Summary
Despite the fact that cataracts constitute the leading cause of blindness worldwide, the mechanisms of lens opacification remain unclear. We recently mapped the aculeiform cataract to the γ-crystallin locus (CRYG) on chromosome 2q33-35, and mutational analysis of the CRYG-genes cluster identified the aculeiform-cataract mutation in exon 2 of γ-crystallin D (CRYGD). This mutation occurred in a highly conserved amino acid and could be associated with an impaired folding of CRYGD. During our study, we observed that the previously reported Coppock-like–cataract mutation, the first human cataract mutation, in the pseudogene CRYGE represented a polymorphism seen in 23% of our control population. Further analysis of the original Coppock-like–cataract family identified a missense mutation in a highly conserved segment of exon 2 of CRYGC. These mutations were not seen in a large control population. There is no direct evidence, to date, that up-regulation of a pseudogene causes cataracts. To our knowledge, these findings are the first evidence of an involvement of CRYGC and support the role of CRYGD in human cataract formation.
Introduction
The genetic approach toward a better understanding of cataract formation has led to the identification of 13 cataract-gene loci and the characterization of seven genes (Brakenhoff et al. 1994; Cartier et al. 1994; Litt et al. 1997, 1998; Hejtmancik 1998; Kannabiran et al. 1998; Shiels et al. 1998; Mackay et al. 1999; Stephan et al. 1999). The γ-crystallin–genes cluster constitutes a family of seven genes encoding major structural proteins of the lens (Papaconstantinou 1967; Piatigorsky 1981, 1984). Of these, the γ-crystallin genes (CRYG) represent an interesting group of candidate genes for aculeiform cataract (MIM 115700), because of (1) their location in the disease-gene interval that we have previously mapped on chromosome 2q33-35 (Héon et al. 1998), (2) their role in maintenance of lens transparency and development, and (3) their association with cataract formation in mice and humans (Cartier et al. 1994; Hejtmancik 1998). Specifically, mutations in the CRYG genes (CRYGA, -B, and -E) have been associated with various murine cataract models (Cartier et al. 1992; Santhiya et al. 1995; Klopp et al. 1998). In humans, a group of sequence changes in the promoter region of CRYGE has been associated with the Coppock-like–cataract phenotype (Brakenhoff et al. 1994) (see table 1). Unlike the situation in the mouse, the human γE is a pseudogene by virtue of an in-frame stop codon in exon 2. However, these sequence changes have been referred to as the “CCL mutation,” since they were found to be associated with a 10-fold increase in the expression of γE. This is the first human cataract mutation that has been identified, but it also is the first suggestion that the up-regulation of a pseudogene could be disease causing.
Table 1.
Mutation Analysis of the CRYGE-Promoter Region in Controls
|
No. of Controls with |
|||||
| Ethnic Group/Haplotype | Haplotype 1a | Haplotype 2b | Haplotype 3c | Haplotype 4d | No. of Alleles |
| Swiss | 23 | 17 | 7 | 1 | 48 |
| Western European | 20 | 14 | 12 | 0 | 46 |
| Eastern European | 3 | 5 | 2 | 0 | 10 |
| Afro-Carribean | 3 | 1 | 4 | 0 | 8 |
| East Indian | 2 | 6 | 0 | 0 | 8 |
| Chinese | 2 | 0 | 4 | 0 | 6 |
| Total | 53 (42.1%) | 43 (34.1%) | 29 (23%) | 1 (.8%) | 126e |
tataTa/cCg.
tataCa/cCg.
tataTa/cTg (proposed Coppock-like–cataract mutation, 5′ to exon 1 [Brakenhoff et al. 1994]).
tataCa/cTg.
An additional 9 control individuals were sequenced and found to be double heterozygotes.
