The recognition, approximately a decade ago, that mutations in mtDNA can impair oxidative phosphorylation (OXPHOS) capacity and lead to human diseases (Holt et al. 1988; Wallace et al. 1988) heightened interest in the regulation and maintenance of this multicopy, extranuclear genome. Since then, mitochondrial dysfunction resulting from mtDNA mutation, instability, or copy-number deregulation has been implicated in numerous pathological conditions and the normal aging process (Wallace 1992; Larsson and Clayton 1995). The preservation of a functional mitochondrial genome over an individual's lifetime requires not only proper replication, segregation, and expression of functional genetic units during development and all subsequent mitotic cell divisions but also protection from and efficient repair of mtDNA damage. As I outline below, our current understanding of basic principles underlying many of these processes has come from studies of the budding yeast, Saccharomyces cerevisiae. Beyond the many fundamental similarities of mtDNA replication in human and yeast cells, however, there are aspects of the process that differ between our two species. These differences, which may, in some cases, limit the utility of S. cerevisiae as a model for human mtDNA replication, need to be considered in the discussion of the etiology of mitochondrial diseases.
Human mtDNA has a number of characteristics that confound our understanding of mitochondrial genetics. First, multiple copies of mtDNA are present in most cell types and within each organelle. The copy number is tightly regulated and typically is 102–104 copies/cell, depending on cell type. Second, a mixture of wild-type and mutated mtDNA molecules can coexist in the same cell, tissue, or individual. At a cellular level, such a heteroplasmic state complicates the segregation pattern of mtDNA mutations, because an apparently random subset of the total mtDNA population is inherited by each daughter cell during cell division. Depending on the percentage of wild-type and mutated mtDNA molecules delivered to and propagated in each daughter cell, the degree of heteroplasmy can drift dramatically, even from undetectable levels of mutant mtDNA to predominantly mutant mtDNA, in the course of a single cell division. Because this phenomenon can also occur in the female germline during either mitotic or meiotic divisions, large shifts in the degree of heteroplasmy are also often observed between mothers and their children. Finally, phenotypic expression of mtDNA mutations exhibit a threshold effect, whereby the percentage of mutated mtDNA molecules in a cell or tissue must reach a critical high percentage (60%–90%) before a defect in OXPHOS is manifest. At percentages of mutant mtDNA that are below this threshold, mtDNA mutations appear to be phenotypically silent. These factors contribute to the complicated tissue-specific and variably penetrant phenotypic-expression patterns of mtDNA mutations observed in individuals affected by mitochondrial genetic disease (Grossman and Shoubridge 1996).
Mitochondrial genomes from different species vary dramatically in size, structure, and coding capacity. In all cases, mtDNA encodes a small subset of the ∼80 protein subunits of the OXPHOS enzyme complexes in the mitochondrial inner membrane, which together generate cellular ATP. Human mtDNA is a 16.5-kb double-stranded circular molecule (fig. 1A) that encodes 13 OXPHOS-related mRNAs, as well as 22 tRNAs and 2 rRNAs required to translate these messages on ribosomes in the mitochondrial matrix. The genome is remarkably compact, with all 37 genes efficiently organized into two polycistronic transcription units. A unique feature of these primary transcripts is that each rRNA and most of the mRNAs are flanked by at least one tRNA. This gene arrangement leads to a mode of gene expression that requires a large number of RNA-processing events in order to liberate the mature mRNA, tRNA, and rRNA species from these complex precursor transcripts. As with gene expression, replication of human mtDNA depends on RNA transcript processing, because the mtDNA polymerase polγ requires specific RNA oligonucleotides to prime initiation. Other than two related ribonucleoproteins, RNase MRP and RNase P, the factors that mediate these processing events in human mitochondria have remained elusive, and the regulatory system that coordinates mtRNA processing with other intracellular and physiological processes remains obscure. Suitable model systems that faithfully reproduce mitochondrial disease phenotypes are needed to approach these regulatory issues. Because the basic machinery of mtDNA transcription and replication appears to be well conserved among diverse eukaryotes, the experimentally tractable yeast cell provides a valuable system in which to characterize key mitochondrial regulatory molecules.
