Summary
Myxomatous mitral-valve prolapse (MMVP), also called Barlow disease, is a common cardiac abnormality and affects up to 5% of the population. It is characterized by an excess of tissue that leads to billowing of the mitral leaflets, sometimes complicated by prolapse. Typical histological findings include myxomatous degeneration and degradation of collagen and elastin. Previous reports have proposed an autosomal dominant inheritance of the trait, with age- and sex-dependent expression. By systematic echocardiographic screening of the first-degree relatives of 17 patients who underwent mitral-valve repair, we have identified four pedigrees showing such an inheritance. Genomewide linkage analysis of the most informative pedigree (24 individuals, three generations) showed a significant linkage for markers mapping to chromosome 16p, with a two-point maximum LOD score for D16S3068 (Zmax=3.30 at θ=0). Linkage to D16S3068 was confirmed in a second family (Zmax=2.02 at θ=0) but was excluded for the two remaining families, thus demonstrating the genetic heterogeneity of the disease. Multipoint linkage analysis performed, with nine additional markers, on the two families with linkage gave maximum multipoint LOD scores of 5.45 and 5.68 for D16S3133, according to a conservative and a stringent model, respectively. Haplotype analysis defined a 5-cM minimal MMVP-1 locus between D16S3068 (16p11.2) and D16S420 (16p12.1) and a 34-cM maximal interval between D16S404 and D16S3068 when recombination events were taken into account only in affected individuals. The identification of this locus represents a first step toward a new molecular classification of mitral-valve prolapse.
Introduction
Mitral-valve prolapse (MIM 157700) is a widely used term that includes a variety of valvular diseases in which the mitral leaflets fail to appose normally (Barlow and Pocock 1979; Carpentier et al. 1980). This dysfunction is frequently encountered, occurring in 2%–8% of the adult population (Proccaci et al. 1976; Levy and Savage 1987). It generally corresponds to a nonsyndromic idiopathic entity, myxomatous mitral-valve prolapse (MMVP), or Barlow disease. It is characterized by a series of clinical and electrocardiographic features, ranging from no symptoms (in the vast majority of individuals) to mitral regurgitation, ruptured chordae, endocarditis, cerebral embolism, and arrhythmias (Devereux 1976; Bourdarias et al. 1991). The echocardiographic findings are also variable, including mild to marked systolic displacement of the mitral leaflets, with varying mitral-leaflet thickening; dilatation of the mitral annulus; and symptoms ranging from mild telesystolic mitral insufficiency to severe dysfunction, with elongated and/or ruptured chordae needing surgical repair (Alpert et al. 1998). Anatomopathological examination of the mitral leaflets is characterized by tissue redundancy with myxomatous changes (Frable 1969; Kern and Tucker 1972; Virmani et al. 1987). Histological and ultrastructural analyses show nonspecific alterations of collagen and elastin and accumulations of proteoglycans but do not identify the primary cause of the disease (Tamura et al. 1995).
Although most cases appear to be sporadic, echocardiographic screening of families for MMVP has suggested autosomal dominant inheritance of the trait (Shappel et al. 1973; Weiss et al. 1975; Bareiss 1976; Cooper and Abinader 1981) with an age- and sex-dependent expression (Devereux et al. 1982). No systematic search for a locus responsible for MMVP has yet been performed, probably because of the small size of the pedigrees reported and the likely heterogeneity of the trait. Mitral-valve prolapse has also been associated with cardiomyopathies, coronary artery disease, atrial septal defect, and various heritable disorders such as Marfan syndrome, Ehler-Danlos syndrome, and osteogenesis imperfecta (Glesby and Pyeritz 1989). This association with inherited connective-tissue disorders, plus the myxomatous degeneration observed in the mitral valves of patients showing MMVP, has suggested that an abnormality of some component of the connective tissue may be involved (Whittaker et al. 1987; Tamura et al. 1995). Previous linkage studies performed on a few affected pedigrees detected by two-dimensional echocardiograms have excluded the involvement of four collagen genes (Henney et al. 1989; Wordworth et al. 1989). Another rare and severe myxomatous valvular dystrophy inherited in an X-linked fashion (XMVD) has recently been mapped to chromosome Xq28-qter in a large French pedigree (Kyndt et al. 1998).
Here we describe four pedigrees including 27 affected members, showing autosomal dominant transmission of MMVP. We have performed a genomewide linkage analysis on the most informative pedigree and have demonstrated significant linkage in this kindred to marker D16S3068 on chromosome 16. Analysis of the three additional pedigrees has resulted in the confirmation of linkage in one family and exclusion in the remaining families. Refined mapping, by haplotype and multipoint linkage analysis, done in the families with linkage now defines a 5-cM minimal or 34-cM maximal interval for MMVP-1 locus.