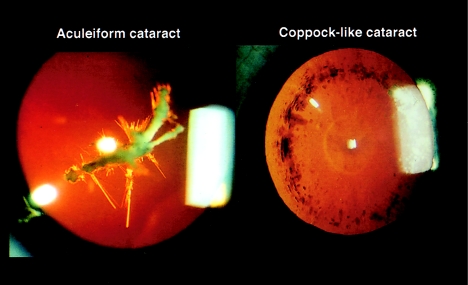
Coppock-like cataract (MIM 116200 and MIM 123660) and aculeiform cataract are distinct clinical entities, in terms of type of lens opacity and their respective natural histories (fig. 1) (Nettleship and Ogilvie 1906; Vogt 1922). They both have an autosomal dominant mode of transmission, but the aculeiform-cataract phenotype is characterized by needlelike lens opacification involving the superficial layer of the fetal nucleus and perhaps the embryonic nucleus, whereas Coppock-like cataract refers to a dustlike opacity of the fetal nucleus with frequent involvement of the zonular lens. A third phenotype, progressive polymorphic cataract (MIM 601286), also has been mapped to the 2q33-35 locus (Rogaev et al. 1996). This entity also is clinically distinct from Coppock-like cataract and aculeiform cataract, and its genetic defect remains unknown. In an attempt to identify the aculeiform-cataract–gene defect, a mutational analysis of CRYG genes was performed in three unrelated affected families whose defect previously had been mapped to the 2q33-35 region (Héon et al. 1998).
Figure 1.
Clinical example of an aculeiform cataract (left) and a Coppock-like cataract (right), shown by transillumination slit-lamp photography.
Subjects and Methods
Patients and Controls
This study was approved by the Ethics Committee of the Faculty of Medicine of the University of Lausanne. Genomic DNA was isolated from peripheral blood by means of methods described elsewhere (Miller et al. 1988).
Direct Sequencing of CRYGB, -C, -D, and -E
Individuals affected with aculeiform cataract, as well as unaffected family members, of previously described pedigrees (Héon et al. 1998) were screened for mutations in CRYGB, -C, -D, and -E, by direct cycle sequencing (Mitchell and Warshawsky 1998). Gene-specific PCR primers were designed to amplify the three exons and flanking intron sequences of crystallin γB, -C, -D, and -E.
CRYGE
Because of the high sequence homology of exon 3 of CRYGE and CRYGF, TaqI restriction-enzyme digestion of genomic DNA was required prior to amplification, to ensure primer specificity. Each PCR reaction contained 100 ng DNA, 200 μM of each dNTP, 5% dimethyl sulfoxide (DMSO), 10 pmol of each primer (one of which was 5′ biotin labeled), GeneAmp® PCR Buffer II, 1.0–1.5 mM MgCl2, and 1 U AmpliTaq polymerase (Applied Biosystems), in 20 μl. Reactions were performed on an MJ Research PTC-100 or Stratagene Robocycler for 35 cycles, after an initial denaturing step (94°C for 5 min) and “hot start” addition of AmpliTaq, variable annealing temperatures, and a final 8-min extension at 72°C. PCR and sequence primers are available on request.
CRYGD
Amplification of CRYGD (exons 1 and 2), required “touchdown” PCR. A 737-bp PCR fragment covering exons 1 and 2 of CRYGD was amplified by use of the following primers: forward, 5′-gccgttttacaaacattctc-3′; reverse, 5′-gttattgtgactgatcgctac-3′ (5′ biotin labeled). Exon 2 sequencing primers were as follows: forward, 5′-gaaggtgagcccagcctgcg-3′; reverse, 5′-tctaatgtttaacttttgcttg-3′. PCR products were purified by use of Dynabeads® M-280 Streptavidin, were resuspended in 20 μl H2O, and were sequenced by use of the Thermo Sequenase™ Cycle Sequencing Core Kit (model US79610; Visible Genetics). The exon 2 sequencing reactions, which included 2 μl template, 10% DMSO, and a nested forward or reverse Cy5.5-labeled primer, were performed for 35 cycles after an initial denaturing step (94°C for 3 min), variable annealing temperatures, and a final 8-min extension at 70°C. The products were analyzed by a MicroGene Blaster™ automated DNA sequencing unit (Visible Genetics). Sequence variants were analyzed in both directions.
CRYGC
A 784-bp fragment covering exons 1 and 2 of CRYGC was amplified with the following primers: forward, 5′-tcaatcatatagacagagcca-3′; and reverse, 5′-atgtccatctaacccttaggt-3′ (5′ biotin labeled). The sequencing reactions for exon 2 were similar to those used for CRYGD—with primers 5′-gagcagaacacaaattaaat-3′ (forward) and 5′-aatgcaaacctccctccctg-3′ (reverse).