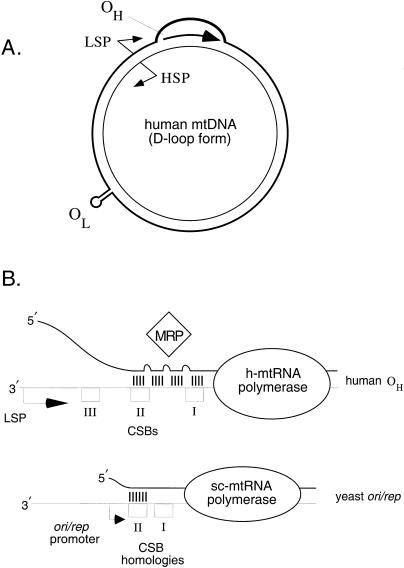
Figure 1.
A, Human mtDNA, a 16.5-kb double-stranded circular molecule, with the two DNA strands designated as H or L strands, on the basis of their relative buoyant densities in denaturing cesium-chloride gradients. OH is located immediately downstream of the LSP, which serves a dual role in gene expression and mtDNA replication. OL is physically separated from OH on the molecule. The HSP, LSP, and OH are all located in the main regulatory locus on the molecule, called the “D-loop region.” The D-loop itself (bulge at top of molecule) results from a terminated H-strand replication event (the arrested nascent H-strand is indicated by the large arrow; for details, see the text). B, Proposed early events in the initiation of mtDNA replication at human OH (top) and at S. cerevisiae ori/rep sequences (bottom). At human OH, transcription initiation at the LSP (bent arrow) by human mtRNA polymerase (oval) begins the process. As h-mtRNA polymerase traverses CSB III, CSB II, and CSB I (open boxes), a complex RNA/DNA hybrid structure is formed between the RNA transcript (thicker line) and the template L-strand (thinner line); RNA-DNA base pairs are indicated (vertical hatch marks). This RNA/DNA hybrid is the putative in vivo substrate for mtRNA-processing enzymes, such as RNase MRP (diamond), which cleave the hybridized transcript at specific sites to form RNA primers for initiation of H-strand replication. A similar mechanism has been proposed for S. cerevisiae mtRNA polymerase–driven initiation of leading-strand mtDNA synthesis at yeast ori/rep sequences (bottom [diagrammed in a manner identical to that described for OH]). Potentially important differences noted at the yeast mtDNA origins include the greater proximity of the ori/rep promoter (bent arrow) to the CSB II-like sequence and an RNA/DNA hybrid that is less complex than that at human OH.
Lessons from Petite Yeast
In S. cerevisiae, as in all mitochondrial eukaryotes, a large number of biochemical reactions occur in mitochondria that make the organelles themselves indispensable for life. However, because yeast are facultative anaerobes, they are not entirely dependent on OXPHOS for the production of ATP and NAD+. In other words, yeast can survive without a functional mitochondrial respiratory chain, provided that an appropriate carbon source (e.g., glucose) is made available to permit continued ATP production via glycolysis and fermentation. Since the mitochondrial genetic system functions only to provide key OXPHOS enzyme subunits, yeast grown on fermentable carbon sources are viable even if mtDNA is mutated or absent. Respiration-deficient mutants of yeast, referred to as “petite” because of their slow growth and small colony size, can arise if either mtDNA or nuclear-DNA mutations compromise OXPHOS. The ability to analyze petite mutants has made yeast an attractive experimental system for isolation and characterization of nuclear-gene products required for mitochondrial function and biogenesis.
Three examples of conservation between human and yeast mtDNA metabolism will suffice to demonstrate the utility of this system. First, the initial gene encoding a dedicated mtRNA polymerase (RPO41) was identified because mutations in this gene confer a petite phenotype, and this gene has been shown to be required not only for mitochondrial transcription but also for maintenance of mtDNA in cells (Greenleaf et al. 1986). Perhaps surprisingly, nucleotide-sequence analysis has revealed that yeast mtRNA polymerase is homologous to the simple RNA polymerases encoded by the Escherichia coli bacteriophage genomes of T7, T3, and SP6 (Masters et. al. 1987). It is now clear that most, if not all, eukaryotic mtRNA polymerases, including human (Tiranti et al. 1997), conform to this bacteriophage model. Second, the first gene encoding an mtDNA polymerase was also discovered in yeast (MIP1) (Foury 1989), revealing that it is a member of the DNA Pol I family (family A) of DNA polymerases (Braithwaite and Ito 1993). In addition, the MIP1 gene sequence revealed the presence of three exonuclease domains in the protein, providing the first compelling evidence that mtDNA polymerases do indeed have intrinsic proofreading capacity. Finally, genes encoding eukaryotic homologues of the bacterial DNA mismatch-repair protein MutS were identified initially in yeast. One of these proteins, Mshlp, is located in the mitochondrial matrix and is required for proper maintenance of mtDNA (Reenan and Kolodner 1992), which suggests that yeast mitochondria have a critical DNA mismatch-recognition and/or -repair capacity. Bogenhagen (1999) recently has reviewed evidence for several forms of mtDNA repair that occur in mammalian cells as well. These examples highlight only a few of the advances in our understanding of mtDNA regulation that have resulted from the study of the yeast model system. For a more extensive discussion of the contribution of yeast studies to the understanding of mitochondrial-gene expression, mitochondrial inheritance, and nuclear-mitochondrial communication, see the work of Costanzo and Fox (1990), Shadel and Clayton (1993), Dieckmann and Staples (1994), Grivell (1995), Poyton and McEwan (1996), Hermann and Shaw (1998), and Yaffe (1999).
