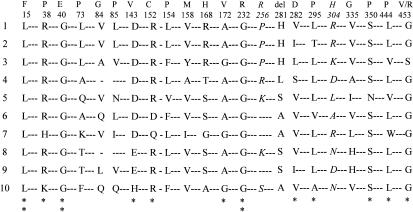Summary
Variegate porphyria (VP) is a low-penetrance, autosomal dominant disorder characterized clinically by skin lesions and acute neurovisceral attacks that occur separately or together. It results from partial deficiency of protoporphyrinogen oxidase encoded by the PPOX gene. VP is relatively common in South Africa, where most patients have inherited the same mutation in the PPOX gene from a common ancestor, but few families from elsewhere have been studied. Here we describe the molecular basis and clinical features of 108 unrelated patients from France and the United Kingdom. Mutations in the PPOX gene were identified by a combination of screening (denaturing gradient gel electrophoresis, heteroduplex analysis, or denaturing high-performance liquid chromatography) and direct automated sequencing of amplified genomic DNA. A total of 60 novel and 6 previously reported mutations (25 missense, 24 frameshift, 10 splice site, and 7 nonsense) were identified in 104 (96%) of these unrelated patients, together with 3 previously unrecognized single-nucleotide polymorphisms. VP is less heterogeneous than other acute porphyrias; 5 mutations were present in 28 (26%) of the families, whereas 47 mutations were restricted to 1 family; only 2 mutations were found in both countries. The pattern of clinical presentation was identical to that reported from South Africa and was not influenced by type of mutation. Our results define the molecular genetics of VP in western Europe, demonstrate its allelic heterogeneity outside South Africa, and show that genotype is not a significant determinant of mode of presentation.
Introduction
Variegate porphyria (VP [MIM 176200]) is a disorder of heme biosynthesis and results from partial deficiency of protoporphyrinogen oxidase (PPOX [E.C.1.3.3.4]) (Brenner and Bloomer 1980; Kirsch et al. 1998). As implied by its name, it presents in different ways—either with skin fragility and bullae, with acute neurovisceral crises, or with both together (Kirsch et al. 1998). Symptoms are very rare before puberty. The disorder is inherited in an autosomal dominant pattern with low clinical penetrance. Approximately three-quarters of those affected remain asymptomatic but are at risk for development of acute neurovisceral crises if they are exposed to certain drugs, alcohol, or other provocative factors (Jenkins 1996; Elder et al. 1997; Kirsch et al. 1998). Detection of asymptomatic affected individuals—so that they can be advised to avoid provoking agents—is an important part of the management of families with VP. Biochemical methods for their detection are technically complex or insensitive (Long et al. 1993; Da Silva et al. 1995).
The 5-kb PPOX gene on chromosome 1q 22-23 contains 1 noncoding and 12 coding exons (Nishimura et al. 1995; Roberts et al. 1995; Taketani et al. 1995; Puy et al. 1996). Mutations in this gene have been shown to cause VP (Deybach et al. 1996; Lam et al. 1996, 1997; Meissner et al. 1996; Warnich et al. 1996; de Rooij et al. 1997; Frank et al. 1997a, 1997b, 1998a, 1998b, 1998c, 1998d; Kauppinen et al. 1997; Corrigall et al. 1998; also see Human Gene Mutation Database) and the rare related condition, homozygous VP (Meissner et al. 1996; Frank et al. 1998e; Roberts et al. 1998). VP is common in South Africa, where an estimated 20,000 descendants of a Dutch couple who married at the Cape in 1688 have inherited the same mutation (R59W) in the PPOX gene (Dean 1971; Meissner et al. 1996; Warnich et al. 1996; Groenewald et al. 1998). Outside South Africa, the disease is less common but is increasingly becoming recognized, as confusion with porphyria cutanea tarda or other acute porphyrias is resolved by application of more-precise diagnostic methods (Poh-Fitzpatrick 1980; Long et al. 1993). Apart from one series of 57 patients from Finland (Mustajoki 1980), of whom only 18 had symptoms, only small numbers of cases have been reported outside South Africa (Armas et al. 1992; Kirsch et al. 1998). PPOX mutations that cause VP have recently been identified in a few families (Deybach et al. 1996; Lam et al. 1996, 1997; de Rooij et al. 1997; Frank et al. 1997a, 1997b, 1998a, 1998b, 1998c, 1998d; Kauppinen et al. 1997) from Europe and the United States, but neither the extent of allelic heterogeneity, the distribution of mutations within the PPOX gene, nor genotype-phenotype correlations have yet been studied in a large group of patients. Here we report an investigation of the PPOX gene in 108 apparently unrelated patients with VP who are from France and the United Kingdom.
Patients and Methods
Patients
We studied 108 apparently unrelated patients with VP who presented sequentially for mutational analysis. Of these, 103 patients had current or past symptoms of VP; 5 patients had never had symptoms but were referred because of a family history of either acute or cutaneous porphyria and were found, by biochemical investigation, to have VP. In all 108 patients, the diagnosis was established by fecal porphyrin measurement, PPOX assay, or fluorescence-emission spectroscopy of plasma (Long et al. 1993; Da Silva et al. 1995). The mode of presentation (skin lesions alone, acute attack alone, or both together) was recorded on all symptomatic patients. Unrelatedness was determined by family inquiries; none of the patients were known to be related. Of the 108 patients, 51 (34 female and 17 male) came from France and 57 (42 female, 15 male) came from the United Kingdom. Patients resided throughout the United Kingdom and France; all were white.
Identification of Mutations
DNA was extracted from peripheral blood leukocytes or Epstein-Barr virus–transformed lymphoblasts. Exons 1–13 of the PPOX gene, along with 45–160 bp of their flanking regions, were amplified by means of the primers and conditions listed in table 1 and were screened for mutations, by either heteroduplex analysis (HDA) (exons 1–7, 12, and 13) or denaturing gradient gel electrophoresis (DGGE) (exons 8–11). Preliminary computer analyses of genomic PPOX sequence were performed with the MELT 87 and SQHTX programs, written and kindly provided by Lerman and Silverstein (1987). They predicted optimal experimental conditions for DGGE only in exons 8–11. For these exons, DGGE analyses were performed according to the method of Myers et al. (1987). HDA was used as an alternative mutation-screening method for those exons not analyzable by DGGE. The gels used for HDA were prepared with a polyacrylamide-derived commercial matrix, MDE™ (mutation-detection enhancement; FMC Bioproducts). Regions showing abnormal patterns were sequenced to identify mutations and polymorphisms.
