is distributed as a 1/2χ21: 1/2χ20 mixture.
Figure 1.
Multipoint LOD scores on chromosome 4, in univariate linkage analysis of COGA, DSM-IV, and ICD-10 diagnoses of alcoholism
Table 1 gives the location and value of the maximum LOD score in each univariate analysis shown in figure 1; also given are the value of the likelihood-ratio statistic λ for the test of linkage, along with its corresponding P value. Under the null hypothesis of no linkage, the likelihood-ratio statistic corresponding to the univariate LOD score is asymptotically distributed as a 1/2: 1/2 mixture of a χ21 distribution and a unit point mass at the origin, denoted by χ20 (Hopper and Mathews 1982; Beaty et al. 1987; Self and Liang 1987; Liang et al. 1996).
Bivariate Genome Scan of Alcoholism Diagnosis and ERP
In view of the linkage signal on chromosome 4 in univariate analyses of the discrete alcoholism diagnoses, bivariate qualitative-quantitative genome scans were undertaken for the COGA, DSM-IV, and ICD-10 diagnoses, with the P300 component of the Cz, O2, and Pz ERPs. Results for these analyses are summarized in table 2.
Table 2.
Maximum Multipoint LOD Scores on Chromosome 4, in Bivariate Linkage Analyses of Three Alcoholism Diagnoses and the P300 Component of Cz, O2, and Pz ERPs
| Diagnosis | ERP | Position(cM) | LOD Score | LOD[1] | λ | Pa |
| COGA | Cz | 101 | 3.17 | 2.65 | 14.58 | 2.38×10-4 |
| DSM-IV | Cz | 101 | 4.75 | 4.16 | 21.89 | 5.86×10-6 |
| ICD-10 | Cz | 100 | 2.47 | 2.00 | 11.38 | 1.22×10-3 |
| COGA | O2 | 100 | 1.69 | 1.27 | 7.76 | 7.83×10-3 |
| DSM-IV | O2 | 101 | 3.16 | 2.64 | 14.53 | 2.43×10-4 |
| ICD-10 | O2 | 100 | 1.58 | 1.18 | 7.30 | 9.95×10-3 |
| COGA | Pz | 100 | 2.35 | 1.88 | 10.83 | 1.61×10-3 |
| DSM-IV | Pz | 101 | 4.01 | 3.46 | 18.45 | 3.34×10-5 |
| ICD-10 | Pz | 100 | 1.98 | 1.54 | 9.13 | 3.85×10-3 |
Under the null hypothesis, the likelihood-ratio statistic λ is distributed as a 1/4χ22: 1/2χ21: 1/4χ20 mixture.
The bivariate LOD scores in table 2 each have 2 df and are not directly comparable to the univariate LOD scores presented in table 1. Under the null hypothesis of no linkage of either alcoholism or ERP phenotypes to a QTL, the likelihood-ratio statistic corresponding to the bivariate LOD score is asymptotically distributed as a 1/4: 1/2: 1/4 mixture of χ22, χ21, and χ20 distributions (Self and Liang 1987). Consequently, in table 2, two LOD scores are given for each bivariate analysis: the true bivariate LOD score having 2 df and the bivariate LOD score after adjustment to 1 df, denoted as “LOD[1].” The adjusted LOD score is determined as the 1-df LOD score required to give the same P value as is given by the true bivariate LOD score and can be compared directly with the univariate LOD scores in table 1.
Similarly, the Zmax-1 support interval for a univariate LOD score (Ott 1991) has its bivariate equivalent. One LOD-score unit under the univariate null distribution ( 1/2χ21: 1/2χ20) corresponds to a P value of .0319. To obtain the same P value under the bivariate null distribution ( 1/4χ22: 1/2χ21: 1/4χ20) requires a LOD score of 1.39; thus, the equivalent bivariate support interval is Zmax-1.39. (Under the naive assumption that the likelihood-ratio statistics corresponding to the univariate and bivariate LOD scores follow χ21 and χ22 distributions, respectively, the corresponding bivariate support interval would be Zmax-1.50.)
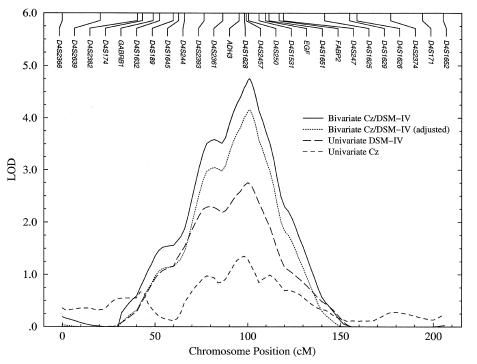
With Cz and Pz, the linkage signal on chromosome 4 increases under all diagnostic definitions and remains strongest with DSM-IV. The greatest evidence for linkage is obtained with the P300 component of the Cz ERP electrode, independently of the alcoholism-diagnostic system. The peak signal location is estimated consistently at 100–101 cM and is effectively at anonymous marker D4S250 at 100.4 cM on the COGA map (5.2 cM qter of ADH3).
One additional significant LOD score and one nearly significant LOD score were also detected in the bivariate genome scan. These signals are at 212 cM on chromosome 6, with Cz/ICD-10 (LOD=3.72, LOD[1]=3.18), and at 214 cM on chromosome 2, with O2/DSM-IV (LOD=3.34, LOD[1]=2.98), and they correspond closely with the results reported by Begleiter et al. (1998). No significant LOD scores were observed in bivariate analyses with Pz.
The multipoint LOD scores on chromosome 4 for the univariate analyses of DSM-IV and Cz, as well as for the bivariate analysis of Cz/DSM-IV, are plotted on a common set of axes in figure 2. Also shown is the bivariate LOD score after adjustment to 1 df. The curves for DSM-IV and for Cz/DSM-IV each peak near marker D4S250 at 100.4 cM. The Zmax-1 support interval (Ott 1991) for the univariate DSM-IV LOD-score curve is 44.6 cM at 1.76. Signal definition is improved in the bivariate analysis, however, and the corresponding Zmax-1.39 support interval decreases to 36.6 cM at 3.36.
Figure 2.