Basic Features of Human mtDNA Replication
The mechanism of human mtDNA replication was determined in broad outline by analysis of replication intermediates isolated from cultured cells (reviewed by Clayton 1982). These early studies led to the discovery of a major mtDNA molecular form that contains a displacement-loop (D-loop), a stable three-stranded DNA structure containing a short nascent mtDNA strand at the origin of heavy (H)-strand replication (OH; see fig. 1). This D-loop itself occurs within a larger noncoding region of the mtDNA molecule (referred to as the “D-loop regulatory region”) that harbors not only OH but also the mitochondrial transcription promoters (fig. 1), making it the main control site for mtDNA replication and transcription. Vertebrate mtDNA also has a second origin of replication for the light (L)-strand (OL). OH and OL are unidirectional origins (i.e., DNA synthesis proceeds only in one direction) and are physically separated from each other on the mtDNA molecule (fig. 1). Thus, human mtDNA replicates by an unusual asynchronous mechanism that is initiated at OH (Clayton 1982).
Because the entire mtDNA replication process is initiated at OH, events that lead to priming of H-strand replication in the D-loop region have been and continue to be a focus of intensive investigation. Initial insight into the mechanism of H-strand replication priming came with the determination that the mitochondrial L-strand and H-strand promoters (LSP and HSP, respectively) are located immediately adjacent to OH, suggesting that transcription and mtDNA replication might be linked. Indeed, detailed mapping and molecular characterization of D-loop RNA and DNA species revealed that RNA transcripts initiated at the LSP serve to prime H-strand DNA synthesis (Clayton 1991). Considerable progress has been made in the understanding of this transcription-dependent mtDNA-replication mechanism, including the isolation and characterization of key, nucleus-encoded factors that interact at the D-loop region to promote transcription and replication of the mitochondrial genome. These data support the general model for initiation of H-strand mtDNA replication in humans that has been described by Shadel and Clayton (1997). In this model, an RNA transcript is initiated at the LSP by human mtRNA polymerase. This event requires the nucleus-encoded transcription factor h-mtTFA (Parisi and Clayton 1991), which binds to the region immediately upstream of the LSP and activates specific transcription initiation by human mtRNA polymerase. Next, as mtRNA polymerase traverses a series of three conserved-sequence blocks (CSB I–CSB III) at OH, the elongating LSP transcript forms an RNA/DNA hybrid that serves as a substrate for processing reactions that generate mature RNA primers (fig. 1). On the basis of in vitro analysis (Lee and Clayton 1998) and subcellular-localization studies (Li et al. 1994), the endoribonuclease RNase MRP appears to be a key player in the processing of a complex precursor RNA/DNA hybrid into mature RNA primers. These primers are recognized by DNA polγ and accessory proteins to initiate H-strand DNA synthesis, thus beginning a productive mtDNA-replication event. Although analysis of mtDNA from other vertebrates has revealed some species-specific sequence variability, the D-loop–region configurations suggest that, in all cases, the transcription-primed mechanism is conserved (Shadel and Clayton 1997).