Mutation-Detection Assay for CRYGD and CRYGC
CRYGD
Allele-specific amplification (also known as “ARMS”) (Newton et al. 1989) was used to screen all family members and controls, for the proposed mutation at 411G→A in exon 2 of CRYGD. The forward primer had a 3′ nucleotide mismatch—5′-gcctccagtacttcctgca-3′—designed specifically to amplify the mutant allele and, together with the reverse-PCR primer, amplified a 194-bp mutant allele–specific fragment in affected individuals. Primers for the STRP marker D1S1663 (409–425 bp) were included in each reaction, as a positive control for PCR.
The standard PCR reaction contained 25 ng genomic DNA and 1.75 mM MgCl2 and had an annealing temperature of 59°C. Products were electrophoresed on 1% agarose gel, were stained with ethidium bromide, and then were visualized by UV transillumination.
CRYGC
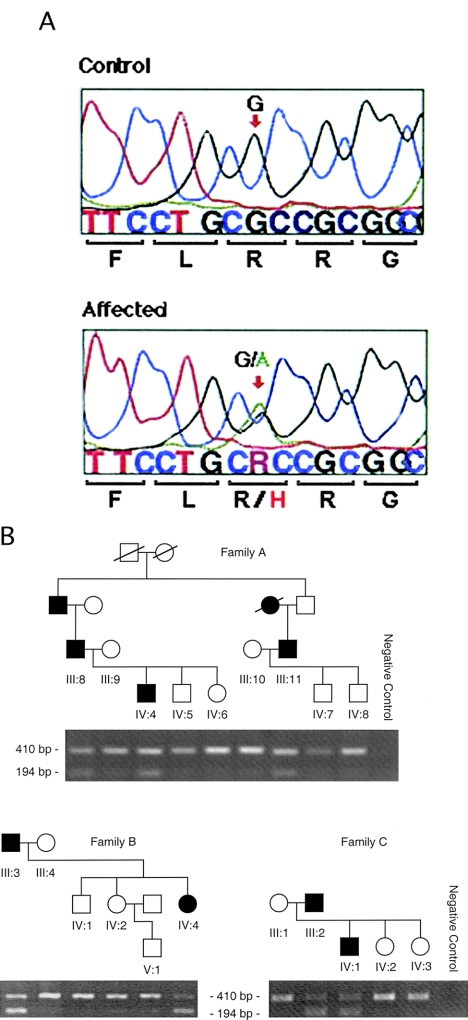
The wild-type DNA contains HphI restriction-enzyme sites that cut the 784-bp PCR product into fragments of 344, 82, and 358 bp, which are seen as one band on a stained agarose gel. The mutation disrupts the second restriction-enzyme site, and products of size 344 and 440 bp are observed as two bands on a stained agarose gel (fig. 2B). HphI restriction-enzyme digestion after PCR amplification was used to screen all family members (nine affected, seven unaffected, and seven spouses) and controls, for the proposed mutation in exon 2 of CRYGC.
Figure 2.
Mutational analysis of CRYGC. a, Sequence chromatograms showing the single-base-pair substitution 225A→C of CRYGC exon 2, found in individuals affected with the Coppock-like–cataract phenotype. b, HphI restriction-enzyme digestion of CRYGC exon 2 of a branch of the original Coppock-like–cataract family. The wild-type fragment contains HphI restriction-enzyme sites that cut the product into fragments of 344, 82, and 358 bp, which would show one band on the agarose gel. The 225A→C mutation disrupts the second restriction-enzyme site, and products of 344 and 440 bp are observed as two bands on a stained agarose gel. Blackened and unblackened symbols represent affected and unaffected individuals, respectively. Symbols are numbered on the basis of the generation identifiers in the original publication (Lubsen et al. 1987), with all individuals numbered from left to right in ascending order.
The standard 20-μl PCR reaction contained 50 ng genomic DNA and 1.5 mM MgCl2, had an annealing temperature of 59°C, and comprised 38 cycles. A 5-μl portion of amplified product was digested in a 10-μl total volume for 3 h. Products were electrophoresed on 2% agarose gel with ethidium bromide staining and then were visualized by UV transillumination.