Comparison of multipoint LOD scores on chromosome 4, in univariate and bivariate linkage analyses of DSM-IV alcoholism diagnosis and P300 component of Cz ERP.
Although the LOD-score curves for the univariate and bivariate analyses in figure 2 have similar profiles, the LOD-score curve for the bivariate analysis is not simply the summed contribution of the curves for the individual univariate analyses of DSM-IV and Cz. The increased linkage evidence obtained with the joint analysis derives in part from the underlying correlational structure of the discrete and continuous traits, even if one of the traits exhibits little or no evidence for linkage at the same location when it is studied by means of univariate methods.
Pleiotropy or Close Linkage?
When multiple traits are analyzed, the mechanisms of pleiotropy and of close linkage of independent major genes are of particular biological interest (Jiang and Zeng 1995; Almasy et al. 1997; Mangin et al. 1998) and can be tested easily by means of likelihood-ratio criteria for nested hypotheses (Williams et al. 1999 [in this issue]). In the bivariate analysis, the mechanisms of complete pleiotropy and coincident linkage are special cases of a general two-QTL genetic linkage model. To test for complete pleiotropy, in which the same gene influences variation in both alcoholism and ERP, the likelihood of a two-QTL linkage model in which the correlation between the QTLs is estimated is compared with the likelihood of a restricted model in which the correlation is constrained to unity. To test for coincident linkage of the traits to two independent but closely spaced genes that happen to be linked, the likelihood of the general linkage model is compared with that of a restricted model in which the correlation is constrained to be zero.
Likelihood-ratio tests for pleiotropy and coincident linkage, made at the location of the peak LOD score (101 cM) on chromosome 4, for Cz/DSM-IV, are summarized in table 3. For each test, the table gives the likelihood-ratio statistic λ, the asymptotic null distribution, and the corresponding P value. The test against complete pleiotropy is not significant, but the test against coincident linkage is moderately so (P<.01), suggesting that variation in alcoholism and variation in the P300 component of the Cz ERP are indeed mediated by common genetic factors.
Table 3.
Likelihood-Ratio Tests for Pleiotropy and Coincident Linkage at 101 cM on Chromosome 4, in Bivariate Linkage Analysis of DSM- IV Alcoholism Phenotype and P300 Component of Cz ERP
| Mechanism | Distribution | λ | P |
| Complete pleiotropy | 1/2χ21: 1/2χ20 | .00 | .5000 |
| Coincident linkage | χ21 | 7.44 | .0064 |
Discussion
The demonstration of ethnic variability in physiological responses to alcohol ingestion was one of the earliest indications that some observed populational differences were not solely culturally determined (Fenna et al. 1971; Wolff 1972, 1973; Ewing et al. 1974; Goedde et al. 1979; Shriver 1997). Numerous studies have since established that certain liver enzymes modulate the rates of oxidation of ethanol and its by-products. Allelic variation at the genetic loci coding for these enzymes is believed to underlie much of the observed two- to threefold interindividual variability in the rate of ethanol oxidation—and, thereby, to influence drinking behavior and the risk of development of alcoholism (Stamatoyannopoulos et al. 1975; Goedde et al. 1979; Teng et al. 1979; Bosron and Li 1986; Shibuya and Yoshida 1988b; Shibuya et al. 1989; Thomasson et al. 1991, 1995; Muramatsu et al. 1995; Higuchi et al. 1996; Edenberg and Bosron 1997; Wall et al. 1997; Osier et al. 1999). To better understand the relevance of the evidence for linkage to the ADH region, however, some background information on alcohol metabolism is useful.
Approximately 90% of ethanol oxidation occurs in the liver, in a two-stage metabolic process. Ethanol is initially oxidized to acetaldehyde by alcohol dehydrogenase (ADH), and acetaldehyde is subsequently oxidized to acetate by aldehyde dehydrogenase (ALDH). Multiple isozymes of both ADH and ALDH have been characterized, but the forms primarily involved in ethanol elimination are ALDH2 and ADH2 (Bosron and Li 1986; Smith 1986; Yoshida et al. 1991; Thomasson et al. 1995; Edenberg and Bosron 1997). The gene encoding ALDH2 is located on chromosome 12q24 (Hsu et al. 1986), and a structural mutation in the gene (a single-base-pair difference) encodes a mitochondrial isozyme of low catalytic activity (Impraim et al. 1982; Yoshida et al. 1983, 1984; Hsu et al. 1988). The low-activity allele ALDH2*2 is dominant over the normal ALDH2*1 allele, and individuals homozygous or heterozygous for ALDH2*2 are ALDH2 deficient and oxidize acetaldehyde at a reduced rate (Schwitters et al. 1982; Crabb et al. 1989).
In response to even small amounts of alcohol, ALDH2-deficient individuals typically experience an acute, unpleasant “flushing response” that can include facial flushing, sweating, palpitations, hyperventilation, tachycardia, nausea, dizziness, and faintness. This alcohol-induced flushing response is very similar to the aversive reaction to alcohol in alcoholic patients being treated with the ALDH inhibitor disulfiram (Wilkin 1981; Zeiner 1981). The atypical ALDH2*2 allele is extremely rare in European populations, but it can reach a frequency of ⩾15%–20% in Asian populations, and the prevalence of ALDH2 deficiency reaches 30%–50% in Japanese, Korean, and Chinese populations (Goedde et al. 1979, 1986, 1992; Teng 1981; Shibuya and Yoshida 1988a, 1988b; Thomasson et al. 1991; Shen et al. 1997). The flushing response is correspondingly common (frequency 50%–80%) in Asian populations but is much less frequent (5%–10%) in European populations (Wolff 1972, 1973).
Until recently, the low-activity form of mitochondrial aldehyde dehydrogenase (i.e., ALDH2) was the only clearly defined genetic factor known to affect drinking behavior and the risk of alcoholism, presumably by reducing the rate of hepatic acetaldehyde oxidation. However, several studies have since established that allelic variation at the alcohol dehydrogenase loci ADH2 and ADH3 also influences drinking behavior and risk of alcoholism, by modulating the rate of acetaldehyde production subsequent to intake of ethanol (Thomasson et al. 1991, 1995; Muramatsu et al. 1995; Chen et al. 1996; Higuchi et al. 1996; Shen et al. 1997).