The degree of conservation of mtDNA replication in lower eukaryotes has been addressed most extensively in S. cerevisiae, and substantial evidence indicates that transcription priming also operates in yeast (fig. 1). Nonetheless, such models remain controversial because of significant differences that exist between human and yeast mitochondrial genetic systems. For example, yeast mtDNA is larger (∼80 kb) than human (16.5 kb) and, instead of two unidirectional origins, contains four putative origins of replication (termed “ori/rep” sequences) that are bidirectional in nature. The greater complexity of yeast mtDNA replication has slowed progress in this system, but this disadvantage may be offset by the existence of a remarkable class of spontaneous mtDNA deletion mutants, the ρ− petites, which are easily identified and studied in yeast. These mutant strains often retain multiple copies of particular ori/rep sequences in mitochondria and have been used extensively to address mtDNA-replication and -segregation issues in yeast. Unfortunately, it remains unclear to what extent the behavior of these repetitive mutated mtDNAs reflects that of the wild-type mitochondrial genome. As discussed below, these issues not only complicate the analysis of wild-type mtDNA replication in yeast but also may limit to some degree the applicability of yeast as a model system for the understanding of human mitochondrial genetic phenomena.
mtDNA Replication in S. cerevisiae
Under normal growth conditions, all laboratory yeast strains spontaneously generate petite mutants at a frequency of ∼1%/generation, largely because yeast mtDNA is prone to massive deletion and rearrangement. The mtDNA sequences retained in the resulting ρ− petite mutants often consist of amplified head-to-tail repeats of a portion of the genome, almost always containing <1% of the normal sequence. These repeat genomes may segregate in unusual ways. For example, when the so-called hypersuppressive (HS) ρ− mutants are mated to a strain harboring a wild-type mitochondrial genome, the HS ρ− genome is inherited preferentially to the wild-type genome by the daughter cells emerging from the cross.
In the vast majority of HS mutants, the repeated region includes an ori/rep sequence (Blanc and Dujon 1980; de Zamaroczy et al. 1981), an ∼280 bp sequence that is structurally similar to the vertebrate OH. Wild-type yeast mitochondrial genomes contain as many as eight such sequences, of which four (ori1–ori3 and ori5) harbor an intact transcriptional promoter and likely represent bona fide origins of DNA replication. In studies of various HS strains, Baldacci et al. (1984) found that RNA transcripts initiated at the ori/rep promoters are used as primers for mtDNA synthesis; more-recent work has shown that the essential oligoribonucleotide primers are 9–17 nucleotides long and hybridize at various sites encompassing the conserved CSB II element (Graves et al. 1998). In at least two of the four ori/rep sequences, CSB II is also the site at which a transcription-dependent RNA/DNA hybrid forms in vitro, suggesting that a longer RNA species that is transcribed in this region serves as the precursor of the oligoribonucleotide primers that are used in replication (Xu and Clayton 1995). Furthermore, cells lacking either mtRNA polymerase or the mitochondrial transcription-initiation factor sc-mtTFB generate ρ− and ρ0 daughters (the latter of which entirely lacks mtDNA) at an increased rate (Greenleaf et al. 1986; Lisowsky and Michaelis 1988; Wang and Shadel 1999). All of these data are consistent with an ori/rep-dependent, transcription-primed replication mechanism that is required for leading-strand synthesis of yeast mtDNA, similar to the mechanism proposed for vertebrate OH (fig. 1). Nonetheless, several features of yeast mtDNA both complicate the interpretation of the data that lead to this model and make wild-type mtDNA replication difficult to address experimentally.
Hypersuppressive Petites: Friends or Foes?
Despite the wealth of information that HS petite mutants have provided regarding mtDNA replication and segregation, complications persist about the proper analysis of these mutants. For instance, Fangman et al. (1989) identified stable ρ− genomes that do not contain ori/rep sequences, and they later demonstrated that certain ori/rep-containing HS genomes can be maintained in the absence of mtRNA polymerase (Fangman et al. 1990). Furthermore, the involvement of recombination intermediates in the preferential inheritance patterns observed for HS genomes (i.e., the hypersuppressive phenomenon) also has been documented (Lockshon et al. 1995). Altogether, these observations have led to the realization that HS genomes can propagate by mechanisms that are independent of ori/rep sequences and mtRNA polymerase-driven transcription. In theory, the highly repetitive nature of ρ− genomes would increase the potential for genetic recombination within and between these mtDNA molecules. Thus, a mode of replication for HS genomes that is dependent on the formation of recombination intermediates is one formal possibility that has been proposed (MacAlpine et al. 1998, and references within). In this regard, it should be stressed that aggressive genetic recombination, as is observed in yeast mitochondria, is not found in human mitochondria. Despite the identification of human mitochondrial activities potentially involved in recombination, homologous recombination between mtDNA molecules has yet to be demonstrated unambiguously in human cells (reviewed by Howell 1997). A mode of replication that involves recombination intermediates, such as that proposed for yeast HS genomes, is therefore almost certainly not involved in the replication of vertebrate mtDNA, highlighting yet another salient difference between yeast and human mtDNA metabolism.