Three-Dimensional Protein Prediction
When the tertiary structure of several γ-crystallins was delineated by x-ray crystallography, it was found to be highly conserved. Bovine γ-B, -D, and -F have 77%–87% identity to human CRYGD and were used as a reference for homology modeling. Models were viewed in a Swiss-Pdb Viewer, version 3.1. Homology modeling was done by SWISS-MODEL, version 2.0.
Results
Mutation Identification of Aculeiform Cataract
Mutation analysis of the γ-crystallin–genes cluster started with CRYGE. A variety of sequence changes were observed in the promoter region of CRYGE (table 1), none of which appeared to be disease specific. The sequence changes associated with the Coppock-like–cataract phenotype (CCL mutation) were seen in one affected individual and in one unaffected spouse. Furthermore, screening of a control population, characterized by individuals with no lens opacification and of various ethnic backgrounds (126 informative chromosomes), showed that the promoter region of CRYGE was very polymorphic. The CCL mutation was observed in 23% of cases. Direct sequencing of the coding sequence of CRYGE, CRYGB, and CRYGC failed to document any disease-causing mutation. The single-nucleotide polymorphisms identified in these genes are listed in table 2.
Table 2.
Single-Nucleotide Polymorphisms Observed in CRYGB, -C, -D, and -E
| Crystallin Gene (Accession Number)and Nucleotide | Variation | Amino Acid |
| γB (M19364): | ||
| 2104 | t/c | Noncoding |
| 2437 | ccC/ccT | Pro |
| γC (M19364): | ||
| 18229 | a/g | Noncoding |
| γD (K03005): | ||
| 286 | taT/taC | Tyr |
| γE (S72943): | ||
| 165 | c/t | Noncoding |
| 179 | c/t | Noncoding |
| 205 | t/g | Noncoding |
| 244 | c/t | Noncoding |
| 406 | caG/caA | Gln |
| 432 | gCg/gTg | Ala/Val |
| 455 | Gtg/Atg | Val/Met |
| 506 | g/t | Noncoding |
| 164 | c/a | Noncoding |
| 188 | t/c | Noncoding |
| 214 | g/c | Noncoding |
| 318 | a/g | Noncoding |
Direct sequencing of CRYGD showed a G→A transition at nucleotide position 411 in exon 2, which substituted the highly conserved amino acid arginine for a histidine (R58H) (fig. 3A). This sequence change cosegregated with the aculeiform-cataract phenotype in the three families (fig. 3B) and was not observed in 210 control individuals of various ethnic backgrounds, in 44 samples from patients affected with age-related cataracts, or in 39 samples from patients with sporadic congenital cataracts of various subtypes (data not shown). This amino acid is found in a region that is highly conserved in the different CRYGs (CRYGA, -B , -C, -D, -E, and -F) (data not shown) and is invariant through various species (table 3).
Figure 3.
Mutational analysis of CRYGD. a, Sequence chromatograms showing the single-base-pair substitution 411G→A of CRYGD (exon 2; nucleotides 404–418) found in individuals affected with the aculeiform-cataract phenotype. b, Mutational analysis (i.e., ARMS) of CRYGD exon 2. The ARMS assay shows cosegregation of the 411G→A mutation with the aculeiform-cataract phenotype in three Swiss families (families A–C). Selective amplification of a 194-bp PCR product for the 411G→A mutation is seen in affected individuals only. The 410-bp fragment amplified with primers for the marker D1S1663 is an internal control for the PCR reaction and is present in all lanes.
Table 3.
