Summary
Niemann-Pick type II disease is an autosomal recessive disorder characterized by a defect in intracellular trafficking of sterols. We have determined the intron/exon boundaries of eight exons from the conserved 3′ portion of NPC1, the gene associated with most cases of the disease. SSCP analyses were designed for these exons and were used to identify the majority of mutations in 13 apparently unrelated families. Thirteen mutations were found, accounting for 19 of the 26 alleles. These mutations included eight different missense mutations (including one reported by Greer et al. [1998]), one 4-bp and two 2-bp deletions that generate premature stop codons, and two intronic mutations that are predicted to alter splicing. Two of the missense mutations were present in predicted transmembrane (TM) domains. Clustering of these and other reported NPC1 mutations in the carboxy-terminal third of the protein indicates that screening of these exons, by means of the SSCP analyses reported here, will detect most mutations. The carboxy-terminal half of the Npc1 protein shares amino acid similarity with the TM domains of the morphogen receptor Patched, with the largest stretch of unrelated sequence lying between two putative TM spans. Alignment of this portion of the human Npc1 protein sequence with Npc1-related sequences from mouse, yeast, nematode, and a plant, Arabidopsis, revealed conserved cysteine residues that may coordinate the structure of this domain. That 7 of a total of 13 NPC1 missense mutations are concentrated in this single Npc1-specific domain suggests that integrity of this region is particularly critical for normal functioning of the protein.
Introduction
Niemann-Pick type II disease is a fatal lipid-storage disorder with autosomal recessive inheritance (Spence and Callahan 1989; Pentchev et al. 1995). The underlying defect is a disruption in the intracellular trafficking of sterols, which leads to an accumulation of unesterified cholesterol within the lysosomal compartment of the cell (Liscum and Klansek 1998; Morris and Carstea 1998; Watari et al. 1999). This perturbation of sterol homeostasis particularly affects the functioning of cells in the brain, liver, and spleen. Although the age at onset of Niemann-Pick type II disease is variable, affected individuals normally present clinically, at preschool age, with ataxic gait and hepatosplenomegaly, with eventual progression to dementia, dysarthria, dystonia, and seizures. Affected individuals typically die during their teen years. Traditionally, Niemann-Pick type II disease has been divided into two entities: type C (NPC [MIM 257220]) and type D (NPD [MIM 257250]). NPD, the Nova Scotia variant, was distinguished by its less severe phenotype and by the Acadian ancestry of affected individuals (Winsor and Welch 1978; Greer et al. 1999).
Complementation studies, in which somatic-cell hybrids of fibroblasts from different patients with Niemann-Pick type II disease were assayed for correction of their biochemical defect, provided evidence for at least two genes, mutations in either of which result in clinically and biochemically indistinguishable disease (Steinberg et al. 1994; Vanier et al. 1996). The NPC1 gene, which is associated with ∼80% of cases, has recently been isolated from the proximal region of chromosome 18q, by use of a strategy of linkage analysis and positional cloning (Carstea et al. 1993, 1997). NPD was subsequently demonstrated to be an allelic variant of NPC1 (Greer et al. 1997, 1998).
Although the function of the Npc1 protein is unknown, the sequence of the NPC1 gene predicts a 1,278-amino-acid protein with 13–16 possible transmembrane (TM) domains; a signal sequence that would direct its synthesis at the endoplasmic reticulum; a leucine zipper motif, located in the amino-terminal region, that may mediate interactions with additional protein(s); and a carboxy-terminal dileucine (LLNF) lysosomal targeting signal (Carstea et al. 1997) (fig. 1). Orthologues sharing sequence conservation with the human Npc1 protein throughout their length have been identified in nematode, yeast, and mouse (Carstea et al. 1997; Loftus et al. 1997), which suggests that the function of this protein has also been conserved through evolution. The Npc1 protein shares significant similarity, throughout most of its TM spans, with the TM domains of a plasma membrane–localized protein, Patched1 (Ptch1) (Johnson et al. 1996; Carstea et al. 1997) (fig. 2). The PTCH1 gene has also been conserved, through evolution, with homologues and a paralogue, the PTCH2 gene, which has been identified in Drosophila, mice, and humans (Goodrich et al. 1996, reviewed in Hammerschmidt et al. 1997). Ptch1 functions both in embryonic development and in tumor suppression, acting as part of the receptor for Sonic hedgehog, a secreted protein originally identified, in Drosophila, by its role in the determination of segment polarity. A region with homology to five of the consecutive TM spans shared between Ptch1 and Npc1 is also found in two other proteins, SCAP (sterol regulatory element–binding protein [SREBP] cleavage-activating protein) and 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (Carstea et al. 1997), both of which are regulated by sterols. This common domain has been implicated in the ability of these latter two endoplasmic reticulum–localized proteins to sense sterol levels. Higher cholesterol levels increase the susceptibility of HMG-CoA reductase, a key enzyme in the sterol biosynthetic pathway, to proteolysis (Goldstein and Brown 1990), whereas lower cholesterol levels trigger activation of SCAP, which leads to proteolytic release of the transcription factors required for activation of genes encoding proteins involved in sterol synthesis and uptake (Brown and Goldstein 1997). That this sterol-sensing domain is present in Npc1 suggests that it may also be regulated by the level of sterols in the cell.