The genes encoding the three class I ADH subunits ADH1, ADH2, and ADH3 are closely linked on chromosome 4q21–23 (Tsukahara and Yoshida 1989; Yasunami et al. 1990; Osier et al. 1999). Polymorphic alleles at the ADH2 and ADH3 loci encode isozyme variants having markedly different kinetic properties (Bosron and Li 1986; Yoshida et al. 1991; Thomasson et al. 1995; Edenberg and Bosron 1997). In particular, the β2β2 homodimeric isozyme encoded by ADH2*2 has a nearly 40-fold-higher activity (Vmax) than is seen in the β1β1 homodimer encoded by ADH2*1. In contrast, the isozyme encoded by ADH3*1 exhibits only 2.5 times the activity of the ADH3*2 isozyme (Bosron and Li 1986; Edenberg and Bosron 1997). Population studies of ADH have found that the ADH2*2 allele is rare (frequency <5%) in most European populations but that it can reach frequencies of 30%–70% in some Asian populations (Thomasson et al. 1991; Shen et al. 1997).
A statistical association of the ADH2*1 and ALDH2*1 alleles with alcoholism has been clearly demonstrated in several Asian populations (Harada et al. 1982, 1985; Shibuya and Yoshida 1988b; Thomasson et al. 1991; Takeshita et al. 1994; Muramatsu et al. 1995; Higuchi et al. 1996; Shen et al. 1997). In populations of Taiwanese and Japanese individuals, alcoholics were found to have significantly lower frequencies of the ADH2*2 and ADH3*1 alleles than were seen in nonalcoholics, and the difference was independent of the ALDH2 genotype (Thomasson et al. 1991). The frequency of the ALDH2*2 allele, as well as the frequencies of individuals homozygous and heterozygous for the allele, were also found to be significantly lower in alcoholics than in nonalcoholics (Harada et al. 1982, 1985; Shibuya and Yoshida 1988b; Thomasson et al. 1991; Muramatsu et al. 1995; Higuchi et al. 1996; Shen et al. 1997). Similar results have also been reported in a comparison of Japanese flushers and nonflushers (Shibuya et al. 1989).
Recent studies in different populations have found evidence for linkage of alcoholism diagnoses to the ADH region of chromosome 4 and, further, suggest that the linkage is contributed largely by unaffected individuals. Two-point and multipoint linkage analyses of alcoholism in sibships from the COGA study (Reich et al. 1996, 1998) found that allele sharing in the ADH region is increased among concordant unaffected pairs and is decreased among pairs discordant for diagnosis. In a study of 172 sib pairs from an American Indian population containing 18 concordant unaffected sib pairs, Long et al. (1998), in two-point linkage analysis, found only nominal evidence for linkage to the ADH region (.01<P<.05) and, in multipoint analysis, found no evidence for linkage to the ADH region. The study by Vallejo et al. (1998) of 135 nuclear families from Finnish pedigrees, using both Haseman-Elston and multipoint methods, also failed to find any evidence of linkage to the ADH region. Vallejo et al. (1998) did not report the number of concordant unaffected sib pairs in their study, but the nature of the probands (criminal offenders) suggests that concordant unaffected individuals might have been uncommon in their sample.
Because the class I ADH genes on chromosome 4 are such plausible candidates for genetic factors influencing susceptibility to alcoholism, it is interesting that the peak linkage signal at 100–101 cM in figures 1 and 2 is offset slightly from the ADH3 locus at 95.2 cM. This displacement may be a consequence of several more highly informative markers clustered with ADH3 in a 5.2-cM region on the COGA map. Located just q-terminal of ADH3 at 95.2 cM are D4S1628 (98.1 cM), D4S2457 (99.0 cM), and D4S250 (100.4 cM). The diallelic ADH3 locus is not very informative (heterozygosity .3496), but the neighboring markers are considerably more so, with, respectively, 6, 8, and 16 alleles and heterozygosities .5670, .7498, and .8131. The COGA marker map is also relatively dense in the region of ADH3, increasing the chances of map inaccuracies that could influence the linkage results.
The LOD-score curves for univariate DSM-IV (fig. 1) and bivariate Cz/DSM-IV (fig. 2) each exhibit two additional relative maxima. The larger of these forms a slight peak, in the bivariate analysis and in both univariate analyses, near marker D4S2393. The smaller, secondary peak is less well defined but is located near the GABRB1 receptor gene, which is also the location of the strongest linkage finding reported by Long et al. (1998) for chromosome 4. GABA (gamma-aminobutyric acid) is the major inhibitory neurotransmitter in the vertebrate CNS, and GABA receptors are known to be modulated by a number of pharmacological agents, including alcohol (Dean et al. 1991).
There is less evidence of linkage of alcoholism diagnoses or ERP traits to chromosomal regions containing loci encoding aldehyde dehydrogenase isozymes. The genes for ALDH1 and ALDH2 are located on human chromosomes 9 and 12, respectively (Hsu et al. 1986; Smith 1986; Yoshida et al. 1991). Univariate linkage analyses of chromosomes 9 and 12 for the COGA, DSM-IV, and ICD-10 diagnoses yielded no LOD scores >0.16. When these diagnoses were reexamined in bivariate analyses (unconditioned on the chromosome 4 signal) with P300 amplitude recorded at Cz, we found nonsignificant LOD scores reaching 1.16–1.78 at 8 cM on chromosome 9 (between D9S917 and GATA62F03) and 1.11–1.27 at 170 cM on chromosome 12 (near D12S1045). The genes ALDH1 and ALDH2 are not on the COGA marker map used for our analyses, but, using the LDB+ database (Collins et al. 1996), we determined that each of them is >50 cM from the LOD-score peaks on chromosomes 9 and 12, respectively, and therefore are unlikely to be linked to a QTL for either alcoholism or ERP. The COGA genetic maps for both chromosomes are sparse, however, and analysis with additional typed markers will be required before firm conclusions can be drawn as to the existence of other linked regions for alcoholism and ERPs. It will also be of interest to examine the evidence for linkage at these locations after conditioning on the signal on chromosome 4 in an oligogenic analysis (Blangero and Almasy 1997; Comuzzie et al. 1997).