Sequence Homology of CRYGD Amino Acid Residues 48–66 and 170–172
|
Sequencea |
|||||
| Species | Crystallin | 5556605805 | // | 111177770123 | Homologyb(%) |
| Man | γD | PNYSGLQYFLRRGDYADHQ | // | IDFS | 100 |
| Bovine | γD (ΙΙΙΒ) | ...L.P.........P.Y. | // | ..IY | 87 |
| Rat | γD (2−2) | ..FT.C.........P.Y. | // | ..IY | 85 |
| Mouse | γD (1) | ..FA.C.........P.Y. | // | M..Y | 83 |
| Cat shark | γ−Μ1 | ..FM.M.F.....E.H.M. | // | T.MC | 49 |
| Common carp | γ−Μ1 | NS.M.N.F.....E.H.M. | // | T.MC | 47 |
| γ−Μ2 | ...M.N...F...E...YM | // | M.SW | 50 | |
| γ−Μ3 | ...M.N.F.M...E...YM | // | M.LC | 50 | |
| Teleostean fish | γΜ1−1 | // | T.MC | 51 | |
| Catfish | γcry | ..FM.M.F.....E.H.M. | // | T.IC | 51 |
| Common frog | γ1 | ...T.H.......E.P.F. | // | QEMF | 57 |
| a From GenBank. Amino acid residue 58 is the site of the aculeiform-cataract mutation. | |||||
| b Of total protein sequence. | |||||
Mutation Identification of Coppock-Like Cataract
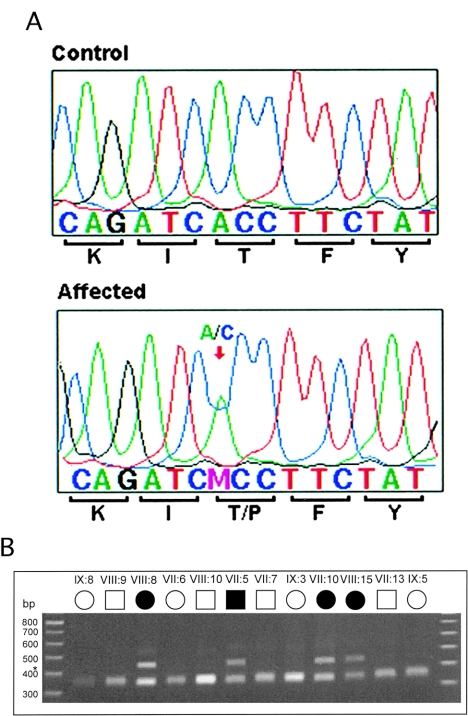
In an attempt to clarify the role of CRYG in the Coppock-like cataract, further mutational analysis of the gene cluster was performed in the original family diagnosed with Coppock-like cataract (Lubsen et al. 1987). Sequencing of CRYG genes confirmed the previously published observations of the sequence changes in CRYGE and that CRYGD did not carry the disease-causing mutation. Direct sequencing of CRYGC identified an A→C transversion in exon 2 (nucleotide position 225 [GenBank K03003]), which changed the amino acid threonine to a proline (T5P). This change cosegregated perfectly with the disease phenotype (i.e., it was present in 9 affected individuals and not in 14 unaffected family members) (fig. 2); nor was it observed in 215 control individuals of various ethnic backgrounds, in 39 patients with congenital cataract, or in 44 patients with age-related cataracts (data not shown). This mutation lies in a highly conserved region. The affected amino acid is invariant in all the γ-crystallins of humans, bovine, rat, and mouse (data not shown).
Discussion
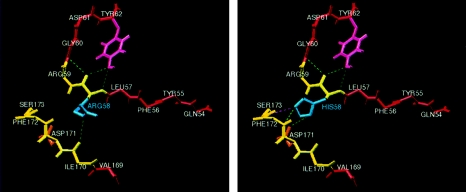
The γ-crystallin proteins are tightly folded in two domains with no free loops. Each domain consists of two Greek-key motifs, each one having a folded hairpin that provides stability between two β-sheets (Meakin et al. 1985). The mutation R58H induces a change in charge and is predicted to introduce hydrogen bonds with residues 171, 172, and 173 (fig. 4). These are crystallographic contact sites (Sergeev et al. 1998), which, when altered, may impair the proper folding of the protein, by either increasing its rigidity or altering its stability. In addition, this mutation is adjacent to an RGD motif known to play a role in cell adhesion. It is possible that the R58H mutation destabilizes the contact between lens-fiber cells, which is critical for the maintenance of lens transparency (White et al. 1989). Improper folding of CRYGD, the most abundantly expressed γ-crystallin in the lens, could well cause protein aggregation and lens opacification. A different mutation in CRYGD recently has been reported in a family affected with a pulverulent cataract, a clinically distinct phenotype (Stephan et al. 1999). This further supports the newly documented role of CRYGD in cataract formation.
Figure 4.