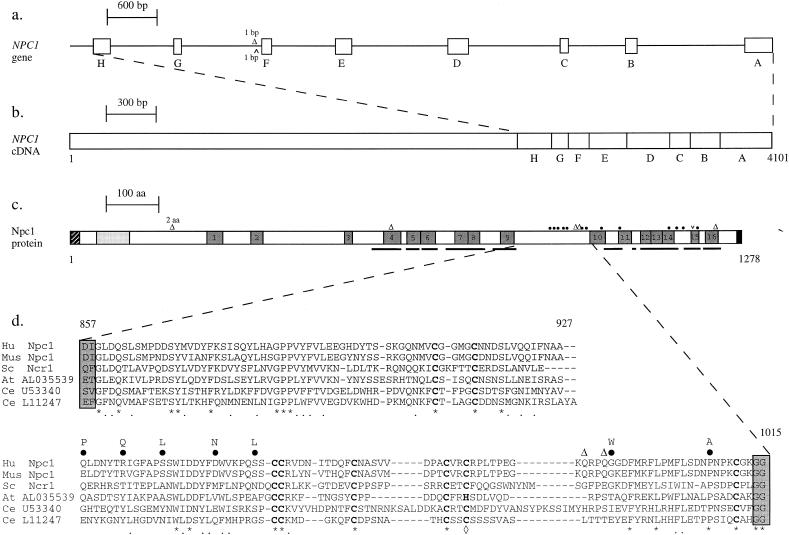
Figure 1.
NPC1 mutations. Maps of (a) the 3′ end of the NPC1 gene and (b) the cDNA are shown. The positions of exons A–H are represented by squares, and introns are represented by lines. The positions of two intronic mutations are indicated. c, Diagram of the Npc1 protein aligned with the cDNA, showing the correspondence between the exons and their encoded amino acid sequence. Predicted domains within the Npc1 protein are indicated by squares shaded as follows: squares with diagonal stripes represent signal sequence; light gray–shaded squares, leucine zipper; dark gray–shaded squares, TM regions; and blackened squares, lysosomal targeting signal (Carstea et al. 1997). Regions of the Npc1 protein sharing >25% amino acid identity with the human Ptch1 protein are underlined. The positions of all reported NPC1 mutations in the coding sequence are represented along the protein as follows: blackened circles, missense mutations; unblackened triangles, deletions; and carats, insertions. Unless otherwise indicated, deletions and insertions result in frameshifts that introduce premature stop codons. d, Alignment of the amino acid sequence for the human (Hu) Npc1 domain between TM spans 9 and 10 with the corresponding regions from Npc1-related sequences from mouse (Mus Npc1), S. cerevisiae (Sc Ncr1), A. thaliana (GenBank accession number AL035539), and C. elegans (GenBank accession numbers U53340 and L11247). Asterisks indicate an identical residue at that position in all six sequences; dots, conservative amino acid substitutions; diamond, a position where a cysteine occurs in all sequences except that of A. thaliana, for which a histidine is substituted; dashes, gaps that are introduced to maximize the alignment; blackened circles above the alignment (with the letter, above the blackened circle, indicating the amino acid substitution), positions of missense mutations identified in this region in patients with Niemann-Pick type II disease; and unblackened triangles, positions of frameshifts created by deletions.
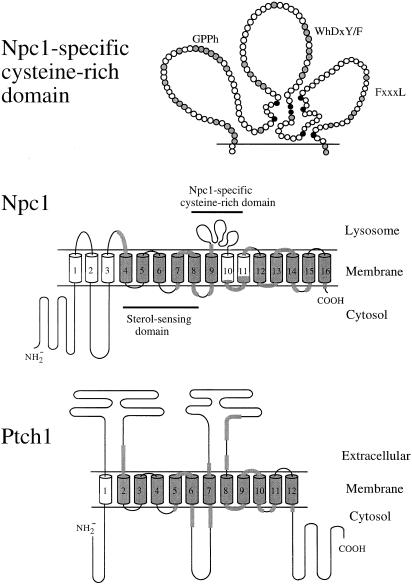
Figure 2.