Material and Methods
Clinical Evaluation
A double-blind analysis of echocardiograms was performed on the families of 17 probands. Informed consent was obtained from all the phenotyped individuals (approval of the Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale). The propositus member of each family underwent mitral-valve replacement and MMVP was diagnosed at surgery (Carpentier et al. 1980). In relatives, MMVP was diagnosed by two-dimensional echocardiography in the left parasternal long-axis view (Levine et al. 1988). Investigations were recorded on videotapes and twice were examined blindly by two experienced echocardiographists (E.A. and A.B.). The echographic diagnostic criteria included displacement of the leaflet edges, thickness and redundancy of the valve, and the diameter of the mitral annulus. The displacement of each leaflet was measured in the parasternal long-axis view above a line connecting the midportions of the annular hinge points (fig. 1a). The thickness of the mitral valve was measured by M-mode recording (Nishimura et al. 1985) (fig. 1b). To take into account the great variability in the expression of the disease (Levy and Savage 1987), subjects were classified according to the following criteria: (1) a positive diagnosis was supported when subjects had a redundant valve with leaflets >5 mm thick, a total leaflet displacement >8 mm, and evidence of annular dilation; (2) a strong suspicion occurred when subjects had valve thickness of 4–5 mm or total leaflet displacement of 3–8 mm, annular dilatation, and frequently late systolic mitral insufficiency on color flow or continuous wave Doppler; (3) a weak suspicion was recorded when subjects had 3–4 mm displacement without any mitral regurgitation; and (4) unaffected subjects were those with a thickness ⩽4 mm and total leaflet displacement ⩽2 mm. To fit subjects into the three phenotypic classes allowed in linkage analysis, strongly suspect subjects were later considered to be affected, and the weakly suspect subjects to be phenotypically undetermined.
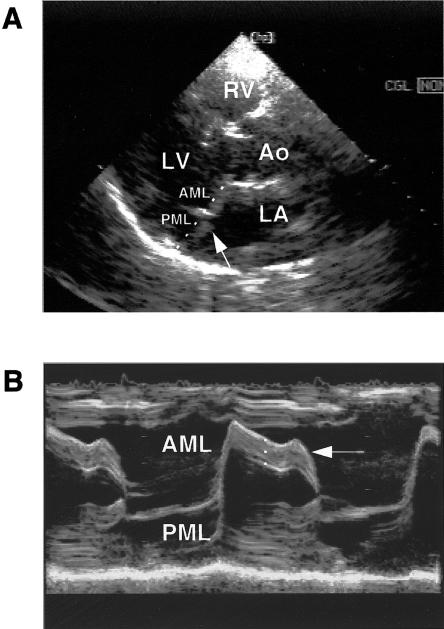
Figure 1.
Echocardiogram of one subject (individual IV.1, kindred 1) with MMVP. a, Two-dimensional echocardiogram in the parasternal long-axis view. The arrow indicates the displacement of the mitral leaflets above the line connecting the midportions of the annular hinge points (shown as a dashed line). RV=right ventricle; LV=left ventricle; Ao=aorta; LA=left atrium; and PML=posterior mitral leaflet. b, M-mode recording in the parasternal long-axis view. The arrow indicates the increased thickness of the AML. Its measurement is performed in the midportion of the leaflet and is shown as a dashed line.
Marker Analysis
Genotyping was performed for 344 microsatellite markers from chromosomes 1 to 16 (kit ABI PRISMTM Linkage Mapping Set, version 2, [PE Biosystems]). The selected markers were polymorphic (heterozygosity = .6–.9), informative, and evenly distributed over the human genome (mean distance between two adjacent markers of 10 cM). Genomic DNA was amplified by PCR with primers that were designed to produce fragments over a broad size range and was labeled with three fluorescent dyes—[FAM], [HEX], and [NED]—to distinguish between overlapping alleles (standard protocol, as indicated by the supplier). Allele sizes were compared between gels, after normalization to control DNA sample CEPH 1347-02. Signal analysis and genotype determination were partly automated by use of GENESCAN and GENOTYPER software.
A similar protocol was used to genotype additional microsatellite markers to obtain a smaller linkage interval. Nine markers (D16S403, D16S420, D16S499, D16S3022, D16S3036, D16S3041, D16S3080, D16S3093, and D16S3133) that mapped to chromosome 16 and that were designed by Généthon were analyzed by use of fluorescent dye–labeled primers. PCR reactions were performed in 7.5-μl volumes containing 50 ng genomic DNA, 1 × GeneAmp® PCR buffer II (Roche Molecular System), 2.5 mM MgCl2, 0.25 mM each dNTP, 0.33 μM each of both primers, and 0.3 U AmpliTaq® Gold DNA polymerase (Roche Molecular Systems). The mix underwent 30 cycles of amplification, under suitable annealing conditions, with a GeneAmp® PCR System 9700 thermal cycler (PE Biosystems).
Statistical Analysis
As a correct description of the trait is critical for detecting linkage (Clerget-Darpoux et al. 1986), and as we observed an incomplete penetrance in young adults in our families, we excluded unaffected subjects aged <40 years (individuals IV.8, IV.9, IV.10, IV.11, V.2, and V.3 of kindred 1; III.1, III.2, III.13, IV.1, IV.3, IV.4, and IV.5 of kindred 3; IV.1 of kindred 4) from statistical analyses and assumed a conservative model of inheritance of the disease. Transmission of the trait was assumed to be autosomal dominant, with a disease-allele frequency of .005 and phenocopies at 1% to take into account the high percentage of sporadic cases. Penetrance was set at 95% for adults aged >40 years. The maximum expected LOD score was estimated by genotype simulation. By use of the SLINK program (Ott 1991), 900 replicates were generated for each pedigree. Two-point LOD scores were then simulated with the MSIM program (Ott 1991). Simulated allele frequencies were set at .3, .2, .2, .1, .1, and .1 (heterozygosity equals .8). Two-point LOD scores for the 344 markers were computed by MLINK from version 5.2 of the LINKAGE package (Lathrop et al. 1985; Ott 1991). Allele frequencies for each marker were obtained online from the Genome Database and were similar to those observed in our white families. Recombination fractions were set as identical for males and females. An admixture test was performed on the four families, by use of the HOMOG1 program (Terwilliger and Ott 1994), with two-point LOD scores obtained at seven different recombination fractions. Multipoint LOD scores were computed by means of the GENEHUNTER program, version 1.1 (Kruglyak et al. 1996), with the assumption of two different models of inheritance: the conservative model previously described and a stringent model characterized by a complete penetrance and no phenocopies. Calculations were performed every 0.5 cM between adjacent markers and 2 cM beyond both frontier markers. Haplotype analysis and pedigree drawing were done with Cyrillic version 2.1 (Cherwell Scientific).