In their sample from an American Indian population, Long et al. (1998), using a two-point analysis of 172 sib pairs and the DSM-III-R phenotype, reported strong evidence (P=.00007) for linkage to D11S1984 located at chromosome 11p15.5. Our analyses of chromosome 11 yielded nonsignificant LOD scores of 0.79 (COGA), 1.06 (DSM-IV), and 0.71 (ICD-10) in the region between D11S1999 (41.1 cM) and D11S902 (55.1 cM), ∼22.6–36.6 cM q-terminal of D11S1984 (18.5 cM). The gene for tryptophan hydroxylase, the rate-limiting enzyme in the biosynthesis of the neurotransmitter serotonin and a potential marker for serotonergic behaviors, is located in this region (Nielsen et al. 1992). The similarity of our results to the two-point results of Long et al. (1998) is suggestive and merits further investigation. At least two factors could account for the slight difference, in location of the maximum LOD score, between the two studies. First, the COGA genetic map for chromosome 11 used for these analyses is inflated (total length 255.3 cM), which could introduce location bias. Also, the p-terminal location of D11S1984 on chromosome 11 in the genetic map of Long et al. (1998), as well as the corresponding lack of flanking markers on one side, may have biased their estimate of maximum-LOD-score location.
Our analyses of the COGA stage IV extended pedigrees, with and without correction for ascertainment, did not detect the recently reported nominally significant (P<.01) linkage of the COGA phenotype to regions of chromosomes 1, 2, and 7 in multipoint sib-pair analyses of 987 individuals in 177 nuclear families derived from the COGA stage IV pedigrees (Reich et al. 1997, 1998). A multipoint linkage result (P<.01) suggesting linkage to a chromosome 8 region near D8S549 at 31.2 cM has also been reported (Reich et al. 1998). In uncorrected analyses, we found a small peak (LOD=0.95) in this region, but the signal was markedly reduced (LOD=0.32) in the ascertainment-corrected analysis, and the location of the signal under the COGA diagnosis did not closely correspond with the locations of the peaks under the DSM-IV and ICD-10 diagnoses.
The dopamine D2 receptor gene on chromosome 11q22–23 has also been implicated in alcoholism (Blum et al. 1990; Blum and Noble 1996), but we found no significant evidence of linkage (LOD=0.58–0.64) in the region of DRD2 at 179.8 cM on chromosome 11 (Edenberg et al. 1998; Phillips et al. 1998). Similarly, despite our evidence that the GABRB1 cluster on chromosome 4 is linked to the alcoholism phenotype, there were no indications of linkage to either of the clusters of GABA receptor genes on chromosomes 5 and 15 (Sinnett et al. 1993; Russek and Farb 1994). No markers in the GABRR1–2 region of chromosome 6 (Cutting et al. 1992) were available for linkage analysis, but this region should be scrutinized further, in view of the evidence for linkage of some ERP signals to this chromosome (Begleiter et al. 1998).
The failure to detect some of these previously reported linkage results is not without precedent (Long et al. 1998; Vallejo et al. 1998) and, in our case, may be related to a number of important differences between the two studies, in linkage methodology and in data structure (Wijsman and Amos 1997; Williams and Blangero 1999). For example, the present study uses complete extended-pedigree information, whereas Reich et al. (1998) analyzed only the nuclear families derived from the COGA extended pedigrees and Long et al. (1998) examined only sib pairs derived from extended pedigrees in an American Indian population. For highly prevalent disorders such as alcoholism, the unaffected individuals can contribute substantially to the overall pedigree likelihood, yet, whereas the threshold model for discrete traits that is described by Duggirala et al. (1997) and Williams et al. (1999 [in this issue]) makes full use of affected and unaffected individuals, affected-sib-pair methods use only affected individuals in making a test for linkage.
The particular choice of a weighting scheme to attempt to correct for the nonindependence of sib pairs in a sib pair–based analysis can also markedly influence linkage results (Van Eerdewegh et al. 1998). In our variance-component approach, however, the nonindependence of relative pairs in a pedigree is completely accounted for by maximizing the joint likelihood of each pedigree, conditional on all its members (Amos et al. 1996; Williams and Blangero 1999), and ad hoc model-dependent weights are unnecessary. We have, however, applied an approximate correction for the complex COGA ascertainment scheme, which may possibly have increased the rate of type II errors (Comuzzie and Williams, in press).
Most significantly, the methods used here and in the studies by Long et al. (1996, 1998) and Reich et al. (1996, 1998) differ radically in their approach to linkage analysis (Williams and Blangero 1999). The power of the sib pair–based method is a function of the sib-pair relative risk (Risch 1990), whereas the power of the variance-component method is a function of the QTL heritability (Almasy and Blangero 1998). Consequently, the power of each linkage method to detect genes is optimal for different QTL effect sizes and trait prevalences.
The statistical problems associated with multiple testing are not an issue in the present study. First, we have intended primarily to present a specific application illustrating the utility of the method described by Williams et al. (1999 [in this issue]). Thus, whereas none of the alcoholism diagnoses individually give significant evidence for linkage (table 1), the joint linkage analysis of DSM-IV and the biologically related endophenotype Cz does reach significance. Second, this is at least the third report of linkage of an alcoholism phenotype to the ADH region; if our results are regarded as a replication of previously reported linkages, then multiple testing is again not an issue, and nominal significance values apply. Finally, we argue that any correction for examination of multiple, highly correlated phenotypes will be extremely difficult to determine, because of their nonindependence—clearly, a correction based simply on the raw number of phenotypes would be overly conservative.
Finally, although strong evidence for linkage of the alcoholism phenotype to the ADH region of chromosome 4 is detected by each diagnostic system, it is interesting to note that neither the most narrowly biological definition of alcoholism (i.e., that of ICD-10) nor the rigorous and broadly psychosocial COGA diagnosis (which requires both the DSM-III-R criteria for alcohol dependence and the Feighner criteria for alcoholism at the “definite” level) generates the greatest evidence for linkage. Considering the difficulties inherent in the definition of phenotypes relating to alcohol-use disorders (Hill 1998), confirmed linkage on the basis of existing diagnostic measures, such as DSM-IV, to a particular chromosomal region could prove valuable in the evaluation and refinement of future diagnostic criteria for use in the study of alcoholism and related disorders.