Comparison of human Npc1 and Ptch1 proteins. A model for the organization of Npc1 within the lysosomal membrane is contrasted with the proposed topology of human Ptch1 in the plasma membrane (Goodrich et al. 1996; Johnson et al. 1996). The model is a revision of that presented by Beachy et al. (1997). In the bottom portion of this figure, predicted TM domains for Npc1 (Carstea et al. 1997) and Ptch1 are numbered and are shown as cylinders. Stretches of amino acid sequence sharing >25% identity between the two proteins are gray shaded. A region with homology to sterol-sensing domains in Ptch1 (TM spans 2–6), HMG-CoA reductase, and SCAP (Carstea et al. 1997) is indicated. The cysteine-rich Npc1-specific domain is overlined and is shown in an expanded form at the top of the figure. To allow the conserved cysteine residues to be in close proximity to one another, the domain is depicted as three hypothetical loops. Blackened circles represent cysteine residues, and gray-shaded circles represent other positions conserved between human Npc1 and the five Npc1-related sequences. Conserved amino acid–sequence motifs in this domain are indicated by the single-letter code for the amino acid—by h for hydrophobic residues or by x for any amino acid.
Carstea et al. (1997) previously reported eight NPC1 mutations in nine unrelated patients. With the exception of a 6-bp deletion and a frameshift mutation, all identified mutations were in the carboxy-terminal third of the protein. That the NPD mutation is also located in the 3′ end of the coding region (Greer et al. 1998) suggests that a large proportion of patients with Niemann-Pick type II disease have mutations affecting the latter third of the gene. Here, we report the intron/exon boundaries of the eight exons from this region, the development of SSCP analyses for use in the detection of mutations in this portion of NPC1, and 12 new NPC1 mutations.
Patients, Material, and Methods
Patients
All patients evaluated in this study were referred either with a diagnosis of Niemann-Pick type II or for evaluation of the possibility of this diagnosis. These patients included one individual from Nova Scotia (3114), two from New Brunswick (3081 and 3093), seven from central or western Canada (3118–3122, 3124, and 3125), and one from Maryland (3123). Diagnosis was confirmed biochemically, at the Izzak Walton Killam (IWK) Grace Health Centre, by measurement of LDL-stimulated cholesterol esterification and by filipin fluorescence (Pentchev et al. 1995). Biochemical defects were scored either as “severe” (characterized by intense perinuclear filipin staining and absence of cholesterol esterification, relative to that seen in NPC control cell line GM3123) or as “moderate” (characterized by intermediate filipin staining and/or partial LDL stimulation, for 6–18 h, of cholesterol esterification, as noted elsewhere [Sidhu et al. 1993] for NPD fibroblasts). DNA samples were also obtained from the parents of an affected NPC patient whose paternal grandparents (3034 and 3035) lived in Nova Scotia and from affected siblings (3056 and 3066) with reported Nova Scotian ancestry. The 13 families were apparently unrelated. DNA was extracted from peripheral blood lymphocytes or from fibroblast cell cultures that were collected with ethical approval.
Characterization of Intron/Exon Boundaries
The entire NPC1 cDNA was amplified, in three sections, by means of PCR; it was then cloned in a TA cloning vector (pCR 2.1 Invitrogen) and was sequenced to confirm that it matched the sequence reported elsewhere (Carstea et al. 1997). Primers generated from the cDNA sequence (GenBank AF002020) were used to obtain genomic amplicons, which were also cloned and sequenced. Intron/exon boundaries flanking exons A–H (fig. 1) from the 3′ end of the gene were identified in a comparison of the sequence of genomic DNA clones with that of cDNA. The size of the intervening sequences was estimated on the basis of the size of PCR amplicons generated from genomic DNA that span adjacent exons.
SSCP Analysis
The sequence of the introns flanking exons A–H was used to generate primers for use in SSCP analysis of each exon. Primers are described in table 1. All eight exons were amplified under the following conditions: initial denaturation at 95°C for 5 min and 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. Reaction mixtures (20 μl) contained 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 500 μg BSA/ml, 250 μM each dNTP, 5 μCi α[32P]-dCTP, 0.5 U Taq polymerase (Gibco BRL), 1 μM each primer, and 500–1,000 ng genomic DNA. (Exons B and C were amplified with an alternate buffer: 25 mM Tris [pH 8.3], 2 mM MgCl2, 1 mM DTT, and 50 mM KCl.) Amplicons from all exons were analyzed by means of SSCP, according to methods described elsewhere (Hayashi 1991). In brief, mutations were detected by separation of denatured amplicons on 8% nondenaturing polyacrylamide gels in both Tris-borate EDTA buffer and varied glycerol concentrations—0%, 5%, or 10%. These gels were run for 17 h at 3W, 5W, or 7W, respectively.