Results
Clinical and Echocardiographic Characteristics
Since the clinical and echocardiographic classifications probably reflect multiple pathological conditions (Glesby and Pyeritz 1989), we restricted our analysis to first-degree relatives of patients who had undergone mitral-valve repair and had surgically proven MMVP. Echocardiographic screening of the families of 17 probands who underwent cardiac surgery identified four pedigrees as having an autosomal dominant transmission, each with 5–9 affected members spanning 3–4 generations (fig. 2). In addition to the probands, who needed mitral-valve repair for complications of MMVP (severe mitral regurgitation caused by chordal rupture, in three cases), most of the affected subjects had either no or only minor symptoms, such as atypical chest pain, dyspnea, or history of palpitations or arrhythmias (table 1). In these four kindreds, there was no morbid association with either thoracic or skin abnormalities that might have evoked a connective-tissue disease or signs of autoimmune disease, except for subject III-3 of kindred 1 (Crohn disease) and subject II-1 of kindred 2 (rheumatoid arthritis). Systematic measurement of the plasma concentration of the thyroxin-stimulating hormone was performed in all subjects and revealed no abnormality. All kindreds were white: kindred 1 was of Ashkenazi Jewish origin; kindred 2 originated in western France, with an ancestor of the 4th generation from Africa; and kindreds 3 and 4 were from the eastern and western parts of France, respectively. In this limited number of subjects and families, there was no clear evidence for sex-dependent penetrance, because 16 of the 25 affected subjects were men and 9 were women. We confirmed an age-dependent penetrance (Weiss et al. 1975; Devereux et al. 1982), because only 6 (33%) of 18 young offspring of the affected subjects expressed the disease, whereas 14 (50%) of 28 subjects of the preceding generation were affected. By definition, a systematic leaflet displacement above the left atrioventricular annulus was observed in the 25 affected relatives of the four kindreds. By contrast, there was an important interfamilial variability of the thickness of the left anterior mitral leaflet (AML): it was >5 mm in all positive patients of kindred 2 (mean 6.8 ± 0.6 mm) but was <5 mm in half (7/14) of the affected relatives of the three remaining families (mean 4.3 ± 1.2 mm, P<.001) (table 1). This increased thickness of the left AML of affected patients in kindred 2 was associated with a tendency for a greater dilatation of the left annulus (35.4 ± 2.2 versus 30.8 ± 3.2 mm, P<.05).
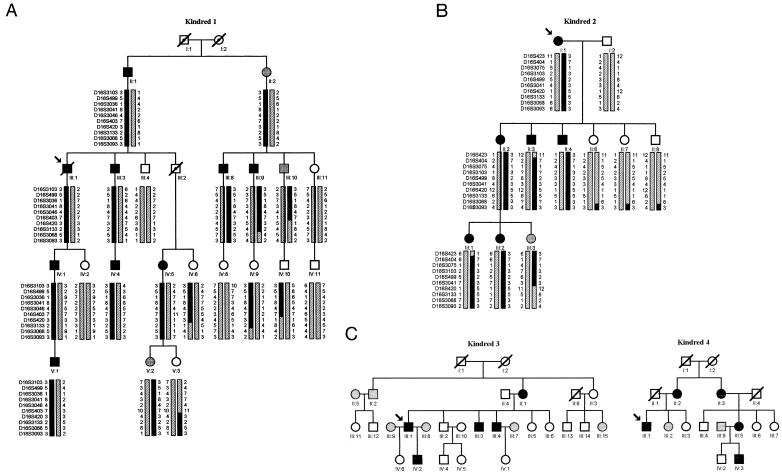
Figure 2.
Pedigrees of four families with MMVP. Females are designated by circles and males are designated by squares. Affected subjects are shown as blackened symbols; individuals whose phenotype was uncertain are shown as gray symbols; unaffected individuals are shown as unblackened symbols; unphenotyped obligate carriers are indicated by a dot within the symbol; and deceased individuals are indicated by a diagonal line across the symbol. Paternal and maternal haplotypes are indicated on the right and left sides, respectively. a, Haplotypes of kindred 1 formed with 10 markers between D16S3103 and D16S3093. b, Haplotypes of kindred 2 formed with 10 markers between D16S423 and D16S3093. c, Genetic status of individuals from kindreds 3 and 4.
Table 1.