Protein-structure prediction of mutated γ-crystallin D: human γ-D crystallin prediction by SWISS-MODEL, illustrating the ARG58HIS mutation (blue) and surrounding region, protein backbone (yellow), and beta sheets (red). Note that amino acid 58 occurs between two β sheets. The R groups involved with putative hydrogen bonding are displayed; dashed green lines denote strong bonds, and dashed purple lines denote weak bonds. Left, Wild-type γ-D crystallin showing ARG58 and putative hydrogen bond between its R group (blue) and the oxygen of the carbonyl group of ILE170 near the C terminus. Right, Mutated γ-D crystallin showing HIS58 and new putative hydrogen bonds between its R group (blue) and R group of ASP171 (orange). Also predicted are hydrogen bonding with the backbone of PHE172 (at the nitrogen of the amide backbone) and weaker hydrogen bonding with the backbone of SER173 (at the nitrogen of the amide backbone).
The role of CRYGC in the process of lens opacification is documented here for the first time. The 225A→C mutation may disturb the protein function and/or the protein's interaction with neighboring proteins, in part because of the amino acid change. Proline, the mutant amino acid, is established as a potent breaker of β-sheet structures in soluble proteins (Wood et al. 1995; Li et al. 1996). This amino acid change to proline could destabilize the β-strand, which may impair the folding of the protein and lead to lens opacification.
The T5P mutation ofCRYGC represents the true Coppock-like–cataract mutation and the first association of this gene with human cataracts. This finding supports our suggestion that previously reported sequence changes in the CRYGE promoter region are not causally related to a cataract phenotype. The R58H mutation supports the previously proposed role of CRYGD in the lens-opacification process. Further study of the related proteins should contribute to a better understanding of the process of cataract formation.
Acknowledgments
The authors are grateful to the families and patients, for their enthusiastic participation, and to Cristina Batek, Nicole Martin, and Robert Klose, for their excellent technical work. This work has been supported by Swiss National Science Foundation grant 32-053750.98 (to E.H., F.L.M., and D.F.S.) and by the Glaucoma Research Society of Ontario, Physicians' Services Incorporated Foundation grant 97-02, the Weston Foundation, and the Imperial Oil Research Fund (all in support of E.H.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Searching GenBank, http://www.ncbi.nlm.nih.gov/Genbank/GenbankSearch.html (for sequence information on crygb and CRYGC [M19364 and K03003, respectively], CRYGD [K03005 and K03006], and CRYGE [S72943 and K03008])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for aculeiform cataract [MIM 115700], Coppock-like cataract [MIM 123660 and MIM 116200], and progressive polymorphic cataract [MIM 601286])
- SWISS-MODEL, http://www.expasy.ch/swissmod/SWISS-MODEL.html (for protein-homology modeling)
References
- Brakenhoff R, Henskens H, van Rossum M, Lubsen N, Schoenmakers G (1994) Activation of the γE-crystallin pseudogene in the human hereditary Coppock-like cataract. Hum Mol Genet 3:279–283 [DOI] [PubMed]
- Cartier M, Breitman M, Tsui L-C (1992) A frame-shift mutation in the gammaE-crystallin gene of the Elo mouse. Nat Genet 2:42–45 [DOI] [PubMed]
- Cartier M, Tsui L, Ball S, Lubsen N (1994) Crystallins genes and cataract. In: Wright A, Jay B (eds) Modern genetics. Vol 2: Molecular genetics of inherited eye disorders. Harwood Academic, Edinburgh, pp 413–443 [Google Scholar]
- Hejtmancik JF (1998) The genetics of cataract: our vision becomes clearer. Am J Hum Genet 62:520–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Héon E, Liu S, Billingsley G, Bernasconi O, Tsifildis C, Shorderet DF, Munier FL (1998) Gene localization for aculeiform cataract, on chromosome 2q33-35. Am J Hum Genet 63:921–926 [DOI] [PMC free article] [PubMed]