Table 1.
Primer Sequences for SSCP Analyses of Exons A–H
| Exon | cDNA Positionof Exona | Primer Sequences | AmpliconSize (bp) | GenBank Accession Number |
| A | 3755–4101 | Forward, TCC AAA GTG GGA TTA CAGGReverse, CCT TAG ACA CAG TTC AGT CAGG | 209 | AF125033 |
| B | 3592–3754 | Forward, GGT TGG TAA AAG TGG GTTCReverse, GAA AAG AAT GCC TCA GGATA | 284 | AF125034 |
| C | 3478–3591 | Forward, GGG TGC CCT GGG TAATReverse, CTT TGT GGT GCG ACT CTG | 250 | AF125035 |
| D | 3246–3477 | Forward, GCG AGC TTT AAT GAG GCC TCCReverse, CCA AAC TGA GAC TGT ATG AGG A | 294 | AF125036 |
| E | 3042–3245 | Forward, TGC TTA GCC TCA AGT GCTCReverse, CCC TTT GCT GGT AAA CCC | 324 | AF125037 |
| F | 2912–3041 | Forward, CTT CTA ACA GTC CTC CCT GCAReverse, CTG TCT TAG CCC AGT CCT CTC | 248 | AF125038 |
| G | 2796–2911 | Forward, GTG GAG CAG GTC TGT AAC CReverse, CAG CAA AAC GGG AAA GAT | 190 | AF125039 |
| H | 2605–2795 | Forward, TGT TCA CAC TCT CTC CTA TCT TCTAReverse, CCT CCG CTG CTT CTG A | 268 | AF125040 |
On the cDNA, where the A of the initiation codon is taken as number 1.
Sequence Analysis
The sequence of amplified products with aberrant SSCP patterns was determined by use of a Thermo Sequence radiolabeled terminator cycle sequencing kit (Amersham Life Science) and by separation of the labeled reactions on denaturing polyacrylamide gels and by exposure of the gels to x-ray film for 24 h. Sequences related to human Npc1 (GenBank accession number AAB63982) were identified by means of BLASTP searches (Altschul et al. 1997) and were aligned with the use of CLUSTAL W (1.7) (Thompson et al. 1994). Related sequences that were used in the alignments are as follows: mouse Npc1 (GenBank accession number AAB63372 ), yeast (Saccharomyces cerevisiae) Ncr1 (GenBank accession number S52525), human Ptch1 (GenBank accession number AAC50550), and three putative proteins identified in genome sequences from Arabidopsis thaliana (GenBank accession number AL035539) and Caenorhabditis elegans (GenBank accession numbers U53340 and L11247). The amino acid sequences that we derived for the putative proteins from AL035539 and L11247 differ from those determined by use of the Genefinder program (authors' unpublished data). Throughout its length, the A. thaliana sequence is as closely related to the human and mouse Npc1 proteins as it is to the yeast Ncr1 protein, and it is more similar to the vertebrate sequences than are either of the C. elegans sequences (authors' unpublished data).
Results
NPC1 Genomic Structure
Overlapping genomic DNA clones were generated with the use of PCR primers previously used for sequencing NPC1 cDNA. By comparison of the genomic DNA and cDNA sequences, eight exons (designated “A”–“H”) were identified in the carboxy-terminal end of NPC1, which is where the majority of mutations had been identified in a previous study (Carstea et al. 1997). The position of each exon in the cDNA is indicated in table 1. Base-pair positions are numbered, starting at the initiating ATG codon of the NPC1 open reading frame. The genomic structure of this region and the approximate size of intervening sequences are shown in figure 1a.
Mutation Analysis
To allow the examination of the NPC1 coding region and splice sites for mutations associated with each patient, primers were generated from intronic sequence. Each of the eight carboxy-terminal exons and their flanking intervening sequences were PCR amplified, with the use of the primers shown in table 1 and with genomic DNA used as a template; they were then assayed for SSCP. Amplicons with abnormal migration were sequenced.
Thirteen patients (two brothers represented one “patient,” and both parents of a deceased patient represented another “patient”), comprising a total of 26 alleles, were analyzed. Nineteen mutant alleles were identified and are described in table 2. Nine alleles had unique mutations, two alleles were found for each of three further mutations, and one mutation accounted for four alleles. Only one patient was found to be a homozygote.
Table 2.