Main Clinical and Echocardiographic Characteristics of the Affected Members of the Four MMVP Pedigrees
|
Clinical Findings |
Echocardiographic Parameters |
||||||||||||
| Symptomsb |
Mitral Regurgitationc |
||||||||||||
| Kindred and Individual | Age | Sexa | D | CP | A | Click | Systolic Murmur | Time | Severity | Leaflet Prolapsed | AML Thickness (mm)e | Annulus Diameter (mm)e | Other Localization |
| Kindred 1: | |||||||||||||
| II.1 | 94 | M | + | + | − | No | Yes | H | Severe | Fail A leaflet | ND | ND | No |
| III.1f | 69 | M | + | + | CE | No | Yes | H | Severe | P + fail A | ND | ND | Tricuspid |
| III.3 | 56 | M | + | − | − | No | Yes | M | Mild | A | 2.5 | 32 | No |
| III.8 | 70 | M | − | + | − | No | Yes | L | Mild | P | 3.0 | 25 | No |
| III.9 | 60 | M | + | + | + | No | Yes | L | Mild | P | 4.5 | 32 | No |
| IV.1 | 40 | M | − | + | − | Yes | Yes | M | Moderate | A+P | 6.5 | 34 | No |
| IV.4 | 25 | M | − | − | − | No | No | L | Mild | A | 5.1 | 28 | No |
| IV.5 | 45 | F | + | + | + | No | Yes | L | Mild | P | 3.7 | ND | No |
| V.1 | 15 | F | − | + | − | No | No | No | No | A | 5.0 | 29 | No |
| Kindred 2: | |||||||||||||
| II.1f | 74 | F | + | + | AF | No | Yes | H | Severe | A + fail P | ND | ND | Tricuspid |
| II.2 | 51 | F | + | − | − | Yes | Yes | H | Moderate | A+P | 6.7 | 38 | No |
| II.3 | 39 | M | − | − | − | No | Yes | L | Mild | A+P | 7.4 | 37 | No |
| II.4 | 53 | M | − | − | − | No | No | L | Mild | A | 7.1 | 36 | No |
| III.1 | 25 | F | − | − | − | No | No | No | No | A+P | 7.1 | 36 | No |
| III.2 | 16 | F | + | + | − | Yes | No | L | Mild | A+P | 5.9 | 32 | No |
| Kindred 3: | |||||||||||||
| II.1 | 69 | F | + | + | − | Yes | No | M | Mild | A | ND | ND | No |
| III.1f | 43 | M | + | + | − | Yes | Yes | H | Severe | A+P | 5.4 | 33 | No |
| III.3 | 41 | M | − | − | + | No | No | L | Mild | A | 5.0 | 28 | No |
| III.4 | 42 | M | − | − | + | No | No | L | Mild | A | 3.5 | 31 | No |
| IV.2 | 27 | M | − | − | − | No | No | L | Mild | A | 5.7 | 30 | No |
| Kindred 4: | |||||||||||||
| II.2f | 69 | F | + | + | AF | No | Yes | H | Severe | A+P | ND | ND | No |
| II.3 | 77 | F | − | + | − | No | No | L | Mild | P | 4.0 | 29 | No |
| III.1 | 46 | M | + | − | − | Yes | Yes | M | Moderate | A | 6.1 | 36 | No |
| III.5 | 60 | F | − | + | − | No | No | L | Mild | P | 2.5 | 31 | No |
| IV.2 | 21 | M | + | − | − | Yes | Yes | M | Mild | A+P | 5.1 | 27 | No |
M = male, and F = female.
D = dyspnea, CP = chest pain, A = history of arrythmias, CE = cerebral embolism, AF = atrial fibrillation, + = symptom present, and − = symptom absent.
H = holosystolic, M = midsystolic, and L = late systolic.
A = anterior leaflet, and P = posterior leaflet. In all cases, presence of a fail leaflet was due to chordal rupture.
ND = Not determined.
Index patients who required mitral-valve repair.
Linkage Analysis
Prior evaluation of the maximum expected LOD score for each pedigree showed that kindred 1 alone could lead to significant linkage (expected Zmax=4.66 at recombination fraction [θ] 0), whereas kindreds 2, 3, and 4 could only lead to suggestive linkage (expected Zmax=2.06, 2.54, and 1.43, respectively, at θ=0). We therefore performed a first-stage genomewide scan of 21 subjects in kindred 1. More than 344 microsatellite markers were tested, representing chromosomes 1–16; 327 markers gave negative two-point LOD scores, whereas 6 markers gave LOD scores >1: D2S206 (Zmax=1.16 at θ=0), D16S3075 (Zmax=1.41 at θ=.1), D16S3103 (Zmax=1.99 at θ=0), D16S3046 (Zmax=2.00 at θ=0), D16S3068 (Zmax=3.30 at θ=0), and D16S503 (Zmax=1.35 at θ=.1). The last five markers were all adjacent on chromosome 16 and thus defined a 54-cM region of interest with a significant maximum two-point LOD score for marker D16S3068 (table 2).
Table 2.