Table 1.
Sequence of Oligonucleotide Primers and Conditions Used in PCR and DGGE or HDA Analysis of All PPOX Exons
| Conditions |
||||||
| PCR |
||||||
| Exon andPrimer | 5′-Sequencea | Size(bp) | MgCl2(mM) | Temperature(°C) | DGGEb | HDAb |
| 1: | ||||||
| PV 617 S | CCAAGTCCCGCCAATCCAGAT | 360 | 1.5 | 61 | … | 500 V for 10 h |
| PV 956 AS | ACGGATGGGCTGGTGGAACAG | |||||
| 2: | ||||||
| PV-MDE-2S | CTGGACGGGGACCTCGCTGTT | 355 | 1.0 | 60 | … | 500 V for 10 h |
| PV-MDE-2AS | GTAGGGAGGGGTAAGAGGCATATT | |||||
| 3: | ||||||
| PV-MDE-3S | CCCTCTGAATATGCCTCTTAC | 305 | 1.0 | 56 | … | 500 V for 10 h |
| PV-MDE-3AS | AACATACTTCCTCCCCTAAAC | |||||
| 4: | ||||||
| PV 1544-S | GGGAATAGAGTTTAGGGGAGGA | 272 | 1.5 | 56 | … | 500 V for 10 h |
| PV 1785-AS | GAAGAAAGAAATGGAAGGCATA | |||||
| 5: | ||||||
| PV-MDE-5S | TGGAGCTGGGGAGGTATGTC | 234 | 1.0 | 56 | … | 500 V for 10 h |
| PV-MDE-5AS | CTGGTTTAGGAGGGAAGAGG | |||||
| 6: | ||||||
| PV-MDE-6S | CCCACCCTCATTCCCTACCA | 307 | 1.0 | 56 | … | 500 V for 10 h |
| PV-MDE-6AS | AGCACCCCTTGTCCCCACTC | |||||
| 7: | ||||||
| PV 2196-S | CCTAAAGTGCTGGGATTACA | 316 | 1.5 | 56 | … | 500 V for 10 h |
| PV 2485-AS | TGGATAAAGATGGAAACACTGA | |||||
| 8: | ||||||
| PV 3774-S-Cl | (GC)50CAATCTCTTCATCCTGGGTCAG | 348 | 1.5 | 56 | 40%–90%, 150 V for 6 h | … |
| PV 4051-AS | AGTTCAGGGGCATGGTGTGGT | |||||
| 9: | ||||||
| PV 4051-S-Cl | (GC)50ACCACACCATGCCCCTGAAC | 370 | 1.0 | 59 | 40%–90%, 150 V for 6 h | … |
| PV 4370-AS | GCCAGGCTGGTCTCGAACTC | |||||
| 10: | ||||||
| PV 4538-S-Cl | (GC)50ATTACACTCCAGCCTGGGTGA | 383 | 1.5 | 56 | 40%–90%, 150 V for 6 h | … |
| PV 4849-AS | AAGGAGGGAATATAGCACTGAT | |||||
| 11: | ||||||
| PV 4762-S | TCAGAGTGACTGTGAGGAGGAG | 433 | 1.5 | 56 | 40%–90%, 150 V for 6 h | … |
| PV 5121-AS-Cl | (GC)50AGGATGCAGGGAAAAGTTTATTAT | |||||
| 12 and 13: | ||||||
| PV 5074-S | AGACTGGAACATTTGTCACTGT | 401 | 1.5 | 54 | … | 500 V for 10 h |
| PV 3851-AS | AAGCCAAGCCAAGCAATTTT | |||||
| 1–4: | ||||||
| V3169-S | GAGGTTATGTACTGGGAGGA | 1565c | 1.5 | 64 | ||
| A6166-AS | CACGTAGAGGAACCTGTTCTGGGCAGC | |||||
| 4–7: | ||||||
| A6167-S | TTGTGGGATGTCTAGGAGAGGTTG | 2014 | 1.5 | 64 | ||
| A6168-AS | GTAGCCCATGTCTAAGTAGCTTCTAAG | |||||
| 7–9: | ||||||
| P2052-S | AGTGGTCACTTCGTGGAG | 1071 | 1.5 | 64 | ||
| V8508-AS | CGAACTCCTGACCTTGTTATC | |||||
| 10–13: | ||||||
| T2713-S | GAGAGACAGCCTCAGCTAG | 851 | 1.5 | 61 | ||
| W7097-AS | TATTTTCATGAATGAGAGTTGGGGATC | |||||
The sequence of GC clamp is as follows: (GC)50 = CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGCCCCCGCCCG.
Percentages are of denaturant (100% = 7 M urea and 40% [v/v] formamide), and time is for electrophoresis at 60°C.
Includes 520 bases of 5′ UTR.
When no abnormality was detected by DGGE or HDA, exons 4, 7, 8, and 10–13, along with 55–256 bp of their flanking regions, were screened by denaturing high-performance liquid chromatography (RP-HPLC), as described elsewhere (O'Donovan et al. 1998), and 520 bp 5′ to exon 1, exons 1–3, 5, 6, and 9 and their flanking regions were sequenced. For those patients in whom no abnormality was found by HPLC, exons 4, 7, 8, and 10–13 and their flanking regions were sequenced.
For DNA sequencing, the PPOX gene was amplified in four fragments—exons 1–4 with 520 bp 5′ to exon 1 (1,565 bp), exons 3–7 (2,014 bp), exons 7–9 (1,071 bp), and exons 10–13 (851 bp)—by means of the primers and conditions listed in table 1. PCR-amplified double-stranded DNA was purified from agarose gels by means of a Qiaquick gel-extraction kit (Qiagen) and was cycle sequenced by means of fluorescent ddNTPs (AmplitaqFS, Rhodamine, or BigDye) and an ABI 377 automated sequencer (PE Biosystems). Sequencing primers were chosen to include 58–250 bp of flanking regions. Sequences were analyzed by visual inspection. The presence of mutations was confirmed either by sequencing of both DNA strands or by digestion, with the appropriate restriction enzyme, of amplified DNA.