Mutations in Families with NPC
| Family | Patient (Sex) | Exon | Mutation | Predicted Protein Alteration | Biochemical Defecta |
| 1 | 3114 | EF | T3182CG2974T | Ile1061ThrGly992Trp | Moderate |
| 2 | 3081 | BG | T3637GG2842A | Leu1213ValAsp948Asn | Severe |
| 3 | 3093 | E | T3182C | Ile1061Thr | Severe |
| 4 | 3125 | EG | T3182CG2801A | Ile1061ThrArg934Gln | Severe |
| 5 | 3124 | G | C2861T | Ser954Leu | Severe |
| 6 | 3121 | … | None detected | None | Severe |
| 7 | 3120 | …… | Intronic insertionbIntronic deletionb | Splicing defect?Splicing defect? | Severe |
| 8 | 3123 | G | C2819T | Ser940Leu | Moderate |
| 9 | 3118 | FF | C3019GC3019G | Pro1007AlaPro1007Ala | Moderate |
| 10 | 3119 | F | ΔAG (2972–2973) | Frameshift | Severe |
| 11 | 3122 | F | ΔAG (2963–2964) | Frameshift | Severe |
| 12 | 3034 (Female)3035 (Male) | GB | G2842AΔACTC (3741–3744) | Asp948AsnFrameshift | ND |
| 13 | 3056/3066 | EG | T3182CG2801A | Ile1061ThrArg934Gln | ND |
ND = no data.
For details, see text.
One of the mutations identified in family 12, a 4-bp deletion (ΔACTC3741–ΔACTC3744), and the 2-bp deletions detected in families 10 and 11 (ΔAG2972–ΔAG2973 and ΔAG2963–ΔAG2964, respectively) generated frameshifts and premature stop codons. Eight missense mutations were identified. One of these missense mutations, found in patient 3114, was a G2974→T transversion in exon F, described elsewhere as the NPD mutation (Greer et al. 1997). The same patient's second allele showed a T3182→C transition that replaces a hydrophobic isoleucine at position 1061 by a hydrophilic threonine residue in a predicted TM domain. This latter allele was also observed in patients 3056, 3093, and 3125. A second predicted TM domain was altered by a T3637→G transversion that changed Leuc1213Val in patient 3081. The remaining five missense mutations (Arg934Gln, Ser940Leu, Asp948Asn, Ser954Leu, and Pro1007Ala), along with the NPD mutation Gly992Trp, generate nonconservative amino acid changes and are located in a single region flanked by the putative TM spans 9 and 10. The two remaining mutations potentially alter splicing in both alleles of patient 3120. One allele has a 1-bp deletion 10 bp upstream from exon F, whereas the second allele has a 1-bp insertion 9 bp upstream from exon F. As indicated in table 2, all mutations were identified in exons E, F, or G, with the exception of two mutations in exon B and the two intronic mutations that were 3′ to exon F. Mutations were not identified in any of the exons analyzed in patient 3121. Fifty unaffected individuals were screened, by SSCP, for the mutations reported here, none of which were found.
Distribution of Identified Mutations in NPC1
The positions of all NPC1 mutations identified in the present study, plus the positions of those previously reported (Carstea et al. 1997; Greer et al. 1998), are shown, with respect to the Npc1 protein sequence, in figure 1. To determine the likely significance of the apparent clustering of the missense mutations, particularly in the region between Npc1 TM regions 9 and 10, the amino acid sequences of the five Npc1 orthologues and putative homologues that were identified from BLASTP searches were aligned with the human Npc1 sequence, to ascertain whether the altered residues represented conserved positions (fig. 1d). Only two of the eight missense mutations (Pro1007Ala and Leu1213Val) affected conserved amino acids.
Discussion
The recent identification of the NPC1 gene (Carstea et al. 1997) has provided an opportunity to elucidate the nature of the mutations responsible for Niemann-Pick type II disease. We have identified the intron/exon boundaries of eight exons from the conserved 3′ portion of the coding sequence. On the basis of this finding, SSCP analysis was designed and was used to identify 19 of 26 NPC1 mutant alleles from 13 families. This may be an underestimation of the number of mutations in this region, since some mutations may not result in altered electrophoretic mobility of the DNA, in SSCP analysis. This may explain the inability to detect an NPC1 mutation in one patient (3121) referred for diagnosis. Alternatively, this individual may have mutations either in a more proximal portion of the NPC1 gene or in another gene, such as NPC2, that would create a similar phenotype.
The two mutant alleles reported in patient 3114 were identified by sequencing the entire coding region of the respective cDNAs. Since both mutations result in nonconservative amino acid substitutions and since no other sequence change was noted, they almost certainly represent the causative defects. Indeed, one allele was previously confirmed to be the NPD mutation (Greer et al. 1998); the other allele alters the charge within a predicted TM span and therefore may affect interaction of the protein with the membrane.