Two-Point LOD Scores for Kindred 1, Obtained at Different Recombination Fractions for 13 Marker Loci Ranked from pter to qter on Chromosome 16
|
LOD Score at θ = |
|||||||
| Marker | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D16S423 | −.65 | −.57 | −.29 | −.04 | .23 | .28 | .20 |
| D16S404 | −2.28 | −2.24 | −2.03 | −1.71 | −1.10 | −.63 | −.27 |
| D16S3075 | .94 | 1.08 | 1.35 | 1.41 | 1.24 | .86 | .37 |
| D16S3103 | 1.99 | 1.98 | 1.90 | 1.75 | 1.36 | .89 | .38 |
| D16S3046 | 2.00 | 1.96 | 1.80 | 1.58 | 1.13 | .66 | .23 |
| D16S3068 | 3.30 | 3.24 | 3.00 | 2.69 | 2.03 | 1.32 | .59 |
| D16S3136 | −.40 | −.26 | −.01 | .07 | .07 | .02 | −.01 |
| D16S415 | −.83 | −.46 | .27 | .62 | .78 | .63 | .34 |
| D16S503 | .51 | .94 | 1.31 | 1.35 | 1.12 | .74 | .33 |
| D16S515 | −2.62 | −1.60 | −.61 | −.20 | .08 | .13 | .09 |
| D16S516 | −2.91 | −2.55 | −1.83 | −1.27 | −.58 | −.22 | −.05 |
| D16S520 | −4.39 | −2.84 | −1.87 | −1.41 | −.74 | −.28 | −.06 |
| D16S3091 | −5.91 | −4.15 | −2.36 | −1.38 | −.47 | −.09 | .05 |
We then analyzed the genotypes of D16S3068 for 48 subjects of kindreds 2, 3, and 4. The maximum two-point LOD score for all families, assuming heterogeneity, was 4.33 at θ=.01. We found significant evidence for locus heterogeneity (χ2=5.91, df 1, P<.02) and an estimated proportion of 60% of families having linkage. When each family was analyzed separately (table 3), with the five markers that gave positive LOD scores in kindred 1 (D16S3075, D16S3103, D16S3046, D16S3068, and D16S503), kindred 2 showed suggestive linkage at marker D16S3068 (Zmax=2.02 at θ=0), kindred 4 gave a positive LOD score at marker D16S3103 (Zmax=1.33 at θ=0) but a nonsignificant negative LOD score at marker D16S3068 (Z=-0.29 at θ=0). Kindred 3 led to exclusion of linkage at marker D16S3068 (Z=-2.44 at θ=0). Given the evidence for linkage of MMVP to a locus on the short arm of chromosome 16 and associated heterogeneity, we therefore propose that this locus be designated MMVP-1.
Table 3.
Detailed Two-Point LOD Scores, for Kindreds 1–4, Obtained for Four Marker Loci on the Linkage Interval[Note]
|
LOD Score at θ = |
|||||||
| Kindred and Marker | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| Kindred 1: | |||||||
| D16S3075 | .94 | 1.08 | 1.35 | 1.41 | 1.24 | .86 | .37 |
| D16S3103 | 1.99 | 1.98 | 1.90 | 1.75 | 1.36 | .89 | .38 |
| D16S3046 | 2.00 | 1.96 | 1.80 | 1.58 | 1.13 | .66 | .23 |
| D16S3068 | 3.30 | 3.24 | 3.00 | 2.69 | 2.03 | 1.32 | .59 |
| D16S503 | .51 | .94 | 1.31 | 1.35 | 1.12 | .74 | .33 |
| Kindred 2: | |||||||
| D16S3075 | 1.77 | 1.74 | 1.61 | 1.45 | 1.10 | .70 | .28 |
| D16S3103 | 1.76 | 1.73 | 1.61 | 1.45 | 1.10 | .70 | .29 |
| D16S3046 | .58 | .57 | .53 | .49 | .39 | .28 | .15 |
| D16S3068 | 2.02 | 1.99 | 1.85 | 1.66 | 1.27 | .82 | .34 |
| D16S503 | −.23 | .04 | .41 | .53 | .52 | .37 | .18 |
| Kindred 3: | |||||||
| D16S3075 | NT | … | … | … | … | … | … |
| D16S3103 | NT | … | … | … | … | … | … |
| D16S3046 | NT | … | … | … | … | … | … |
| D16S3068 | −2.44 | −1.94 | −1.00 | −.48 | −.02 | .13 | .12 |
| D16S503 | NT | … | … | … | … | … | … |
| Kindred 4: | |||||||
| D16S3075 | −.95 | −.64 | −.17 | .04 | .16 | .15 | .08 |
| D16S3103 | 1.33 | 1.30 | 1.18 | 1.02 | .70 | .39 | .12 |
| D16S3046 | −3.52 | −1.80 | −.67 | .24 | .05 | .11 | .07 |
| D16S3068 | −.29 | −.24 | −.10 | −.01 | .04 | .03 | .01 |
| D16S503 | −4.55 | −2.93 | −1.48 | −.80 | −.22 | 0 | .05 |
Note.—NT = not tested.
Multipoint and Haplotype Analyses
We refined the location of the MMVP-1 locus by a multipoint linkage analysis of the two families that showed linkage (kindreds 1 and 2) with nine additional markers spanning 25 cM (fig. 3). With the conservative model, a maximum multipoint LOD score of 5.45 was obtained at the location of marker D16S3133 (adjacent to D16S3068) when data on the two families were combined. The multipoint LOD score was also maximum for D16S3133, with the stringent model (Zmax=5.68). The 1-LOD-unit and 3-LOD-unit support intervals spanned 4.6 cM and 48.3 cM, respectively, with the conservative model and spanned 4.2 cM and 21.9 cM with the stringent model.
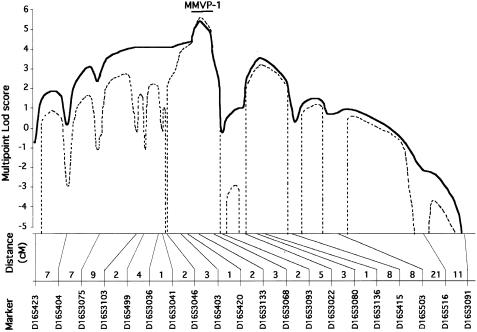
Figure 3.
Refined location of the MMVP-1 locus. A multipoint linkage analysis comparing the segregation of MMVP and 20 marker loci was performed with the assumption of two modes of inheritance. Only families that showed linkage to D16S3068 (kindreds 1 and 2) were included in this analysis. The map of marker loci used and the distances in centimorgans are indicated below the plots of multipoint LOD score; the full curve indicates LOD score calculated with the conservative model and the dotted line indicates LOD score calculated with the stringent model; a bar represents the 1-LOD-unit support interval for the location of MMVP-1.