RNA Analysis
For investigation of splicing of exon 8, RNA was obtained by extraction from phytohemagglutinin (PHA)-transformed lymphocytes and was reverse transcribed as described elsewhere (Roberts et al. 1998). cDNA for PPOX was amplified by PCR with a sense primer in exon 6 (5′-AGGTCCTGCTTTCCCAGTCTCTTC) and an antisense primer in exon 13 (5′-TATTTTCATGAATGAGAGTTGGGGATC). The product was purified from a 2% agarose gel by means of a Qiaquick gel-extraction kit, and both strands were sequenced as described above.
Polymorphisms
The frequency of known and suspected single-nucleotide polymorphisms (SNPs) in the PPOX gene were determined by study of either 105 unrelated subjects (65 French and 40 U.K.) or 78 nonporphyric white families from the Centre d'Etude du Polymorphisme Humain (Paris), by means of either direct sequencing, restriction-enzyme digestion, DGGE, or HDA (table 2). Homozygosity or heterozygosity at intragenic polymorphic sites was determined for all patients with mutations that were present in four or more families; partial haplotypes were deduced by comparison of the profiles of polymorphic variation for individuals who shared a mutation.
Table 2.
SNPs in the Human PPOX Gene
| Exon/Intronand Positiona | Screening Method | Allele(Frequency) |
| Exon 1: | ||
| −253 | Direct sequencing | G (1.00) |
| C (0.0) | ||
| −247 | Direct sequencing | A (.46) |
| C (.54) | ||
| −246 | Direct sequencing | G (1.00) |
| T (.00) | ||
| −151 | Direct sequencing | G (.93) |
| T (.07) | ||
| −118 | Direct sequencing | C (.84) |
| G (.16) | ||
| Intron 2: | ||
| IVS2−47 | HDA | G (.61) |
| C (.39) | ||
| Intron 4: | ||
| IVS4−327 | HinfI digestion | G (.94) |
| A (.06) | ||
| Intron 6: | ||
| IVS6−150 | SfcI digestion | T (.96) |
| C (.04) | ||
| IVS6−237 | Direct sequencing | A (.73) |
| C (.27) | ||
| Exon 7: | ||
| 767 (P256R)b | HDA | C (.95) |
| G (.05) | ||
| Intron 8: | ||
| IVS8+57b | DGGE | A (.96) |
| C (.04) | ||
| Exon 9: | ||
| 911 (R304H) | DGGE | G (.61) |
| A (.39) | ||
| Intron 10: | ||
| IVS10−22b | DGGE | G (.97) |
| C (.03) |
Nucleotides are numbered according to the PPOX cDNA sequence (see “Numbering System” subsection in text).
Previously unrecognized SNPs.
Numbering System
Nucleotides are numbered according to the cDNA sequence derived from the PPOX genomic sequence (Puy et al. 1996) (HGMP GenBank accession number X99450), in which the A of the ATG initiation codon is numbered “+1.”
Results
Identification of Mutations
Initially, exons 1–13 and their flanking regions were amplified from genomic DNA from all 108 unrelated patients with VP. Amplified fragments were screened by either HDA or DGGE, and any showing abnormal patterns were sequenced. This approach identified 54 mutations in 82 patients and 3 SNPs in addition to the 10 SNPs reported elsewhere (Puy et al. 1996; Warnich et al. 1996; Lam et al. 1997; Kotze et al. 1998) (table 2). A total of 13 mutations were identified in 22 of the other 26 patients, by either direct sequencing (9 mutations) or HPLC (4 mutations). Sequencing of all exons, their flanking regions, and 520 bp upstream from exon 1 failed to detect any base change in the four remaining patients. Each of these four patients was heterozygous for one intragenic SNP (−247A/C, −118C/G, IVS2−47C/G, or 911A/G).
Overall, analysis of exons 1–13 by HDA or DGGE identified 54 (77%) of 70 mutations, if it is assumed that the four patients with undetected mutations all had different mutations. All 11 SNPs found in the population that we studied gave abnormal patterns on HDA or DGGE, facilitating screening of sufficient subjects to establish allele frequencies (table 2). Direct automated DNA sequencing detected 66 mutations in 104 (96%) of our 108 patients, giving this method a sensitivity of 94% (66 of 70 mutations detected) (95% confidence interval 90%–99%).
Distribution and Type of Mutation
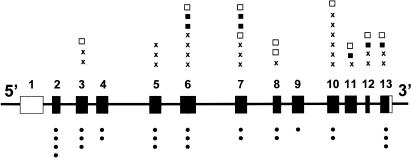
Mutations were distributed throughout the coding region of the PPOX gene, with no exon having >10 or <2 (fig. 1 andtable 3); 24 (36%) of the mutations were small insertions or deletions that introduced a frameshift leading to a stop codon; 25 (38%) were missense, including one codon deletion (841–843delCAC [H281del]) and three that altered the initiation codon; 9 (14%) changed invariant nucleotides at splice sites; and 7 (11%) produced stop codons (fig. 1 andtable 3). In one patient, the only base change identified was adjacent to the splice-acceptor site of exon 8 (IVS7−9T→G). Reverse transcription–PCR of cDNA for PPOX (exons 6–13) from this patient gave a product (∼900 bp) of the size (918 bp) predicted for the normal allele. Sequencing of this product showed a mixture of similar quantities of two cDNA species—one from the normal allele and one with addition of 8 bp to the 5′ end of exon 8—indicating that this mutation creates an additional splice-acceptor site (GTTTTCAG[GCTCTCAG––––––) that is used in preference to the normal site.
Figure 1.
Distribution of mutations in the human PPOX gene. Missense mutations (•) are shown below the gene diagram, with chain termination and splice mutations (frameshift [×], nonsense [▪], and splice defects [□]) above it. Unshaded areas of exons denote noncoding regions.
Table 3.