The relevance of the above-mentioned properties of HS genomes to the mode of replication of wild-type yeast mtDNA also remains speculative. The existence of an alternative mode of replication for ρ− mtDNA need not argue against the proposed transcription-dependent (and ori/rep sequence-dependent) mode of replication for wild-type genomes. Likewise, the presence of a primarily transcription-dependent replication mechanism for wild-type mtDNA does not rule out the possibility that other modes of mtDNA replication may operate under certain circumstances. For example, a decrease or loss of mitochondrial transcription could result in a switch from a transcription-dependent to a transcription-independent (e.g., recombination-mediated) mode of mtDNA replication. Indeed, the complex assortment of mtDNA 5′ and 3′ ends and double-strand breaks at yeast ori1 in vivo may indicate that these sequences are recombinogenic (Van Dyck and Clayton 1998). Further speculation suggests that it is possible that the inherent instability of wild-type yeast mtDNA under normal growth conditions is due in part to the simultaneous occurrence of replication and recombination at ori/rep sequences, which might be expected to promote deletions or rearrangements.
Yeast Mitochondrial-Genome Instability
Perhaps even more surprising than the normal high level of mtDNA instability is the number of nuclear-gene mutations in yeast that enhance this instability (table 1). Many of these loci can be rationalized easily, especially in light of an accepted model of transcription-coupled replication. These include MIP1, which encodes DNA polγ; MTF1, which encodes the transcription-initiation factor sc-mtTFB; and RIM1, which encodes a mitochondrial single-strand DNA-binding protein (mtSSB). Other loci have little obvious connection to mtDNA metabolism and cannot be explained as easily. For example, mutations in genes for a Lon protease homologue (PIM1); an enzyme involved in branched-chain amino acid synthesis (ILV5); and a putative mitochondrial solute carrier (RIM2/MRS12) all promote the formation of ρ− or ρ0 cells. Evidently, the mtDNA-instability phenotype in yeast can result from any number of nuclear- or mitochondrial-gene mutations and therefore must be interpreted with some caution, particularly when studies in yeast are used to ascribe mtDNA-related functions to yeast homologues of human disease genes. For example, disruption of the yeast homologue (YFH1) of the human FRATAXIN gene has been shown to cause mtDNA instability in yeast, a phenotype that has not yet been reported in cells from individuals with Friedreich ataxia (reviewed in Knight et al. 1999).
Table 1.
Yeast Genes Implicated in mtDNA Stability[Note]
| Gene | Protein Information and Mutant Phenotype |
| ABF2 | HMG-box protein sc-mtTFA/mitochondrial nucleoid component |
| ADR1 | Nuclear transcription factor |
| ERV1 | Null mutation lethal, affects mitochondrial transcript levels |
| FZO1 | Putative GTPase involved in mitochondrial membrane fusion |
| ILV5 | Enzyme in branched-chain amino acid biosynthesis |
| MDM10 | Mitochondrial outer-membrane protein involved in mitochondrial inheritance |
| MDM12 | Mitochondrial outer-membrane protein involved in mitochondrial inheritance |
| MGM1 | Dynamin-like GTPase in outer mitochondrial membrane |
| MGM101 | Mitochondrial nucleoid-component/oxidative mtDNA-damage response |
| MGM104 | Function unknown |
| MHR1 | Implicated in mtDNA recombination and/or repair |
| MIP1 | DNA polγ |
| MMM1 | Mitochondrial outer-membrane protein involved in mitochondrial inheritance |
| MSH1 | Bacterial MutS homologue located in mitochondria |
| MTF1 | sc-mtTFB |
| MTF2/NAM1 | Putative RNA-processing and/or -transcription factor |
| OCT1 | Yeast intermediate peptidase/protein import and iron homeostasis |
| PET18 | Function unknown, PET18 mutation affecting multiple genes |
| PET56 | Mitochondrial 21S rRNA methyltransferase |
| PET112 | Posttranscriptional regulator of mitochondrial COX II gene |
| PET127 | Mitochondrial membrane-bound, putative RNA-processing factor |
| PIF1 | mtDNA helicase |
| PIM1 | Bacterial Lon protease homologue located in mitochondria |
| PPA2 | Mitochondrial inorganic pyrophosphatase |
| RIM1 | mtSSB |
| RIM2/MRS12 | Mitochondrial carrier-family homologue |
| RPO41 | Core mtRNA polymerase |
| TTS1 | Cytoplasmic tyrosyl-tRNA synthetase, suppressing MGM104-1 mutant |
| YFH1 | Yeast frataxin homologue/iron homeostasis |
Note.—Many genes known to encode factors involved directly in mitochondrial protein synthesis (e.g., mitochondrial ribosomal proteins, tRNA synthetases, and translation factors) are not included; for details, see the text. Because of a citation limitation, references describing the isolation and characterization of many of the genes listed could not be included.