These results were supported by the haplotype analyses. The MMVP-1 locus was a sex-averaged 5-cM interval between D16S3068 and D16S420 in kindred 1 that included D16S3133 and mapped to chromosome 16p12.1-p11.2, as shown by the recombination events in individuals IV.6 and III.9 (fig. 2a). However, given the age-dependent penetrance of MMVP demonstrated in our families, it must be noted that this minimal interval partly relies on the unaffected individual IV.6. A maximal 34-cM interval can be defined by another recombination event proximal to D16S404 in affected individual III.8. In kindred 2, recombination events in individuals II.3, II.6, II.7, and II.8 defined a larger cosegregation interval (42 cM) that still included the MMVP-1 locus (fig. 2b). Haplotypes were also drawn for the six unaffected subjects from kindred 1 who were aged <40 years (individuals IV.8, IV.9, IV.10, IV.11, V.2, and V.3), although these individuals were not included in the previous statistical analyses; three unaffected subjects aged <30 years had the disease allele, whereas no recombinant was found among older subjects or in the preceding generation. This was compatible with the observation that penetrance of MMVP is low at age <30 years, at least in the families we studied.
The same markers of chromosome 16 were analyzed in kindred 4, since linkage could not be formally excluded in this family. Two-point LOD scores were >1 for two markers, D16S3133 and D16S3103, with the conservative model of inheritance. However, haplotype analysis clearly showed that the MMVP-1 locus described above does not cosegregate with MMVP in kindred 4 (data not shown).
Discussion
This first mapping of a gene for nonsyndromic autosomal dominant mitral-valve prolapse represents a first step toward the identification of genes involved in that heterogeneous group of diseases. Since Barlow's description (1963), a variety of names have been given to mitral-valve prolapse, including “the click-murmur syndrome,” “the floppy mitral-valve syndrome,” “myxomatous mitral valve,” and “idiopathic mitral-valve prolapse.” This reflects the uncertainty about its pathophysiology (Alpert et al. 1998). We decided to use the term “myxomatous mitral-valve prolapse” (MMVP) because our aim was to focus on the abnormality of the mitral-valve structure and not on its possible functional dynamic consequences (i.e., prolapse and/or mitral insufficiency). By excluding patients with connective-tissue disorders (e.g., Marfan and Ehler-Danlos syndromes) by means of a careful clinical examination and by selecting patients related to a proband with surgically proven MMVP and families demonstrating autosomal inheritance, we restricted our analysis to a more homogeneous entity and possibly increased our chance to identify a responsible locus. This strategy proved to be successful and, together with another locus responsible for X-transmitted valvular dystrophy (Kyndt et al. 1998), should help clarify the nosology of mitral-valve prolapse.
The analysis of four pedigrees allowed us to find a positive linkage with the chromosome 16p11.2-12.1 region. A positive two-point linkage was already apparent in kindred 1, with a LOD score of 3.30 at θ=0 with D16S3068. A positive LOD score was also evident in kindred 2, in which we found Z=2.02 at θ=0, for an expected Zmax=2.06. Kindred 4 was more problematic, because we could not exclude linkage. Moreover, with the conservative model, this family had a 40% probability of being linked to D16S3068. We therefore performed a separate haplotype analysis in kindred 4 and found no cosegregation interval that overlapped with the MMVP-1 locus. Even though kindreds 1 and 2 were linked to the same locus, they did not share the same haplotype and had clearly different origins (Central Europe and western France, respectively), thus making the presence of a common ancestral mutation very unlikely.
Penetrance of MVP has been related to age and sex (Devereux et al. 1982). In the largest familial study conducted so far, echocardiographic mitral prolapse was present in 54 (30%) of 179 first-degree relatives of 45 affected probands (Devereux et al. 1982). Its presence was significantly higher in women (41%) than in men (19%) and was more common in adults (35%) than in children (8%). In our limited sample of four families, we could not confirm the effect of sex, as the number of affected men (n=16) was even greater than the number of affected women (n=9). On the contrary, we observed an age-dependent penetrance in our families but could not evaluate it precisely. Therefore, unaffected individuals from the youngest generation were not taken into account in the first-stage genomewide scan. Although less powerful, this method allowed us to demonstrate significant linkage. We also performed various multipoint linkage analyses that included the youngest unaffected subjects, assuming different models of age-dependent penetrance and thus simulating the data observed in our family (low penetrance for young subjects, high penetrance for older subjects). These additional analyses did not alter the previous results, since there was still evidence for linkage to D16S3133 (Z=4.94–5.80, assuming a maximum penetrance after the ages of 30 and 60 years, respectively). Haplotype analyses finally showed that the disease was rarely expressed in young adults aged <30 years.
The mild level of derangement of the mitral-valve structure in the vast majority of subjects (Levy and Savage 1987), together with the possibility of false positive diagnosis by two-dimensional echocardiography (Nishimura et al. 1985; Levine et al. 1988), has led both to various estimations of the prevalence of mitral-valve prolapse and to the need for new surgical (Carpentier et al. 1980) and echocardiographic (Perloff et al. 1986) classifications. In a recent familial echocardiographic study of mitral-valve prolapse, relatives of index cases with AML >5 mm thick were affected significantly more often than were the relatives of other index cases (Zuppiroli et al. 1998). However, leaflet thickening per se was not a heritable characteristic identifying distinct subtypes, because both groups of relatives had similar echocardiographic leaflet characteristics. Our study confirms these findings at the phenotypic and molecular levels. Even though the four probands had typical anatomical features of MMVP at surgery, echocardiographic thickness of the AML varied from one kindred to another, with higher values in affected relatives of kindred 2 compared with those of the other three kindreds. These phenotypic differences did not explain the genetic heterogeneity of MMVP: both kindreds 1 and 2 were linked to the MMVP-1 locus, whereas linkage was excluded in kindred 3 and cosegregation of MMVP-1 locus with MMVP was excluded in kindred 4.