Mutations in the PPOX Gene in VP
| Exon/Intronand Mutation | Effect | Screen | Restriction-Enzyme Site | Country | No. of Families |
| Exon 2: | |||||
| 1A→G | M1V | HDA | NlaIII | France | 1 |
| 1A→C | M1L | HDA | NlaIII | France | 1 |
| 3G→C | M1I | HDA | NlaIII | France | 1 |
| 45G→C | L15F | HDA | EaeI | United Kingdom | 7 |
| Exon 3: | |||||
| 113G→C | R38P | … | BslI | France | 1 |
| 119G→A | G40E | HDA | MnlI | France | 1 |
| 157–160delATCT | Stop+13 | HDA | United Kingdom | 1 | |
| 199–200insT | Stop+10 | HDA | BglI | France | 1 |
| 218T→C | L73P | … | BsaJI | United Kingdom | 1 |
| Intron 3: | |||||
| IVS3−1G→C | Deletion of exon 4 | … | MaeIII | France | 1 |
| Exon 4: | |||||
| 251T→G | V84G | HDA | CviJI | United Kingdom | 1 |
| 254T→C | L85P | HDA | France | 2 | |
| Exon 5: | |||||
| 363–364insC | Stop+21 | HDA | United Kingdom | 2 | |
| 376–377delCT | Stop+16 | HDA | MnlI | United Kingdom | 1 |
| 428A→T | D143V | HDA | France | 1 | |
| 454C→Ta | R152C | … | HhaI | France | 1 |
| 460del23 | Stop+15 | HDA | United Kingdom | 1 | |
| 461T→C | L154P | HDA | EcoRI | France | 1 |
| Exon 6: | |||||
| 472G→A | V158M | HDA | France | 1 | |
| 503G→Ab | R168H | HDA | NcoI | France and United Kingdom | 2 |
| 515C→T | A172V | … | France | 1 | |
| 528–529insT | Stop+1 | HDA | France | 1 | |
| 538–539delATc | Stop+10 | HDA | MvaI | United Kingdom | 2 |
| 542–556del15 | Stop+291 | HDA | France | 1 | |
| 565delC | Stop+44 | HDA | United Kingdom | 1 | |
| 565C→T | E189X | HDA | United Kingdom | 4 | |
| 593T→G | L198X | HDA | France | 1 | |
| Intron 6: | |||||
| IVS6+1G→T | Deletion of exon 6 | HDA | France | 1 | |
| IVS6−1G→T | Deletion of exon 7 | HDA | PstI | France | 1 |
| Exon 7: | |||||
| 672G→A | W224X | HDA and DGGE | NlaIV | France | 2 |
| 694G→Cd | G232R | DGGE | MnlI | France | 2 |
| 695G→C | G232R | HDA and DGGE | MnlI | France | 1 |
| 745–746insCd | Stop+31 | HDA and DGGE | DdeI | France | 1 |
| 745delG | Stop+23 | HDA and DGGE | France and United Kingdom | 2 | |
| 745–746insG | Stop+31 | HDA and DGGE | TaqI | France | 1 |
| 803G→A | W268X | HDA and DGGE | MaeI | United Kingdom | 2 |
| Intron 7: | |||||
| IVS7+2T→C | Deletion of exon 7 | HPLC | United Kingdom | 1 | |
| IVS7−9T→G | 8 bp into cDNA | DGGE | NlaIII | United Kingdom | 1 |
| Exon 8: | |||||
| 841–843delCAC | H281del | HDA and DGGE | France | 3 | |
| 845T→A | V282D | HPLC | MaeIII | United Kingdom | 2 |
| 856delA | Stop+20 | HDA and DGGE | France | 1 | |
| 868G→C | V290L (deletion of exon 8) | DGGE | MaeIII | United Kingdom | 1 |
| Exon 9: | |||||
| 884T→C | L295P | DGGE | United Kingdom | 6 | |
| Intron 9: | |||||
| IVS9−1G→C | Deletion of exon 10 | DGGE | EcoRII | United Kingdom | 1 |
| Exon 10: | |||||
| 1004T→G | V335G | DGGE | France | 1 | |
| 1048T→C | S350P | DGGE | HinfI | United Kingdom | 1 |
| 1053–1054insT | Stop+3 | HDA and DGGE | United Kingdom | 1 | |
| 1081–1082insG | Stop+19 | HDA and DGGE | BglI | United Kingdom | 2 |
| 1082–1083insC | Stop+18 | HDA and DGGE | BglI | France | 6 |
| 1083delTe | Stop+3 | HDA and DGGE | BglI | France | 1 |
| 1083–1084insG | Stop+18 | HDA and DGGE | BglI | France | 1 |
| 1090–1091delAG | Stop+15 | HDA and DGGE | AlwNI | United Kingdom | 1 |
| Intron 10: | |||||
| IVS10−1G→T | Deletion of exon 11 | DGGE | DdeI | France | 1 |
| Exon 11: | |||||
| 1119G→A | W373X | DGGE | ApyI | France | 1 |
| 1147–1148delGT | Stop+50 | HPLC | France | 1 | |
| Exon 12: | |||||
| 1274–1275delGT | Stop+7 | HDA | MaeIII | United Kingdom | 1 |
| 1281G→A | W427X | … | BstI | United Kingdom | 1 |
| 1287delA | Stop | HDA and DGGE | France | 2 | |
| Intron 12: | |||||
| IVS12+1delG | Deletion of exon 12 | HDA | SpeI | France | 2 |
| IVS12−2A→G | Deletion of exon 13 | HDA | MspI | United Kingdom | 1 |
| Exon 13: | |||||
| 1292–1293delAG | Stop+2 | … | HinfI | United Kingdom | 2 |
| 1303C→T | Q435X | HPLC | United Kingdom | 5 | |
| 1331T→C | L444P | HDA | United Kingdom | 1 | |
| 1357G→A | G453R | … | United Kingdom | 1 | |
| 1358G→T | G453V | … | United Kingdom | 1 | |
| 1384–1385delAG | Stop+13 | HDA and DGGE | France | 1 |
Allelic Heterogeneity
Five mutations were present in four to seven families (table 3), accounting for a total of 28 (26%) of our 108 families. All other mutations were present in three or fewer families, with 47 being restricted to a single family (table 3).