Of particular interest in this regard, gene products involved in mitochondrial protein synthesis all appear to be required in the maintenance of yeast mtDNA (Myers et al. 1985). Mutations that promote the unstable phenotype are found in virtually all of the known mitochondrial ribosomal protein genes and in several genes for mitochondrial tRNA synthetases and translation factors. This phenomenon remains unexplained, but it raises the unorthodox suggestion that the mtDNA-instability phenotype in strains lacking the complete mitochondrial transcriptional machinery is the direct result of impaired translation, rather than of impaired transcription per se. For some time, this alternative interpretation appeared to weaken the argument for transcription-coupled mtDNA replication in yeast. However, as is detailed in the accompanying sidebar, we have begun to shed light on this issue by showing that an amino-terminal domain of yeast mtRNA polymerase is required for mtDNA stability, even though this portion of the protein is dispensable for transcription initiation in vivo (Wang and Shadel 1999). This result provides a compelling argument that mtRNA polymerase is, in fact, directly involved in the coupling of mtDNA-replication activities to the transcription process in yeast. Although it is not yet clear whether an amino-terminal domain of the human mtRNA polymerase plays a role identical to that which has been defined for this portion of the yeast protein, it follows from these results that mtRNA polymerase is intimately involved in some form of transcription-coupled mtDNA replication in both species.
ADE-ing Up mtDNA Instability.
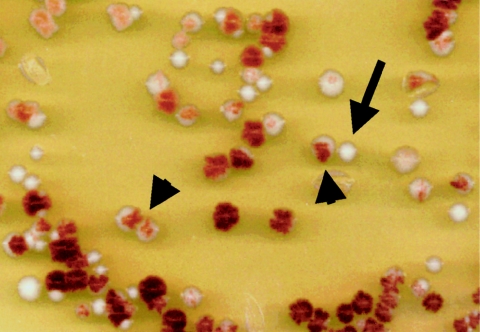
Yeast strains that lack a functioning mitochondrial OXPHOS system are described as petite because they grow slowly and form small colonies. Among the other easily observed features of petite strains are lack of growth on nonfermentable carbon sources and their inability to convert an intermediate in the adenine biosynthesis pathway into a striking red pigment. Normally this intermediate, which accumulates in yeast cells with null mutations in the ADE2 gene, undergoes a series of oxidation and covalent changes and becomes concentrated intracellularly, giving the cell a pink-to-red cast. However, this pigment-formation pathway is defective in ade2− cells that have suffered damage to their mtDNA, because such cells lack OXPHOS. These mtDNA mutant cells, which arise spontaneously in all laboratory yeast strains, form white colonies (indicated by an arrow in the figure below) that stand out in a field of red colonies. If such a mutation occurs during the growth of a single colony, white sectors advertise the presence of mutant cells and allow for a ready estimate of mutation frequency. Two such colonies are indicated by arrowheads in the figure below.
I have used this red-color phenotype during my efforts to better understand the mtDNA instability that results when the mtRNA polymerase (encoded by the nuclear RPO41 gene) is defective. I have shown that rpo41Δ3 yeast cells, which harbor a deletion in the amino-terminal portion of mtRNA polymerase, remain competent for transcription initiation in mitochondria but, nonetheless, exhibit mitochondrial-genome instability (Wang and Shadel 1999). This phenotype can be rescued by supplying the amino-terminal portion of the enzyme in trans, suggesting that this region constitutes a novel functional domain of the enzyme. Thus, an amino-terminal domain of mtRNA polymerase is required to couple mtDNA replication or stability to transcription in mitochondria, making the rpo41Δ3 strain a unique tool for study of the mechanism of transcription-dependent mtDNA replication, a phenomenon that occurs in human cells as well as in yeast. These results hold significance for human mitochondrial genetics, because the human mtRNA polymerase also harbors an amino-terminal extension. Although the yeast and human enzymes show no obvious sequence similarity in their amino-terminal regions, it is likely that an amino-terminal domain of the human enzyme also plays a role in transcription-coupled processes necessary for human mitochondrial-gene expression and mtDNA replication.