The remaining 11 mutations were identified by means of SSCP analysis of eight exons from the 3′ region of the gene. It is possible that some of these changes represent silent polymorphisms and that causative mutations exist elsewhere in the gene; however, at least three of the reported mutations can reasonably be expected to disrupt gene function. The 2- and 4-bp deletions identified in families 10, 11, and 12 result in frameshifts and premature stop codons that would lead to the expression of truncated Npc1 proteins lacking the carboxy-terminal lysosomal targeting motif. Transfection of a Chinese hamster ovary NPC-model cell line (CT60) with a human NPC1 gene in which this motif was deleted resulted in expression of protein that accumulated at the endoplasmic reticulum and that failed to correct the cholesterol-trafficking defect of these cells (Watari et al. 1999). Transport of the protein to the lysosome therefore appears to be essential for expression of its biological activity; at the very least, all three deletions would be expected to disrupt Npc1 function, as a result of mislocalization. In addition, the frameshifts created by the deletions might also have effects on protein structure and function.
The amino acid change (Leu1213Val) identified in patient 3081 is conservative. This type of change would usually represent a nondisruptive polymorphism; however, a leucine at this position in the putative TM domain is invariant among the human, mouse, yeast, Arabidopsis, and C. elegans homologues of Npc1 and at the homologous position in all available vertebrate Ptch1 and Ptch2 sequences. Thus, it may be critical for proper functioning of the protein.
In patient 3120, a mutation in the intronic sequence close to the 3′ end of exon F was identified in each allele. Sequence analysis showed both the deletion of a thymidine nucleotide 10 bp upstream of exon F in one allele and the insertion of a deoxyadenosine nucleotide 9 bp upstream of exon F in the other allele. Both alterations reduce the length of an already vulnerably short tract of pyrimidines adjacent to the consensus sequence at the 3′ splice site and could potentially disrupt splicing of this intron.
Six missense mutations that produce nonconservative amino acid substitutions were identified in a single 155-amino-acid domain flanked by predicted TM spans 9 and 10 (fig. 1). What is striking is that 7 of the 13 reported missense mutations (Carstea et al. 1997; Greer et al. 1998; present study) are clustered within an 80-amino-acid stretch. Although this part of the protein sequence shows similarity between human, mouse, Arabidopsis, C. elegans, and yeast NPC1 homologues, only one of the mutations, Pro1007Ala, alters a residue that is conserved at that position in the six Npc1-related sequences compared here. This region contains eight cysteine residues that are conserved in all six sequences, with the exception of a single site, in the Arabidopsis NPC1 orthologue, where histidine is substituted for cysteine (fig. 2). The conservation of these residues indicates that they may be important in the determination of the folding of this inter-TM domain, either through formation of intramolecular disulfide bonds or by coordination of a metal ion. The latter possibility is supported by the substitution of a histidine for a cysteine in one of the putative orthologues. The cysteine-rich motif C-x(4–5)-C-x(39–43)-C-C-x(8–10)-C-x(5–13)-C-x2-C/H-x(28–40)-C (where x can be any amino acid and where numbers refer to the length of the intervening sequence), which was conserved here, bears some resemblance to a motif known as the “RING finger” (Freemont 1993) and, also, to a cysteine-rich motif in the regulatory domain of protein kinase C (PKC) (Ono et al. 1989). The eight conserved residues (7Cys/1His or 6Cys/2His) of the RING-finger motif and the PKC regulatory domain bind two zinc atoms and fold into a single domain that is similar to that of the better-characterized zinc fingers; however, the sequence and length of the loops between the zinc ligands are less conserved than they are in the zinc-finger family of protein motifs (Hubbard et al. 1991; Freemont 1993). Although RING-finger domains are found in many proteins involved in gene regulation, their postulated role in DNA binding has not been established. These motifs may, instead, mediate protein-protein or protein-lipid interactions. The integrity of the RING-finger domain has been shown to be essential for the function of two yeast proteins, Vps18/Pep3 and Vps11/Pep5, both of which are required for biogenesis of vacuoles (the yeast equivalent of the lysosome) and for sorting and transport of vacuolar enzymes (Robinson et al. 1991). Interestingly, mutation of human Npc1 also leads to a lysosomal defect, although, in this case, trafficking of cholesterol is affected. The cysteine-rich regulatory domain of PKC has been shown to be essential in the binding of phorbol esters to the enzyme (Ono et al. 1989), so it is possible that the cysteine-rich motif in Npc1 may similarly coordinate a structure capable of direct interaction with cholesterol.