Our study demonstrates the genetic heterogeneity of the autosomal dominant forms of MMVP, and several genes are likely to be implicated in that disease. Thus, the presence of myxomatous tissue and the corresponding histological changes observed in MMVP are probably not specific and have already been attributed to different abnormalities, such as variations in either the morphology of the mitral annulus (Hutchins et al. 1986) or the thickness of the zona spongiosa of the mitral leaflet (Olsen and Al-Rufaie 1980). Mitral-valve prolapse has been found in several, but not all, affected members of a large family with dilated cardiomyopathy mapping to chromosome 10q21-23 (Bowles et al. 1996). In this family, it was not possible to distinguish whether the association was coincidental, secondary to the ventricular dysfunction, or part of a genetic defect causing both dilated cardiomyopathy and mitral-valve prolapse. Mitral-valve prolapse has also been associated with connective-tissue disorders and a variety of congenital intracardiac anomalies (Glesby and Pyeritz 1989).
The genes involved in the structure or development of atrioventricular valves are therefore candidate genes for MMVP. However, none of the corresponding genes have been mapped to a region that could correspond to the MMVP-1 locus. Advances in the Human Genome Project should facilitate a positional candidate-gene approach. So far, the Institute of Genome Research has mapped more than 24 genes to the region overlapping the MMVP-1 5-cM interval. It is interesting to note that among these genes, sialophorin, the CD11 integrin cluster, CD19, and the interleukin-4 receptor are involved in cellular adhesion and the immune response and have been suggested as candidate genes for Crohn disease, an inflammatory bowel disease that maps to the pericentromeric region of chromosome 16 (Hugot et al. 1996). Cardiotrophin-1, a member of the family of cytokines, is present in the heart during embryogenesis (Pennica et al. 1995) and may play a role in cardiac hypertrophy. In addition, >10 genes with as-yet-unknown products and a large number of ESTs could provide other reasonable candidates. The gene putatively responsible for pseudoxanthoma elasticum has been mapped to a region overlapping our more secure 35-cM interval (Struk et al. 1997). Although vascular and cardiac lesions have been reported in that disease, they do not resemble those observed in MMVP. Finally, the myosin heavy-chain 11 and other genes coding for transcription factors could also be evaluated.
Acknowledgments
The authors thank all the family members who participated in this study; C. Lucas and R. de Bavinchove, for secretarial work; and E. Genin, F. Clerget-Darpoux, and A. Vuagnat, for helpful discussions on the statistical analyses. This work was supported by PHRC (Programme Hospitalier de Recherche Clinique, Assistance Publique, Hôpitaux de Paris) grant AOA94046.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Généthon, http://www.genethon.fr/ (for microsatellite markers)
- Genome Database (GDB), http://www.gdb.org/ (for allele frequencies)
- Institute of Genome Research, http://www.tigr.org/ (for genomic sequences of the MMVP-1 locus)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for MMVP [MIM 157700])
References
- Alpert JS, Sabik J, Cosgrove DM (1998) Mitral valve disease. In: Topol EJ (ed) Textbook of cardiovascular medicine. Lippincott-Raven, Philadelphia, pp 519–528 [Google Scholar]
- Bareiss P (1976) Familial forms of the mid-end systolic click and murmur syndrome with deviations of left ventricular kinetics. Arch Mal Coeur Vaiss 69:71–81 [PubMed]
- Barlow JB, Pocock WA (1979) Mitral valve prolapse, the specific billowing mitral leaflet syndrome, or an insignificant non-ejection systolic click. Am Heart J 97:277–285 [DOI] [PubMed]
- Barlow JB, Pocock WA, Marchand P, Denny M (1963) The significance of late systolic murmur. Am Heart J 66:443–452 [Google Scholar]
- Boudarias JP (1991) Mitral valve prolapse: a severe abnormality? Arch Mal Coeur Vaiss 84:981–986 [PubMed]
- Bowles KR, Garjaski R, Porter P, Goyta V, Bachinski L, Roberts R, Pignatelli R, et al (1996) Gene mapping of familial autosomal dominant dilated cardiomyopathy to chromosome 10q21-23. J Clin Invest 98:1355–1360 [DOI] [PMC free article] [PubMed]
- Carpentier A, Chauvaud S, Fabiani JN, Deloche A, Relland J, Lessana A, D'Allaines C, et al (1980) Reconstructive surgery of mitral valve incompetence: ten-year appraisal. J Thorac Cardiovasc Surg 79:338–348 [PubMed]
- Clerget-Darpoux F, Bonaïti-Pellier C, Hochez J (1986) Effects of misspecifying genetic parameters in lod score analysis. Biometrics 42:393–399 [PubMed]
- Cooper MJ, Abinader EG (1981) Family history in assessing the risk for progression of mitral valve prolapse. Report of a kindred. Am J Dis Child 135:647–649 [DOI] [PubMed]