Human Mitochondrial-Genome Maintenance
Decreased OXPHOS capacity resulting from mtDNA mutations is a now well-documented cause of human disease. These mutations cover the known spectrum of mtDNA lesions, including point mutations, deletions, duplications, and other rearrangements that, when present in the female germline, can be inherited in a maternal fashion. In addition, mitochondrial respiration defects also can result from nuclear-gene mutations that are inherited in a Mendelian fashion. Nuclear genes that are implicated in mitochondrial dysfunction may encode the OXPHOS components, as well as factors that either are required for assembly or function of the OXPHOS system or participate in expression or maintenance of the mitochondrial genome. Grossman and Shoubridge (1996) have noted that nuclear-gene mutations must be responsible for an mtDNA-depletion syndrome and for a complex assortment of autosomal dominant disorders that result in an increased incidence of multiple mtDNA deletions. Moreover, mtDNA mutations accumulate in common neurological conditions, such as Alzheimer disease and Parkinson disease, and even during the normal aging process (Wallace 1992), suggesting that inability to maintain a functional mitochondrial genome contributes to pathology under certain genetic or physiological conditions. In view of the complete dependence of mitochondrial-genome replication and expression on nuclear-gene products, the number of diseases found to be caused by mutations in nucleus-encoded mtDNA-regulatory factors almost certainly will increase. At present, candidate human disease loci with known or predicted roles in mtDNA transcription or replication include the catalytic subunits of mtRNA and mtDNA polymerase, h-mtTFA, mtSSB, and DNA ligase III. Clearly, additional mtDNA-regulatory factors remain to be identified, including molecules required for mtDNA replication, RNA processing, translation, and mtDNA repair. As in the past, the sophisticated genetic analysis that S. cerevisiae permits will no doubt facilitate progress in this field, perhaps even more so now that whole genome–based analytic strategies can be devised to identify the full complement of mitochondrial proteins in yeast and humans. However, the predictive value of mtDNA-instability phenotypes that result when yeast homologues of human disease genes are disrupted will remain in doubt until mtDNA replication is better understood in both wild-type and ρ− strains. Thus, deciphering the mechanisms of replication of mitochondrial genomes in yeast remains a challenging and a worthwhile goal.
Acknowledgments
I wish to thank Drs. John Ashkenas, Susan Kaech, and Bonnie Seidel-Rogol for critical reading of the manuscript. Support was provided by National Institutes of Health grant HL-59655.
Footnotes
*This article represents the opinion of the authors and has not been peer reviewed.
References
- Baldacci G, Chérif-Zahar B, Bernardi G (1984) The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J 3:2115–2120 [DOI] [PMC free article] [PubMed]
- Blanc H, Dujon B (1980) Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci USA 77:3942–3946 [DOI] [PMC free article] [PubMed]
- Bogenhagen DF (1999) Repair of mtDNA in vertebrates. Am J Hum Genet 64:1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite D, Ito J (1993) Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 21:787–802 [DOI] [PMC free article] [PubMed]
- Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28:693–705 [DOI] [PubMed]
- ——— (1991) Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol 7:453–478 [DOI] [PubMed]
- Costanzo MC, Fox TD (1990) Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet 24:91–113 [DOI] [PubMed]
- de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G (1981) The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature 292:75–78 [DOI] [PubMed]
- Dieckmann CL, Staples RR (1994) Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int Rev Cytol 152:145–181 [DOI] [PubMed]
- Fangman WL, Henly JW, Brewer BJ (1990) RPO41-independent maintenance of [ρ−] mitochondrial DNA in Saccharomyces cerevisiae. Mol Cell Biol 10:10–15 [DOI] [PMC free article] [PubMed]
- Fangman WL, Henly JW, Churchill G, Brewer BJ (1989) Stable maintenance of a 35-base-pair yeast mitochondrial genome. Mol Cell Biol 9:1917–1921 [DOI] [PMC free article] [PubMed]