Acknowledgments
The development of the statistical methods used in solar was supported by National Institutes of Health grants MH59490, DK44297, HL45522, HL28972, GM31575, and GM18897. Information on the analysis package solar is available at from The Southwest Foundation for Biomedical Research. The Collaborative Study on the Genetics of Alcoholism (COGA) (H. Begleiter, SUNY HSCB, Principal Investigator, T. Reich, Washington University, co–principal investigator) includes six different centers where data collection takes place. The six sites and Principal Investigator and Coinvestigators are Indiana University (J. Nurnberger, Jr., T.-K. Li, P. M. Conneally, and H. Edenberg); University of Iowa (R. Crowe and S. Kuperman); University of California at San Diego and Scripps Institute (M. Schuckit and F. E. Bloom); University of Connecticut (V. Hesselbrock); State University of New York, Health Science Center at Brooklyn (H. Begleiter and B. Porjesz); and Washington University, St. Louis (T. Reich, C. R. Cloninger, and J. Rice). This national collaborative study is supported by the National Institute on Alcohol Abuse and Alcoholism, through U.S. Public Health Service grants NIAAA U10AA08401, U10AA08402, and U10AA08403.
Electronic-Database Information
The URL for data in this article is as follows:
- Southwest Foundation for Biomedical Research, The, http://www.sfbr.org
References
- Alexander JE, Polich J, Bloom FE, Bauer LO, Kuperman S, Rohrbaugh J, Morzorati S, et al (1994) P300 from an auditory oddball task: inter-laboratory consistency. Int J Psychophysiol 17:35–46 [DOI] [PubMed]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J (1997) Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol 14:953–958 [DOI] [PubMed]
- Almasy L, Porjesz B, Blangero J, Chorlian DB, O'Connor SJ, Kuperman S, Rohrbaugh J, et al (1999) Heritability of event-related brain potentials in families with a history of alcoholism. Am J Med Genet 88:383–390 [PubMed]
- American Psychiatric Association (1980) Diagnostic and statistical manual of mental disorders, DSM-III, 3d ed. American Psychiatric Press, Washington, DC [Google Scholar]
- ——— (1987) Diagnostic and statistical manual of mental disorders, DSM-III-R, 3d ed, rev. American Psychiatric Press, Washington, DC [Google Scholar]
- ——— (1994) Diagnostic and statistical manual of mental disorders, DSM-IV, 4th ed. American Psychiatric Press, Washington, DC [Google Scholar]
- Amos CI, Zhu DK, Boerwinkle E (1996) Assessing genetic linkage and association with robust components of variance approaches. Ann Hum Genet 60:143–160 [DOI] [PubMed]
- Beaty TH, Liang KY (1987) Robust inference for variance components models in families ascertained through probands. I. Conditioning on the proband's phenotype. Genet Epidemiol 4:203–210 [DOI] [PubMed]
- Beaty TH, Liang KY, Seerey S, Cohen BH (1987) Robust inference for variance components models in families ascertained through probands. II. Analysis of spirometric measures. Genet Epidemiol 4:211–221 [DOI] [PubMed]
- Begleiter H, Porjesz B, Bihari B, Kissin B (1984) Event-related brain potentials in boys at risk for alcoholism. Science 225:1493–1496 [DOI] [PubMed]
- Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, Almasy L, et al (1998) Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroencephalogr Clin Neurophysiol 108:244–250 [DOI] [PubMed]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li T-K, Schuckit MA, Edenberg HJ, et al (1995) The collaborative study on the genetics of alcoholism. Alcohol Health Res World 19:228–236 [PMC free article] [PubMed] [Google Scholar]
- Blackwood DHR, St Clair DM, Muir WJ, Duffy JC (1991) Auditory P300 and eye tracking dysfunction in schizophrenic pedigrees. Arch Gen Psychiatry 48:899–909 [DOI] [PubMed]
- Blangero J, Almasy L (1997) Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol 14:959–964 [DOI] [PubMed]
- Blangero J, Almasy L, Williams JT, Porjesz B, Reich T, Begleiter H, COGA collaborators (1997) Incorporating quantitative traits in genomic scans of psychiatric diseases: alcoholism and event-related potentials. Am J Med Genet 74:602 [Google Scholar]
- Blum K, Noble EP (eds) (1996) Handbook of psychiatric genetics. CRC Press, Boca Raton, FL [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Nogami H, et al (1990) Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA 263:2055–2060 [PubMed]
- Boehnke M, Greenberg DA (1984) The effects of conditioning on probands to correct for multiple ascertainment. Am J Hum Genet 36:1298–1308 [PMC free article] [PubMed]
- Boehnke M, Moll PP, Lange K, Weidman WH, Kottke BA (1986) Univariate and bivariate analyses of cholesterol and triglyceride levels in pedigrees. Am J Med Genet 23:775–792 [DOI] [PubMed]
- Bosron WF, Li T-K (1986) Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases, and their relationship to alcohol metabolism and alcoholism. Hepatology 6:502–510 [DOI] [PubMed]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr, Reich T, et al (1994) A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol 55:149–158 [DOI] [PubMed]
- Chen WJ, Loh EW, Yun-Pung PH, Chiao-Chicy C, Yu J-M, Cheng ATA (1996) Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry 168:762–767 [DOI] [PubMed]
- Cohen HL, Wang W, Porjesz B, Bauer L, Kuperman S, O'Connor SJ, Rohrbaugh J, et al (1994) Visual P300: an interlaboratory consistency study. Alcohol 11:583–587 [DOI] [PubMed]