- Devereux RB (1976) Mitral valve prolapse. Circulation 54:3–14 [DOI] [PubMed]
- Devereux RB, Brown WT, Kramer-Fox R, Sachs I (1982) Inheritance of mitral valve prolapse: effect of age and sex on gene expression. Ann Intern Med 97:826–832 [DOI] [PubMed]
- Frable WJ (1969) Mucinous degeneration of the cardiac valve, the floppy valve syndrome. J Thorac Cardiovasc Surg 58:62–70 [PubMed]
- Glesby MJ, Pyeritz RE (1989) Association of mitral valve prolapse and systemic abnormalities of connective tissue: a phenotypic continuum. JAMA 262:523–528 [PubMed]
- Henney AM, Tsipouras P, Schwartz RC, Child AH, Devereux RB, Leech GJ (1989) Genetic evidence that mutations in the COL1A1, COL1A2, COL3A1, or COL5A2 collagen genes are not responsible for mitral valve prolapse. Br Heart J 61:292–299 [DOI] [PMC free article] [PubMed]
- Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, et al (1996) Mapping a susceptibility locus for Crohn's disease on chromosome 16. Nature 379:821–823 [DOI] [PubMed]
- Hutchins GM, Moore GW, Skoog DK (1986) The association of floppy mitral valve with disjunction of the mitral annulus fibrosus. N Engl J Med 314:535–540 [DOI] [PubMed]
- Kern WH, Tucker BL (1972) Myxoid changes in cardiac valves, pathologic, clinical and ultrastructural studies. Am Heart J 84:294–301 [DOI] [PubMed]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed]
- Kyndt F, Schott JJ, Trochu JN, Baranger F, Herbert O, Scott V, Fressinaud E, et al (1998) Mapping of X-linked myxomatous valvular dystrophy to chromosome Xq28. Am J Hum Genet 62:627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans, detection of linkage and estimation of recombinations. Am J Hum Genet 37:482–498 [PMC free article] [PubMed]
- Levine RA, Stathosgiannis E, Newell JB, Harrigan P, Weyman AE (1988) Reconsideration of echocardiographic standards for mitral valve prolapse: lack of association between leaflet displacement isolated to the apical four chamber view and independent echocardiographic evidence of abnormality. J Am Coll Cardiol 11:1010–1019 [DOI] [PubMed]
- Levy D, Savage DD (1987) Prevalence and clinical features of mitral valve prolapse. Am Heart J 113:1981–1990 [DOI] [PubMed]
- Nishimura RA, McGoon MD, Shub C, Miller FA Jr, Ilstrup DM, Tajik AJ (1985) Echocardiographically documented mitral valve prolapse. N Engl J Med 313:1305–1309 [DOI] [PubMed]
- Olsen EG, Al-Rufaie HK (1980) The floppy mitral valve: study on pathogenesis. Br Heart J 44:674–683 [DOI] [PMC free article] [PubMed]
- Ott J (ed) (1991) Analysis of human genetic linkage. Revised ed. John Hopkins University Press, Baltimore, MD [Google Scholar]
- Pennica D, King KL, Shaw KJ, Luis E, Rullamas J, Luoh SM, Darbonne WC, et al (1995) Expression cloning of cardiotrophin 1, a cytokine that induces cardiac myocyte hypertrophy. Proc Nat Acad Sci USA 92:1142–1146 [DOI] [PMC free article] [PubMed]
- Perloff JK, Child JS, Edwards JE (1986) New guidelines for the clinical diagnosis of mitral valve prolapse. Am J Cardiol 57:1124–1129 [DOI] [PubMed]
- Procacci PM, Savran SV, Schreiter SL, Bryson AL (1976) Prevalence of clinical mitral-valve prolapse in 1169 young women. N Engl J Med 294:1086–1088 [DOI] [PubMed]
- Shappell SD, Marshall CE, Brown RE, Bruce TA (1973) Sudden death and the familial occurrence of mid-systolic click, late systolic murmur syndrome. Circulation 48:1128–1134 [DOI] [PubMed]
- Struk B, Neldner KH, Rao VS, St Jean P, Lindpaintner K (1997) Mapping of both autosomal recessive and dominant variants of pseudoxanthoma elasticum to chromosome 16p13.1. Hum Mol Genet 6:1823–1828 [DOI] [PubMed]
- Tamura K, Fukuda Y, Ishizaki M, Masuda Y, Yamanaka N, Ferrans VJ (1995) Abnormalities in elastic fibers and other connective-tissue components of floppy mitral valve. Am Heart J 129:1149–1158 [DOI] [PubMed]
- Terwilliger JD, Ott J (eds) (1994) Handbook of human genetic linkage. John Hopkins University Press, Baltimore, MD [Google Scholar]
- Virmani R, Atkinson JB, Forman MB, Robinowitz M (1987) Mitral valve prolapse. Hum Pathol 18:596–602 [DOI] [PubMed]
- Weiss AN, Mimbs JW, Ludbrook PA, Sobel BE (1975) Echocardiographic detection of mitral valve prolapse, exclusion of false positive diagnosis and determination of inheritance. Circulation 52:1091–1096 [DOI] [PubMed]
- Whittaker P, Boughner DR, Perkins DG, Canham PB (1987). Quantitative structural analysis of collagen in chordae tendinae and its relation to floppy mitral valves and proteoglycan infiltration. Br Heart J 57:264–269 [DOI] [PMC free article] [PubMed]
- Wordsworth P, Ogilvie D, Akhras F, Jackson G, Sykes B (1989) Genetic segregation analysis of familial valve prolapse shows no linkage to fibrillar collagen genes. Br Heart J 61:300–306 [DOI] [PMC free article] [PubMed]
- Zuppiroli A, Roman MJ, O'Grady M, Devereux RB (1998) A family study of anterior mitral leaflet thickness and mitral valve prolapse. Am J Cardiol 82:823–826 [DOI] [PubMed]