- Foury F (1989) Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem 264:20552–20560 [PubMed]
- Graves T, Dante M, Eisenhour L, Christianson TW (1998) Precise mapping and characterization of the RNA primers of DNA replication for a yeast hypersuppressive petite by in vitro capping with guanylyltransferase. Nucleic Acids Res 26:1309–1316 [DOI] [PMC free article] [PubMed]
- Greenleaf AL, Kelly JL, Lehman IR (1986) Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci USA 83:3391–3394 [DOI] [PMC free article] [PubMed]
- Grivell L (1995) Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit Rev Biochem Mol Biol 30:121–164 [DOI] [PubMed]
- Grossman LI, Shoubridge EA (1996) Mitochondrial genetics and human disease. Bioessays 18:983–991 [DOI] [PubMed]
- Hermann GJ, Shaw JM (1998) Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol 14:265–303 [DOI] [PubMed]
- Holt IJ, Harding AE, Morgan-Hughes JA (1988) Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331:717–719 [DOI] [PubMed]
- Howell N (1997) mtDNA recombination: what do in vitro data mean? Am J Hum Genet 61:19–22 [DOI] [PMC free article] [PubMed]
- Knight SAB, Kim R, Pain D, Dancis A (1999) The yeast connection to Friedreich ataxia. Am J Hum Genet 64:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson N, Clayton DA (1995) Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet 29:151–178 [DOI] [PubMed]
- Lee DY, Clayton DA (1998) Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem 273:30614–30621 [DOI] [PubMed]
- Li K, Smagula CS, Parsons WJ, Richardson JA Gonzalez M, Hagler HK, Williams RS (1994) Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol 124:871–882 [DOI] [PMC free article] [PubMed]
- Lisowsky T, Michaelis G (1988) A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription in Saccharomyces cerevisiae. Mol Gen Genet 214:218–223 [DOI] [PubMed]
- Lockshon D, Zweifel SG, Freeman-Cook LL, Lorimer HE, Brewer BJ, Fangman WL (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81:947–955 [DOI] [PubMed]
- MacAlpine DM, Perlman PS, Butow RA (1998) The high-mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc Natl Acad Sci USA 95:6739–6743 [DOI] [PMC free article] [PubMed]
- Masters BS, Stohl LL, Clayton DA (1987) Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51:89–99 [DOI] [PubMed]
- Myers AM, Pape LK, Tzagoloff A (1985) Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J 4:2087–2092 [DOI] [PMC free article] [PubMed]
- Parisi MA, Clayton DA (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252:965–969 [DOI] [PubMed]
- Poyton RO, McEwen JE (1996) Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem 65:563–607 [DOI] [PubMed]
- Reenan RA, Kolodner RD (1992) Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132:975–985 [DOI] [PMC free article] [PubMed]
- Shadel GS, Clayton DA (1993) Mitochondrial transcription initiation: variation and conservation. J Biol Chem 268:16083–16086 [PubMed]
- ——— (1997) Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66:409–435 [DOI] [PubMed]
- Tiranti V, Savoia A, Forti F, D'Apolito M, Centra M, Rocchi M, Zeviani M (1997) Identification of the gene encoding the human mitochondrial RNA polymerase (hmtRPOL) by cyberscreening of the expressed sequence tags database. Hum Mol Genet 6:615–625 [DOI] [PubMed]
- Van Dyck E, Clayton DA (1998) Transcription-dependent DNA transactions in the mitochondrial genome of a yeast hypersuppressive petite mutant. Mol Cell Biol 18:2976–2985 [DOI] [PMC free article] [PubMed]
- Wallace DC (1992) Diseases of mitochondrial DNA. Annu Rev Biochem 61:1175–1212 [DOI] [PubMed]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AMS, Elsas LJ II, et al (1988) Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242:1427–1430 [DOI] [PubMed]
- Wang Y, Shadel GS (1999) Stability of the mitochondrial genome requires an amino-terminal domain of yeast mitochondrial RNA polymerase. Proc Natl Acad Sci USA 96:8046–8051 [DOI] [PMC free article] [PubMed]
- Xu B, Clayton DA (1995) A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol Cell Biol 15:580–589 [DOI] [PMC free article] [PubMed]
- Yaffe MP (1999) The machinery of mitochondrial inheritance and behavior. Science 283:1493–1497 [DOI] [PubMed]