Five of the NPC1 missense mutations flank a conserved Trp-(Ile/Leu)-Asp-x-(Tyr/Phe) motif, located at position 942–946, that lies between two pairs of cysteine residues within the cysteine-rich domain. The amino acid changes produced by these mutations do not alter conserved residues. However, they may affect the folding of this subdomain and the relative positioning of the conserved motif with respect to the rest of the domain or to other protein(s) that interact with Npc1, thereby disrupting Npc1 function.
The cysteine-rich domain is contained within a region of Npc1 that shares no sequence similarity to the human Ptch1 protein (fig. 2). The analogous region in Ptch1 forms an extracellular domain that interacts with Sonic hedgehog and that may be regulated by the adjacent sterol-sensing domain (Johnson et al. 1996). Although the Npc1 protein is targeted to the lysosome (Neufeld et al. 1999; Watari et al. 1999) rather than to the plasma membrane, the extended sequence similarity of the majority of the TM spans of the two proteins suggests that they should be similarly oriented in the membrane, placing the Npc1 domain lying between TM spans 9 and 10 in the lysosomal compartment, rather than in the cytosol (fig. 2). This topology would give the Npc1 sterol-sensing domain TM spans 4–8 the same orientation, with respect to their cytosolic contacts, as that which is seen in the homologous sterol-sensing domains in both HMG-CoA reductase and SCAP in which the membrane topology has been determined (Olender and Simon 1992; Nohturfft et al. 1998). It is tempting to speculate that the Npc1 sterol-sensing domain regulates the ability of the adjacent cysteine-rich conserved domain to mediate, either directly or indirectly, the transport of cholesterol out of the lysosome, in response to altered sterol levels in the lysosomal membrane.
The 13 NPC1 mutations described here are different from the 8 NPC1 mutations reported elsewhere (Carstea et al. 1997). From this small sample, it appears that a wide diversity of mutations are associated with Niemann-Pick type II disease; however, since the majority (19 of 21) of those mutations now reported are located in the distal third of the gene, an initial mutational analysis of the eight exons described here may be the most rapid approach toward establishing a diagnosis and carrier status within newly identified families. In addition, these observations suggest a critical role for this portion of the protein, particularly the cysteine-rich conserved domain within this region, in the mediation of Npc1 function.
Acknowledgments
The authors would like to thank the families and patients who participated in this study; Robert Zwicker, for growing cell lines; and Karen Cleveland, for secretarial support. This research was supported by the Queen Elizabeth II Health Sciences Centre and the Medical Research Council of Canada.
Note added in proof.—Morris et al. (1999) have recently reported the genomic organization of the complete NPC1 gene.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for the NPC1 cDNA sequence [accession number AF002020]; sequences related to human Npc1 [accession number AAB63982], mouse Npc1 [accession number AAB63372], yeast Ncr1 [accession number S52525], and human Ptch1 [accession number AAC50550]; genome sequences from A. thaliana [accession number AL035539] and from C. elegans [accession numbers U53340 and L11247]; and sequences of amplicons shown in table 1 [accession numbers AF125033–AF125040])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for NPC [MIM 257220] and NPD [MIM 257250])
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed]
- Beachy PA, Cooper MK, Young KE, von Kessler DP, Park W-J, Hall TM, Leahy DJ, et al (1997) Multiple roles of cholesterol in hedgehog protein biogenesis and signaling. Cold Spring Harb Symp Quant Biol 62:191–204 [PubMed]
- Brown M, Goldstein JL (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 [DOI] [PubMed]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, et al (1997) Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277:228–231 [DOI] [PubMed]
- Carstea ED, Polymeropoulos MH, Parker CC, Detera-Wadleigh SD, O'Neill RR, Patterson MC, Goldin E, et al (1993) Linkage of Niemann-Pick disease type C to human chromosome 18. Proc Natl Acad Sci USA 90:2002–2004 [DOI] [PMC free article] [PubMed]
- Freemont PS (1993) The RING finger: a novel protein sequence motif related to the zinc finger. Ann NY Acad Sci 684:174–192 [DOI] [PubMed]