The disease was less heterogeneous in the United Kingdom than in France. Of the 37 mutations identified in French families, only 1 (1082insC) was found in more than three families, being present in six families (12% of the 47 families) (table 3). In contrast, 4 of the 31 mutations in U.K. patients (i.e., mutations L15F, E189X, L295P, and Q435X) were found in 22 (39%) of the families from that country (table 3). Analysis of intragenic polymorphisms showed that the L15F and Q435X mutations were each associated with at least two different haplotypes (mutation L15P—haplotypes [−247A, −151G, IVS2−47G, 911A] and [−247A, −151G, IVS2−47G, 911G]; and mutation G435X—haplotypes [−151G, −181C, IVS2−47C, 911G] and [−151G, −181C, IVS2−47G, 911G]), indicating that neither mutation is likely to have been inherited from a single common ancestor. For the three other mutations, this analysis did not exclude common ancestry.
Only two infrequent mutations (503G→A [R168H] and 745delG) were found in both countries (table 3), and both occur at potentially hypermutable sites: a CpG dinucleotide on the antisense strand (R168H) and a polyG tract (745delG) (Cooper and Krawczak 1993).
Phenotype-Genotype Relationship
VP presents clinically in three distinct ways: with skin lesions alone, with an attack of acute porphyria without skin lesions, or with both together. The numbers of our patients in each category are shown in table 4. Percentages did not differ significantly between France (59% cutaneous) and the United Kingdom (58% cutaneous) and are very close to those reported elsewhere for a large series from South Africa (Eales et al. 1980; Kirsch et al. 1998).
Table 4.
VP Clinical Features
|
No. (%) in Subjects from |
||
| ClinicalFeature(s) | PresentStudy (n=103)a | South Africab(n=269) |
| Skin lesions alone | 61 (59%) | 156 (58%) |
| Acute attack alone | 20 (20%) | 51 (19%) |
| Both together | 22 (21%) | 62 (23%) |
Figures for France and the United Kingdom were not significantly different. Clinical information was available for 103 of the 108 unrelated patients.
Data are from Eales et al. (1980).
Table 5 shows the relationship between mode of presentation and type of mutation. Although truncating mutations (frameshift, nonsense, or splice site) appear to be associated with presentation with skin lesions alone, the difference was not statistically significant (χ2=5.33; P=.07). Four of the five mutations that were present in five or more families were associated with more than one type of clinical presentation; all patients with the 1082insC mutation presented with skin lesions alone.
Table 5.
VP Clinical Features and Type of Mutation
|
No. of Patients (%) of Total with Mutation Type |
|||
| Clinical Feature(s)a | Missense(n=38) | Splice Site(n=11) | Nonsense or Frameshift(n=51) |
| Skin lesions alone | 18 (47%) | 5 (45%) | 35 (69%) |
| Acute attack alone | 12 (32%) | 1 (10%) | 7 (14%) |
| Both together | 8 (21%) | 5 (45%) | 9 (17%) |
At presentation, in the index case from 100 unrelated families. Differences between groups are not significant (χ2= 5.33; P=.07).
Discussion
Here we have documented the clinical features and molecular basis of VP in 108 unrelated patients from western Europe. Mutations were identified in genomic DNA by a combined screening and sequencing approach. DGGE, which has been shown to be highly effective for the detection of mutations in the other acute porphyrias (Puy et al. 1997; Rosipal et al. 1999), cannot be applied to the whole of the PPOX gene, and HDA is known to lack sensitivity (Tchernitchko et al. 1999). However, all the mutations that we identified could be detected by direct automated sequencing of exons and their flanking regions, and this would appear to be the method of choice for mutational analysis of VP.
Mutations were not identified in four patients. Complete deletions of the PPOX gene were excluded by showing that each of these patients was heterozygous for an intragenic SNP. All had an unequivocal diagnosis of VP, and it is probable that the causative mutations either lie outside the gene regions that we sequenced or were partial deletions or insertions not detectable by our PCR-based methods.
Two patients had both a chain-termination or splice mutation (IVS6+1G→T or 1054insT, respectively) and a missense mutation (767C→G [P256R]) that were shown, by family studies, to be on the same allele. The P256R mutation has previously been reported to be one of two mutant PPOX alleles in a compound heterozygote with homozygous VP (Kauppinen et al. 1997). Investigation of 156 normal alleles showed that this mutation, which affects a residue that is poorly conserved between species (fig. 2), is a polymorphism present in ∼10% of the French population (table 2). Two previously unreported intragenic SNPs were also identified (table 2). Five single-base changes in exon 1 have recently been reported in VP and have been interpreted as evidence that this UTR of the gene is particularly mutation prone (Kotze et al. 1998). We sequenced exon 1 from 45 patients with VP and were unable to confirm this finding; two of the SNPs found by Kotze et al. (1998) in only three normal controls (i.e., mutations −253G/C and −246G/C) were not present in our cohort (table 2).
Figure 2.
Missense mutations and intraexonic SNPs in human PPOX. The amino acid sequence for the human enzyme (1) was compared with those of the mouse (2), Saccharomyces cerevisiae (3), Myxococcus xanthus (4), Bacillus subtilis (5) Mycobacterium tuberculosis (6), Propionibacterium freundenreichii (7), Nicotiana tabacum I (plastid) (8), Nicotiana tabacum II (mitochondria) (9), and Arabadopsis thaliana (10). Comparisons are shown only for mutated residues; numbers refer to the human sequence; letters above numbers denote mutant residues. Intraexonic polymorphisms (P256R [256] and R304H [304]) are in italics. A pair of vertically stacked asterisks (**) denotes a conserved residue, and a single asterisk (*) denotes a highly conserved residue. Sequences were aligned by Clustal W1.7 (Thompson et al. 1994).
We have identified 66 different mutations in the PPOX gene in VP (table 3), 60 of which are novel, bringing the total number of mutations reported in this condition to 79 (Deybach et al. 1996; Lam et al. 1996, 1997; Meissner et al. 1996; Warnich et al. 1996; de Rooij et al. 1997; Frank et al. 1997a; Frank et al. 1997b; Frank et al. 1998a, 1998b, 1998c, 1998d; Kauppinen et al. 1997; Corrigall et al. 1998; also see Human Gene Mutation Database), whereas a further 9 mutations have been identified only in its homozygous variant (Meissner et al. 1996; Kauppinen et al. 1997; Frank et al. 1998e; Roberts et al. 1998). Of the mutations in our patients, 31 (7 nonsense and 24 frameshift) introduce premature stop codons (table 3) and are likely to abolish PPOX activity, primarily by accelerating mRNA decay (Culbertson 1999). The 10 splice-site mutations that we identified (table 3) either change invariant bases at the donor or acceptor site (Cooper and Krawczak 1993) or introduce an alternative splice site that produces a frameshift in mRNA for PPOX (table 3) and are also likely to substantially reduce activity.