- Collins A, Frezal J, Teague J, Morton NE (1996) A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci USA 93:14771–14775 [DOI] [PMC free article] [PubMed]
- Comuzzie AG, Mahaney MC, Almasy L, Dyer TD, Blangero J (1997) Exploiting pleiotropy to map genes for oligogenic phenotypes using extended pedigree data. Genet Epidemiol 14:975–980 [DOI] [PubMed]
- Comuzzie AG, Williams JT. Correcting for ascertainment in the COGA data set. Genet Epidemiol (in press) [DOI] [PubMed] [Google Scholar]
- Crabb DW, Edenberg HJ, Bosron WF, Li T-K (1989) Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity: the inactive ALDH22 allele is dominant. J Clin Invest 83:314–316 [DOI] [PMC free article] [PubMed]
- Cutting GR, Curristin S, Zoghbi H, O'Hara B, Seldin MF, Uhl GR (1992) Identification of a putative γ-aminobutyric acid (GABA) receptor subunit rho2 cDNA and colocalization of the genes encoding rho2 (GABRR2) and rho1 (GABRR1) to human chromosome 6q14–q21 and mouse chromosome 4. Genomics 12:801–806 [DOI] [PubMed]
- Czerwinski SA, Mahaney MC, Williams JT, Almasy L, Blangero J. Genetic analysis of personality traits and alcoholism using a mixed discrete-continuous trait variance component model. Genet Epidemiol (in press) [DOI] [PubMed] [Google Scholar]
- Dean M, Lucas-Derse S, Bolos A, O'Brien SJ, Kirkness EF, Fraser CM, Goldman D (1991) Genetic mapping of the β1 GABA receptor gene to human chromosome 4, using a tetranucleotide repeat polymorphism. Am J Hum Genet 49:621–626 [PMC free article] [PubMed]
- Duggirala R, Williams JT, Williams-Blangero S, Blangero J (1997) A variance component approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol 14:987–992 [DOI] [PubMed]
- Edenberg HJ, Bosron WF (1997) Alcohol dehydrogenases. In: Sipes IG, Gandolfi AJ, McQueen C (eds) Comprehensive toxicology. Pergamon Press, New York, pp 119–131 [Google Scholar]
- Edenberg HJ, Foroud T, Koller DL, Goate A, Rice J, Van Eerdewegh P, Reich T, et al (1998) A family-based analysis of the association of the dopamine D2 receptor DRD2 with alcoholism. Alcohol Clin Exp Res 22:505–512 [PubMed]
- Ewing JA, Rouse BA, Pellizzari ED (1974) Alcohol sensitivity and ethnic background. Am J Psychiatry 131:206–210 [DOI] [PubMed]
- Feighner JP, Robins E, Guze SB, Woodruff RA Jr, Winokur G, Munoz R (1972) Diagnostic criteria for use in psychiatric research. Arch Gen Psychiatry 26:57–63 [DOI] [PubMed]
- Fenna D, Mix L, Schaefer O, Gilbert JAL (1971) Ethanol metabolism in various racial groups. Can Med Assoc J 105:472–475 [PMC free article] [PubMed]
- Freedman R, Coon H, Myles-Worsley M, Orr-Urtreger A, Olincy A, Davis A, Polymeropoulos M, et al (1997) Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus. Proc Natl Acad Sci USA 94:587–592 [DOI] [PMC free article] [PubMed]
- Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, et al (1992) Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet 88:344–346 [DOI] [PubMed]
- Goedde HW, Agarwal DP, Harada S, Rothhammer F, Whittaker JO, Lisker R (1986) Aldehyde dehydrogenase polymorphism in North American, South American and Mexican Indian populations. Am J Hum Genet 38:395–399 [PMC free article] [PubMed]
- Goedde HW, Harada S, Agarwal DP (1979) Racial differences in alcohol sensitivity: a new hypothesis. Hum Genet 51:331–334 [DOI] [PubMed]
- Harada S, Agarwal DP, Goedde HW (1985) Aldehyde dehydrogenase polymorphism and alcohol metabolism in alcoholics. Alcohol 2:391–392 [DOI] [PubMed]
- Harada S, Agarwal DP, Goedde HW, Tagaki S, Ishikawa B (1982) Possible protective role against alcoholism for aldehyde dehydrogenase isozyme deficiency in Japan. Lancet 2:827 [DOI] [PubMed]
- Higuchi S, Matsushita S, Muramatsu T, Murayama M, Hayashida M (1996) Alcohol and aldehyde dehydrogenase genotypes and drinking behavior in Japanese. Alcohol Clin Exp Res 20:493–497 [DOI] [PubMed]
- Hill SY (1998) Alternative strategies for uncovering genes contributing to alcoholism risk: unpredictable findings in a genetic wonderland. Alcohol 16:53–59 [DOI] [PubMed]
- Holzman PS, Proctor LR, Hughes DW (1973) Eye-tracking patterns in schizophrenia. Science 181:179–181 [DOI] [PubMed]
- Hopper JL, Mathews JD (1982) Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet 46:373–383 [DOI] [PubMed] [Google Scholar]
- Hsu LC, Bendel RE, Yoshida A (1988) Genomic structure of the human mitochondrial aldehyde dehydrogenase gene. Genomics 2:57–65 [DOI] [PubMed]
- Hsu LC, Yoshida A, Mohandas T (1986) Chromosomal assignment of the genes for human aldehyde dehydrogenase-1 and aldehyde dehydrogenase-2. Am J Hum Genet 38:641–648 [PMC free article] [PubMed]
- Impraim C, Wang G, Yoshida A (1982) Structural mutation in a major human aldehyde dehydrogenase gene results in loss of enzyme activity. Am J Hum Genet 34:837–841 [PMC free article] [PubMed]
- Jiang C, Zeng ZB (1995) Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140:1111–1127 [DOI] [PMC free article] [PubMed]
- Katsanis J, Iacono WG, McGue MK, Carlson SR (1997) P300 event-related potential heritability in monozygotic and dizygotic twins. Psychophysiology 34:47–58 [DOI] [PubMed]
- Lander ES (1988) Splitting schizophrenia. Nature 336:105–106 [DOI] [PubMed]
- Liang K-Y, Rathouz PJ, Beaty TH (1996) Determining linkage and mode of inheritance: mod scores and other methods. Genet Epidemiol 13:575–593 [DOI] [PubMed]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, et al (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed]