- Goldstein JL, Brown M (1990) Regulation of the mevalonate pathway. Nature 343:425–430 [DOI] [PubMed]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP (1996) Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10:301–312 [DOI] [PubMed]
- Greer WL, Riddell DC, Byers DM, Welch JP, Girouard GS, Sparrow SM, Gillan TL, et al (1997) Linkage of Niemann-Pick disease type D to the same region of human chromosome 18 as Niemann-Pick disease type C. Am J Hum Genet 61:139–142 [DOI] [PMC free article] [PubMed]
- Greer WL, Riddell DC, Gillan TL, Girouard GS, Sparrow SM, Byers DM, Dobson MJ, et al (1998) The Nova Scotia (type D) form of Niemann-Pick disease is caused by a G3097→T transversion in NPC1. Am J Hum Genet 63:52-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer WL, Riddell DC, Murty S, Gillan TL, Girouard GS, Sparrow SM, Tatlidil C, et al (1999) Linkage disequilibrium mapping of the Nova Scotia variant of Niemann-Pick disease. Clin Genet 55:248–255 [DOI] [PubMed]
- Hammerschmidt M, Brook A, McMahon AP (1997) The world according to hedgehog. Trends Genet 13:14–21 [DOI] [PubMed]
- Hayashi K (1991) PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl 1:34–38 [DOI] [PubMed]
- Hubbard SR, Bishop WR, Kirschmeier P, George SJ, Cramer SP, Hendrickson WA (1991) Identification and characterization of zinc binding sites in protein kinase C. Science 254:1776–1779 [DOI] [PubMed]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, et al (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272:1668–1671 [DOI] [PubMed]
- Liscum L, Klansek JJ (1998) Niemann-Pick disease type C. Curr Opin Lipidol 9:131–135 [DOI] [PubMed]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, et al (1997) Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277:232–235 [DOI] [PubMed]
- Morris JA, Carstea ED (1998) Niemann-Pick C disease: cholesterol handling gone awry. Mol Med Today 4:525–531 [DOI] [PubMed]
- Morris JA, Zhang D, Coleman KG, Nagle J, Pentchev PG, Carstea ED (1999) The genomic organization and polymorphism analysis of the human Niemann-Pick C1 gene. Biochem Biophys Res Commun 261:493–498 [DOI] [PubMed]
- Neufeld EB, Wastney M, Patel S, Suresh S, Cooney AM, Dwyer NK, Roff CF, et al (1999) The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J Biol Chem 274:9627–9635 [DOI] [PubMed]
- Nohturfft A, Brown MS, Goldstein JL (1998) Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J Biol Chem 273:17243–17250 [DOI] [PubMed]
- Olender EH, Simon RD (1992) The intracellular targeting and membrane topology of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem 267:4223–4235 [PubMed]
- Ono Y, Fujii T, Igarashi F, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y (1989) Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci USA 86:4868–4871 [DOI] [PMC free article] [PubMed]
- Pentchev PG, Vanier MT, Suzuki K, Patterson MC (1995) Niemann-Pick disease type C: a cellular cholesterol lipidosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 9th ed. McGraw-Hill, New York, pp 2625–2639 [Google Scholar]
- Robinson JS, Graham TR, Emr SD (1991) A putative zinc finger protein, Saccharomyces cerevisiae Vps18p, affects late Golgi functions required for vacuolar protein sorting and efficient α-factor prohormone maturation. Mol Cell Biol 12:5813–5824 [DOI] [PMC free article] [PubMed]
- Sidhu H, Rastogi SAR, Byers DM, Guernsey DL, Cook HW, Palmer FBStC, Spence MW (1993) Regulation of low density lipoprotein receptor and 3-hydroxy-3-methyl-glutaryl-CoA reductase activities are differentially affected in Niemann-Pick type C and type D fibroblasts. Biochem Cell Biol 71:467–474 [DOI] [PubMed]
- Spence MW, Callahan JW (1989) Sphingomyelin-cholesterol lipidoses: the Niemann-Pick group of diseases. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 6th ed. McGraw-Hill, New York, pp 1655–1676 [Google Scholar]
- Steinberg SJ, Ward CP, Fensom AH (1994) Complementation studies in Niemann-Pick disease type C indicate the existence of a second group. J Med Genet 313:317–320 [DOI] [PMC free article] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed]
- Vanier MT, Duthel S, Rodriguez-Lafrasse C, Penchev P, Carstea ED (1996) Genetic heterogeneity in Niemann-Pick C Disease: a study using cell hybridization and linkage analysis. Am J Hum Genet 58:118–125 [PMC free article] [PubMed]
- Watari J, Blanchette-Mackie EJ, Dwyer NK, Glick JM, Patel S, Neufeld EB, Brady RO, et al (1999) Niemann-Pick C1 protein: obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc Natl Acad Sci USA 96:805–810 [DOI] [PMC free article] [PubMed]
- Winsor EJT, Welch JP (1978) Genetic and demographic aspects of Nova Scotia Niemann-Pick disease (type D). Am J Hum Genet 30:530–538 [PMC free article] [PubMed]