Twenty-five mutations altered the coding sequence for PPOX (table 3). Three of these altered the translation-initiation codon (table 3). Similar mutations in other genes are known to decrease steady-state mRNA concentrations, through failure of initiation with premature termination (Cooper and Krawczak 1993). The consequences of the 22 other missense mutations that we have identified are more difficult to predict, because little is known about the structure/function of PPOX. The human PPOX gene encodes a 51-kD protein that functions as a flavin- and oxygen-dependent protoporphyrinogen oxidase associated with the inner mitochondrial membrane (Deybach et al. 1985; Nishimura et al. 1995; Dailey and Dailey 1996). Sequence analysis shows both a dinucleotide-binding motif, in the amino-terminal region, that is highly conserved among flavin adenine dinucleotide (FAD)–binding proteins (Nishimura et al. 1995; Dailey and Dailey 1998) and an adjacent 60-residue-long region that is similar throughout a superfamily of FAD-binding proteins (Dailey and Dailey 1998). There is no identifiable transmembrane domain (Dailey and Dailey 1996), and the absence of a mitochondrial targeting presequence suggests the presence of an internal targeting signal that remains to be identified. Other than the initial methionine, 12 of the 20 residues that are changed by missense mutations are highly conserved between mammals, bacteria, and plants (fig. 2) and are therefore likely to be essential for PPOX function, stability, or transport. One of these (L15F) lies in the FAD-binding site, whereas two others (R38P and G40E) are in the adjacent region shared by the FAD-binding protein superfamily (Dailey and Dailey 1998). Seven (35%) of the residues are substituted by proline, with five of them being leucine-to-proline mutations, and a similar mutation (S450P) has been reported elsewhere (Frank et al. 1998d). The high frequency of mutations of proline is unusual and, since this residue is known to decrease protein flexibility (Tian et al. 1998) and to interrupt α-helical regions, may suggest that PPOX is very sensitive to small structural changes. Two other missense mutations (V282D and 281delH) introduce major changes (charge alteration or deletion) at adjacent residues that are partially conserved (fig. 2).
We have recently shown that missense PPOX mutations that preserve 10%–25% of wild-type activity are present on at least one allele in all patients with the rare homozygous form of VP (Roberts et al. 1998). None of these were found in our study, and it has yet to be shown that such mild mutations ever cause clinically overt VP in heterozygotes. It seems likely that such mutations represent one end of a functional spectrum of PPOX missense mutations, with mutations that abolish activity—and that therefore are lethal in homozygotes and thus only found in VP—at the other end. The frameshift, stop, and at least some of the missense mutations that we have identified are likely to abolish activity. It seems probable that mutations with intermediate effect on catalytic activity also exist and may cause VP, but with a low clinical penetrance. Further studies may show that some of the missense and splice-defective mutations in our patients fall into this category.
VP shows less allelic heterogeneity in western Europe than is shown by other acute hepatic porphyrias. In France and the United Kingdom, all the mutations identified in the HBMS (hydroxymethylbilane synthase) gene in 166 unrelated patients with acute intermittent porphyria have a prevalence of <5%—apart from one mutation (R173W) in the United Kingdom, which has a prevalence of 10% (Puy et al. 1997; Whatley et al. 1999). Hereditary coproporphyria is similarly heterogeneous in both France and the United Kingdom (Rosipal et al. 1999; authors' unpublished data). In contrast, five of the mutations that we have identified in VP have prevalences, in these two countries, of 7%–12%. Analysis of intragenic SNPs showed that at least two of these mutations are likely to have arisen on more than one occasion, so their relatively high frequency cannot be explained solely by their presence in the population of extended families. The apparent difference, in heterogeneity, between France and the United Kingdom (table 3) requires further investigation, but neither country shows the predominance of a single mutation seen in South Africa and, to a lesser extent, Finland (Kauppinen et al. 1997). In addition, even though the frequencies of SNPs were similar, there was an almost complete separation between the types of mutation found in France and those found in the United Kingdom, with only two infrequent mutations (R168H and 745delG) being present in both populations; both of these occur at hypermutable sites, and one of them (R168H) has been found in other countries (de Rooij et al. 1997; Frank et al. 1998b). This separation suggests a relatively recent origin for the mutations in our families. The R59W South African founder mutation was not present in any of our patients. To date, it has been identified only in Afrikans families of Dutch descent (Kirsch et al. 1998) and in the Netherlands (De Rooij et al. 1997).
Previous reports of VP outside South Africa (Mustajoki 1980; Arnas et al. 1992; Kirsch et al. 1998) have included too few symptomatic patients for any firm conclusions to be drawn about whether the clinical features reported from South Africa (Eales et al. 1980; Kirsch et al. 1998) are special to that country or typical of the disease in general. Here we have shown that the clinical features in a large, molecularly heterogeneous series from western Europe are identical to those seen in South Africa, where almost all patients share the same mutation (Meissner et al. 1996; Groenewald et al. 1998). This finding suggests that VP in South Africa is clinically representative of the disease elsewhere and that allelic heterogeneity does not substantially alter the clinical pattern—an observation that would be predicted if the majority of PPOX mutations that cause VP lead to near-complete or total loss of activity, as does the R59W mutation in South Africa (Meissner et al. 1996). Measurements of PPOX activity in South African and other patients with VP have consistently shown a 50% decrease, consistent with complete haplodeficiency (Brenner and Bloomer 1980; Da Silva et al. 1995; Kirsch et al. 1998). More detailed analysis of our findings also fails to identify any major relationship between clinical features and type of mutation. Of the mutations that we identified, 31 (47%) produced premature-termination codons and are likely to have abolished all PPOX activity. Yet the distribution of the associated clinical features was similar to that found in the group as a whole (tables 4 and 5), and there was no significant correlation between type of mutation and clinical presentation (table 5). In addition, four of the five relatively frequent mutations that we found were associated with two or more clinical presentations. Overall, therefore, the genotype does not appear to be a significant determinant of clinical severity, if skin disease alone is regarded as a mild manifestation and if an acute attack is regarded as a severe one. Environmental influences and genetic effects from other loci are likely to be more important in this respect.