- Long JC, Urbanek M, Robin R, Schimpf S, Moore E, Goldman D (1996) Alcoholism genetics in a Native American tribe. Psychiatr Genet 6:162 [Google Scholar]
- Mangin B, Thoquet P, Grimsley N (1998) Pleiotropic QTL analysis. Biometrics 54:88–99 [Google Scholar]
- McGuffin P (1984) Biological markers and psychosis. Psychol Med 14:255–258 [DOI] [PubMed]
- Moldin SO (1994) Indicators of liability to schizophrenia: perspectives from genetic epidemiology. Schizophr Bull 20:169–184 [DOI] [PubMed]
- Muramatsu T, Zu-Cheng W, Yi-Ru F, Kou-Bao H, Heqin Y, Yamada K, Higuchi S, et al (1995) Alcohol and aldehyde dehydrogenase genotypes and drinking behavior of Chinese living in Shanghai. Hum Genet 96:151–154 [DOI] [PubMed]
- Nielsen DA, Dean M, Goldman D (1992) Genetic mapping of the human tryptophan hydroxylase gene on chromosome 11, using an intronic conformational polymorphism. Am J Hum Genet 51:1366–1371 [PMC free article] [PubMed]
- O'Connor S, Hesselbrock V, Tasman A, DePalma N (1987) P3 amplitudes in two distinct tasks are decreased in young men with a history of paternal alcoholism. Alcohol 4:323–330 [DOI] [PubMed]
- O'Connor S, Morzorati S, Christian JC, Li T-K (1994) Heritable features of the auditory oddball event-related potential: peaks, latencies, morphology and topography. Electroencephalogr Clin Neurophysiol 92:115–125 [DOI] [PubMed]
- Osier M, Pakstis AJ, Kidd JR, Lee J-F, Yin S-J, Ko H-C, Edenberg HJ, et al (1999) Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 64:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1991) Analysis of human genetic linkage, rev ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- ——— (1995) Linkage analysis with biological markers. Hum Hered 45:169–174 [DOI] [PubMed]
- Pfefferbaum A, Ford JM, White PM, Mathalon D (1991) Event-related potentials in alcoholic men: P3 amplitude reflects family history but not alcohol consumption. Alcohol Clin Exp Res 15:839–850 [DOI] [PubMed]
- Phillips TJ, Brown KJ, Burkhart-Kash S, Wenger CD, Kelly MA, Rubinstein M, Grandy DK, et al (1998) Alcohol preference and sensitivity are markedly reduced in mice lacking dopamine D2 receptors. Nat Neurosci 1:610–615 [DOI] [PubMed]
- Polich J, Burns T (1987) P300 from identical twins. Neuropsychologia 25:299–304 [DOI] [PubMed]
- Polich J, Pollock VE, Bloom FE (1994) Meta-analysis of P300 amplitude from males at risk for alcoholism. Psychol Bull 115:55–73 [DOI] [PubMed]
- Porjesz B, Begleiter H (1985) Human brain electrophysiology and alcoholism. In: Tarter RE, Van Thiel DH (eds) Alcohol and the brain: chronic effects. Plenum Press, New York, pp 139–182 [Google Scholar]
- ——— (1996) Effects of alcohol on electrophysiological activity of the brain. In: Begleiter H, Kissin B (eds) Alcohol and alcoholism. Vol 2: The pharmacology of alcohol and alcohol dependence. Oxford University Press, New York, pp 207–247 [Google Scholar]
- Porjesz B, Begleiter H, Reich T, Van Eerdewegh P, Edenberg HJ, Foroud T, Goate A, et al (1998) Amplitude of visual P3 event-related potential as a phenotypic marker for a predisposition to alcoholism: preliminary results from the COGA project. Alcohol Clin Exp Res 22:1317–1323 [PubMed]
- Pritchard WS (1981) Psychophysiology of P300. Psychol Bull 89:506–540 [PubMed]
- Regan D (1989) Human brain electrophysiology: evoked potentials and evoked magnetic fields in science and medicine. Appleton & Lange, New York [Google Scholar]
- Reich T, Edenberg HJ, Begleiter H, Cloninger CR (1996) A genomic survey of alcohol dependence and related phenotypes: results from the Collaborative Study on the Genetics of Alcoholism (COGA). Alcohol Clin Exp Res Suppl 20:133A–137A [DOI] [PubMed]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, et al (1998) Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81:207–215 [PubMed]
- Reich T, Williams JT, Goate A, Rice JP, Bierut LJ, Edenberg H, Foroud T, et al (1997) A genome-wide search for susceptibility genes for alcohol dependence in COGA: ascertainment, analysis, and initial survey. Am J Med Genet 74:573 [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed]
- Rogers TD, Deary I (1991) The P300 component of the auditory event-related potential in monozygotic and dizygotic twins. Acta Psychiatr Scand 83:412–416 [DOI] [PubMed]
- Russek SJ, Farb DH (1994) Mapping of the β2 subunit gene (GABRB2) to microdissected human chromosome 5q34–q35 defines a gene cluster for the most abundant GABAA receptor isoform. Genomics 23:528–533 [DOI] [PubMed]
- Schuckit MA, Hesselbrock V, Tipp J, Anthenelli R, Bucholz K, Radziminski S (1994) A comparison of DSM-III-R, DSM-IV and ICD-10 substance use disorders diagnoses in 1922 men and women subjects in the COGA study. Addiction 89:1629–1638 [DOI] [PubMed]
- Schwitters SY, Johnson RC, Johnson SB, Ahern FM (1982) Familial resemblances in flushing following alcohol use. Behav Genet 12:349–352 [DOI] [PubMed]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Shen Y-C, Fan J-H, Edenberg HJ, Li T-K, Cui Y-H, Wang Y-F, Tian C-H, et al (1997) Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–1277 [PubMed]
- Shibuya A, Yasunami M, Yoshida A (1989) Genotypes of alcohol dehydrogenase and aldehyde dehydrogenase loci in Japanese alcohol flushers and nonflushers. Hum Genet 82:14–16 [DOI] [PubMed]
- Shibuya A, Yoshida A (1988a) Frequency of the atypical aldehyde dehydrogenase-2 gene (ALDH22) in Japanese and Caucasians. Am J Hum Genet 43:741–743 [PMC free article] [PubMed]