Acknowledgments
We thank the Welsh Scheme for the Development of Health and Social Research (support to S.D.W., R.R.M., A.G.R., and G.H.E) and INSERM U409 and Université Paris 7 (support to H.P., A.-M.R., Y.N., and J.-C.D.) for financial support, and we thank Dr. M. O'Donovan, Department of Psychological Medicine, University of Wales College of Medicine, for HPLC analyses.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Human Gene Mutation Database, http://www.uwcm.ac.uk/uwcm/mg/hgmd0.html (for mutations in PPOX gene)
- Human Genome Mapping Project, http://www.hgmp.mrc.ac.uk (for accession numbers for PPOX genomic/cDNA sequences: X99450 [human], U25114 [mouse], U18778 [Saccharomyces cerevisiae], M73709 [Myxococcus xanthus], M97208 [Bacillus subtilis], AL021186 [Mycobacterium tuberculosis], D85417 [Propionibacterium freundenreichii], Y13467 [Nicotiana tabacum I], Y13466 [Nicotiana tabacum II], and D83139 [Arabidopsis thaliana])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for VP [MIM 176200])
References
- Armas R, Wolffe C, Krause P, Chana P, Parraguez A, Soto J (1992) The hepatic porphyrias: experience with 105 cases (in Spanish). Rev Med Chil 120:259–266 [PubMed]
- Brenner DA, Bloomer JR (1980) The enzymatic defect in variegate porphyria. N Engl J Med 302:765–769 [DOI] [PubMed]
- Cooper DN, Krawczak M (1993) Human gene mutation. BIOS Scientific, Oxford [Google Scholar]
- Corrigall AV, Hift RJ, Hancock V, Meissner D, Davids L, Kirsch RE, Meissner PN (1998) Identification and characterisation of a deletion (537delAT) in the protoporphyrinogen oxidase gene in a South African variegate porphyria family. Hum Mutat 12:403–407 [DOI] [PubMed]
- Culbertson MR (1999) RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet 15:74–80 [DOI] [PubMed]
- Dailey TA, Dailey HA (1996) Human protoporphyrinogen oxidase: expression, purification, and characterization of the cloned enzyme. Protein Sci 5:98–105 [DOI] [PMC free article] [PubMed]
- ——— (1998) Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases and phytoene desaturase: expression and characterization of phytoene desaturase of Myxococcus xanthus. J Biol Chem 273:13658–13662 [DOI] [PubMed]
- Da Silva V, Simonin S, Deybach J-C, Puy H, Nordmann Y (1995) Variegate porphyria: diagnostic value of fluorometric scanning of plasma porphyrins. Clin Chim Acta 238:163–168 [DOI] [PubMed]
- Dean G (1971) The porphyrias: a story of inheritance and environment, 2d ed. Pitman Medical, London [Google Scholar]
- De Rooij FWM, Minderman G, De Baar E, Wilson JHP, Sinke JRJ, Ploos van Amstel JK, TeVelde K (1997) Six new protoporphyrinogen oxidase mutations in Dutch variegate porphyria patients and the R59W mutation in historical perspective. Acta Haematol 98 Suppl 1:103 [Google Scholar]
- Deybach J-C, da Silva V, Grandchamp B, Nordmann Y (1985) The mitochondrial location of protoporphyrinogen oxidase. Eur J Biochem 149:431–435 [DOI] [PubMed]
- Deybach J-C, Puy H, Robreau AM, Lamoril J, Da Silva V, Grandchamp B, Nordmann Y (1996) Mutations in the protoporphyrinogen oxidase gene in patients with variegate porphyria. Hum Mol Genet 5:407–410 [DOI] [PubMed]
- Eales L, Day RS, Blekkenhorst GH (1980) The clinical and biochemical features of variegate porphyria: an analysis of 300 cases studied at Groote Schuur Hospital, Cape Town. Int J Biochem 12:837–853 [DOI] [PubMed]
- Elder GH, Hift RJ, Meissner PN (1997) The acute porphyrias. Lancet 349:1613–1617 [DOI] [PubMed]
- Frank J, Christiano AM (1997) Genetic research strategies: a review of the acute porphyrias. Retinoids 13:88–92 [Google Scholar]
- Frank J, Jugert FK, Kalka K, Goerz G, Merk HF, Christiano AM (1998a) Variegate porphyria: identification of a nonsense mutation in the protoporphyrinogen oxidase gene. J Invest Dermatol 110:449–451 [DOI] [PubMed]
- Frank J, Jugert FK, Breitkopf C, Goerz G, Merk HF, Christiano AM (1998b) Recurrent missense mutation in the protoporphyrinogen oxidase gene underlies variegate porphyria. Am J Med Genet 79:22–26 [PubMed]
- Frank J, Jugert FK, Kalka K, Goerz G, Merk HF, Christiano AM (1997a) Premature termination codons in the protoporphyrinogen oxidase gene underlie variegate porphyria. Acta Haematol 98 Suppl 1:97 [Google Scholar]
- Frank J, Lam H, Zaider E, Poh-Fitzpatrick M, Christiano AM (1998c) The genetic basis of “Scarsdale gourmet diet” variegate porphyria: a missense mutation in the protoporphyrinogen oxidase gene. Arch Dermatol Res 290:441–445 [DOI] [PubMed]
- Frank J, Lam H, Zaider E, Poh-Fitzpatrick M, Christiano AM (1998d) Molecular basis of variegate porphyria: a missense mutation in the protoporphyrinogen oxidase gene. J Med Genet 35:244–247 [DOI] [PMC free article] [PubMed]
- Frank J, McGrath J, Lam H, Graham RM, Hawk JL, Christiano AM (1998e) Homozygous variegate porphyria: identification of mutations on both alleles of the protoporphyrinogen oxidase gene in a severely affected proband. J Invest Dermatol 110:452–455 [DOI] [PubMed]
- Frank J, Zaider E, Jugert FK, Goerz G, Merk HF, Poh-Fitzpatrick M, Christiano AM (1997b) Variegate porphyria: identification of three novel missense mutations in the protoporphyrinogen oxidase gene. Acta Haematol 98 Suppl 1:96 [Google Scholar]
- Groenewald JZ, Liebenberg J, Groenewald IM, Warnich L (1998) Linkage disequilibrium analysis in a recently founded population: evaluation of the variegate porphyria founder in South African Afrikaners. Am J Hum Genet 62:1254–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins T (1996) The South African malady. Nat Genet 13:7–9 [DOI] [PubMed]
- Kauppinen R, Timonen K, Laitinen E, Kuusisto K, Ahola H, Tenhunen R, Mustajoki P (1997) Molecular genetics and clinical characteristics of variegate porphyria. Acta Haematol 98 Suppl 1:96 [Google Scholar]
- Kirsch RE, Meissner PN, Hift RJ (1998) Variegate porphyria. Sem Liver Dis 18:33–41 [DOI] [PubMed]
- Kotze MJ, De Villiers JN, Groenewald JZ, Rooney RN, Loubser O, Thiart R, Oosthuizen CJ, et al (1998) Molecular analysis reveals a high mutation frequency in the first untranslated exon of the PPOX gene and largely excludes variegate porphyria in a subset of clinically affected Afrikaner families. Mol Cell Probes 12:293–300 [DOI] [PubMed]
- Lam H, Dragan L, Tsou HC, Merk H, Peacocke M, Goerz G, Sassa S, et al (1996) Molecular basis of variegate porphyria: frameshift mutations in the protoporphyrinogen oxidase gene. J Invest Dermatol 107:144 [DOI] [PubMed] [Google Scholar]
- Lam H, Dragan L, Tsou HC, Merk H, Peacocke M, Goerz G, Sassa S, et al (1997) Molecular basis of variegate porphyria: a de novo insertion mutation in the protoporphyrinogen oxidase gene. Hum Genet 99:126–129 [DOI] [PubMed]
- Lerman LS, Silverstein K (1987) Computational simulation of DNA melting and its application to denaturing gel electrophoresis. Methods Enzymol 155:482–501 [DOI] [PubMed]
- Long C, Smyth SJ, Woolf J, Murphy GM, Finlay AY, Newcombe RG, Elder GH (1993) Detection of latent variegate porphyria by fluorescence emission spectroscopy of plasma. Br J Dermatol 129:9–13 [DOI] [PubMed]
- Meissner PN, Dailey TA, Hift RJ, Ziman M, Corrigall AV, Roberts AG, Meissner D, et al (1996) A R59W mutation in protoporphyrinogen oxidase results in decreased enzyme activity and is prevalent in South Africans with variegate porphyria. Nat Genet 13:95–97 [DOI] [PubMed]
- Mustajoki P (1980) Variegate porphyria. Q J Med 49:191–203 [PubMed]
- Myers RM, Mamatis T, Lerman LS (1987) Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol 155:501–527 [DOI] [PubMed]
- Nishimura K, Taketani S, Inokuchi H (1995) Cloning of a cDNA for protoporphyrinogen oxidase by complementation in vivo of a hemG mutant of Escherichia coli. J Biol Chem 270:8076–8080 [DOI] [PubMed]
- O'Donovan MC, Oefner PH, Roberts SC, Austin J, Hoogendoorn B, Guy C, Speight G, et al (1998) Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics 52:44–49 [DOI] [PubMed]
- Poh-Fitzpatrick MB (1980) A plasma porphyrin fluorescence marker for variegate porphyria. Arch Dermatol 116:543–547 [PubMed]
- Puy H, Deybach J-C, Lamoril J, Robreau AM, Da Silva V, Gouya L, Grandchamp B, et al (1997) Molecular epidemiology and diagnosis of PBG deaminase gene defects in acute intermittent porphyria. Am J Hum Genet 60:1373–1383 [DOI] [PMC free article] [PubMed]
- Puy H, Robreau AM, Rosipal R, Nordmann Y, Deybach JC (1996) Protoporphyrinogen oxidase: complete genomic sequence and polymorphisms in the human gene. Biochem Biophys Res Commun 226:227–230 [DOI] [PubMed]
- Roberts AG, Puy H, Dailey TA, Morgan RR, Whatley SD, Dailey HA, Martasek P, et al (1998) Molecular characterization of homozygous variegate porphyria. Hum Mol Genet 7:1921–1925 [DOI] [PubMed]
- Roberts AG, Whatley SD, Daniels J, Holmans P, Fenton J, Owen MJ, Thomson P, et al (1995) Partial characterization and assignment of the gene for protoporphyrinogen oxidase and variegate porphyria to human chromosome 1q23. Hum Mol Genet 4:2387–2390 [DOI] [PubMed]
- Rosipal R, Lamoril J, Puy H, Da Silva V, Gouya L, De Rooij FWM, Te Velde K, et al (1999) Systematic analysis of coproporphyrinogen oxidase gene defects in hereditary coproporphyria and mutation update. Hum Mutat 13:44–53 [DOI] [PubMed]
- Taketani S, Inazawa J, Abe T, Furukawa T, Kohno H, Tokunaga R, Nishimura K, et al (1995) The human protoporphyrinogen oxidase gene (PPOX): organization and location to chromosome 1. Genomics 29:698–703 [DOI] [PubMed]
- Tchernitchko D, Lamoril J, Puy H, Robreau AM, Bogard C, Rosipal R, Gouya L, et al (1999) Evaluation of mutation screening by heteroduplex analysis in acute intermittent porphyria: comparison with denaturing gradient gel electrophoresis. Clin Chim Acta 279:133–143 [DOI] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ (1994) Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position- specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed]
- Tian H, Yu L, Mather MW, Yu CA (1998) Flexibility of the neck region of the rieske iron-sulfur protein is functionally important in the cytochrome bc1 complex. J Biol Chem 273:27953–27959 [DOI] [PubMed]
- Warnich L, Kotze MJ, Groenewald IM, Groenewald JZ, van Brakel MG, van Heerden CJ, de Villiers JN, et al (1996) Identification of three mutations and associated haplotypes in the protoporphyrinogen oxidase gene in South African families with variegate porphyria. Hum Mol Genet 3:981–984 [DOI] [PubMed]
- Whatley SD, Woolf JR, Elder GH (1999) Comparison of complementary and genomic DNA sequencing for the detection of mutations in the HMBS gene in British patients with acute intermittent porphyria: identification of twenty-five novel mutations. Hum Genet 104:505–510 [DOI] [PubMed]