- ——— (1988b) Genotypes of alcohol-metabolizing enzymes in Japanese with alcohol liver diseases: a strong association of the usual Caucasian-type aldehyde dehydrogenase gene (ALDH21) with the disease. Am J Hum Genet 43:744–748 [PMC free article] [PubMed]
- Shriver MD (1997) Ethnic variation as a key to the biology of human disease. Ann Intern Med 127:401–403 [DOI] [PubMed]
- Sinnett D, Wagstaff J, Glatt K, Woolf E, Kirkness EJ, Lalande M (1993) High-resolution mapping of the γ-aminobutyric acid receptor subunit β3 and α5 gene cluster on chromosome 15q11–q13, and localization of breakpoints in two Angelman syndrome patients. Am J Hum Genet 52:1216–1229 [PMC free article] [PubMed]
- Smith M (1986) Genetics of human alcohol and aldehyde dehydrogenases. Adv Hum Genet 15:249–290 [DOI] [PubMed]
- Stamatoyannopoulos G, Chen S-H, Fukui M (1975) Liver alcohol dehydrogenase in Japanese: high population frequency of atypical form and its possible role in alcohol sensitivity. Am J Hum Genet 27:789–796 [PMC free article] [PubMed]
- Takeshita T, Morimoto K, Mao XQ, Hashimoto T, Furuyama J-I (1994) Characterization of the three genotypes of low Km aldehyde dehydrogenase in a Japanese population. Hum Genet 94:217–223 [DOI] [PubMed]
- Teng Y-S (1981) Human liver aldehyde dehydrogenase in Chinese and Asiatic Indians: gene deletion and its possible implications in alcohol metabolism. Biochem Genet 19:107–114 [DOI] [PubMed]
- Teng Y-S, Jehan S, Lie-Injo LE (1979) Human alcohol dehydrogenase ADH2 and ADH3 polymorphisms in ethnic Chinese and Indians of West Malaysia. Hum Genet 53:87–90 [DOI] [PubMed]
- Thomasson HR, Beard JD, Li T-K (1995) ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin Exp Res 19:1494–1499 [DOI] [PubMed]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai X-L, Jerome RE, Li T-K, Wang S-P, et al (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48:677–681 [PMC free article] [PubMed]
- Thompson EA (1993) Sampling and ascertainment in genetic epidemiology: a tutorial review. Tech rep 243, Department of Statistics, University of Washington, Seattle [Google Scholar]
- Tsukahara M, Yoshida A (1989) Chromosomal assignment of the alcohol dehydrogenase cluster locus to human chromosome 4q21–23 by in situ hybridization. Genomics 4:218–220 [DOI] [PubMed]
- Vallejo RL, Wilhelm LA, Hoopes D, High T, Virkkunen M, Linnoila M, Goldman D, et al (1998) Genome-wide search for genes affecting vulnerability to alcohol dependence in Finnish pedigrees. Am J Hum Genet Suppl 63:A45 [Google Scholar]
- van Beijsterveldt CEM, Molenaar PCM, de Geus EJC, Boomsma DI (1996) Heritability of human brain functioning as assessed by electroencephalography. Am J Hum Genet 58:562–573 [PMC free article] [PubMed]
- Van Eerdewegh P, Dupuis J, Santangelo S, Hayward LB, Blacker D (1998) The importance of watching our weights: how the choice of weights for non-independent sibpairs can dramatically alter results. Paper presented at Genetic Analysis Workshop 11, Arcachon, France, September 8–10 [DOI] [PubMed] [Google Scholar]
- Wall TL, Peterson CM, Peterson KP, Johnson ML, Thomasson HR, Cole M, Ehlers CL (1997) Alcohol metabolism in Asian-American men with genetic polymorphisms of aldehyde dehydrogenase. Ann Intern Med 127:376–379 [DOI] [PubMed]
- Wijsman EM, Amos CI (1997) Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet Epidemiol 14:719–735 [DOI] [PubMed]
- Wilkin JK (1981) Flushing reactions: consequences and mechanisms. Ann Intern Med 95:468–476 [DOI] [PubMed]
- Williams JT, Blangero J (1999) Comparison of variance components and sibpair-based approaches to quantitative trait linkage analysis in unselected samples. Genet Epidemiol 16:113–134 [DOI] [PubMed]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J (1999) Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet 65:1134–1147 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff PH (1972) Ethnic differences in alcohol sensitivity. Science 175:449–450 [DOI] [PubMed]
- ——— (1973) Vasomotor sensitivity to alcohol in diverse Mongoloid populations. Am J Hum Genet 25:193–199 [PMC free article] [PubMed]
- World Health Organization (1992) ICD-10: the ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. World Health Organization, Geneva [Google Scholar]
- Yasunami M, Kikuchi I, Sarapata D, Yoshida A (1990) The human class I alcohol dehydrogenase gene cluster: three genes are tandemly organized in an 80-kb-long segment of the genome. Genomics 7:152–158 [DOI] [PubMed]
- Yoshida A, Hsu LC, Yasunami M (1991) Genetics of human alcohol-metabolizing enzymes. Prog Nucleic Acid Res Mol Biol 40:255–287 [DOI] [PubMed]
- Yoshida A, Huang I-Y, Ikawa M (1984) Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA 81:258–261 [DOI] [PMC free article] [PubMed]
- Yoshida A, Wang G, Davé V (1983) Determination of genotypes of human aldehyde dehydrogenase ALDH2 locus. Am J Hum Genet 35:1107–1116 [PMC free article] [PubMed]
- Xu S, Atchley WR (1996) Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics 143:1417–1424 [DOI] [PMC free article] [PubMed]
- Zeiner AR (1981) Are differences in the disulfiram-alcohol reaction the basis of racial differences in biological sensitivity to ethanol? In: Schecter AJ (ed) Drug dependence and alcoholism. Vol 1: Biomedical issues. Plenum Press, New York, pp 573–581 [Google Scholar]