- Kannabiran C, Rogan P, Olmos L, Basti S, Rao G, Kaiser-Kupfer M, Hejtmancik J (1998) Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the βA3/A1-crystallin gene. Mol Vis 4:21–31 [PubMed]
- Klopp N, Favor J, Loster J, Lutz R, Neuhauser-Klaus A, Prescott A, Pretsch W, et al (1998) Three murine cataract mutants (Cat2) are defective in different gamma-crystallin genes. Genomics 52:152–158 [DOI] [PubMed]
- Li S-C, Goto N, Williams K, Deber C (1996) α-helical, but not β-sheet, propensity of proline is determined by peptide environment. Proc Natl Acad Sci USA 93:6676–6681 [DOI] [PMC free article] [PubMed]
- Litt M, Carrero VR, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH (1997) Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet 6:665–668 [DOI] [PubMed]
- Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG (1998) Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet 7:471–474 [DOI] [PubMed]
- Lubsen N, Renwick J, Tsui L-C, Breitman M, Schoenmackers J (1987) A locus of a human hereditary cataract is closely linked to the gamma-crystallin gene family. Proc Natl Acad Sci USA 84:489–492 [DOI] [PMC free article] [PubMed]
- Mackay D, Ionides A, Kibar Z, Rouleau G, Berry V, Moore A, Shiels A, et al (1999) Connexin46 mutations in autosomal dominant congenital cataract. Am J Hum Genet 64:1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meakin S, Breitman M, Tsui L (1985) Structural and evolutionary relationships among five members of the human γ-crystallins gene family. Mol Cell Biol 5:1408–1414 [DOI] [PMC free article] [PubMed]
- Miller S, Dykes D, Polesky H (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 16:1215 [DOI] [PMC free article] [PubMed]
- Mitchell KR, Warshawsky D (1998) Mutational analysis using enriched PCR and cycle sequencing. Biotechniques 24:1028–1031 [DOI] [PubMed]
- Nettleship E, Ogilvie F (1906) A peculiar form of hereditary congenital cataract. Trans Ophthalmol Soc UK 26:191–207 [Google Scholar]
- Newton C, Graham A, Heptinsall I, Powell S, Summers C, Kalsheker N (1989) Analysis of any point mutation in DNA: the amplification refractory mutation system (ARMS). Nucleic Acid Res 17:2503–2515 [DOI] [PMC free article] [PubMed]
- Papaconstantinou J (1967) Molecular aspects of lens differentiation. Science 156:338–346 [DOI] [PubMed]
- Piatigorsky J (1981) Lens differentiation in vertebrates: a review of cellular and molecular features. Differentiation 19:134–153 [DOI] [PubMed]
- ——— (1984) Lens crystallins and their gene families. Cell 38:620–621 [DOI] [PubMed]
- Rogaev EI, Rogaeva EA, Korovaitseva GI, Farrer LA, Petrin AN, Keryanov SA, Turaeva S, et al (1996) Linkage of polymorphic congenital cataract to the gamma-crystallin gene locus on human chromosome 2q33-35. Hum Mol Genet 5:699–703 [DOI] [PubMed]
- Santhiya S, Abd-alla S, Löster J, Graw J (1995) Reduced levels of gamma-crystallin transcripts during embryonic development of murine Cat2nop mutant lenses. Graefes Arch Clin Exp Ophthalmol 233:795–800 [DOI] [PubMed]
- Sergeev YV, David LL, Chen HC, Hope JN, Hejtmancik JF (1998) Local microdomain structure in the terminal extensions of betaA3- and betaB2-crystallins. Mol Vis 4:9 [PubMed]
- Shiels A, Mackay D, Ionides A, Berry V, Moore A, Bhattacharya S (1998) A missense mutation in the human connexin50 gene (GJA8) underlies autosomal dominant “zonular pulverulent” cataract, on chromosome 1q. Am J Hum Genet 62:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan D, Gillanders E, Vanderveen D, Freas-Lutz D, Wistow G, Baxevanis A, Robbins C, et al (1999) Progressive juvenile-onset punctate cataracts caused by mutation of the gammaD-crystallin gene. Proc Natl Acad Sci USA 96:1008–1012 [DOI] [PMC free article] [PubMed]
- Vogt A (1922) Weitere Ergebnisse der Spaltlampenmikroskopie des vorderen Bulbsabschnittes. III. Angeborene und früth erworbene Linsenveränderungen. von Greafes Arch Ophthalmol 107:237–240 [Google Scholar]
- White HE, Driessen HP, Slingsby C, Moss DS, Lindley PF (1989) Packing interactions in the eye-lens: structural analysis, internal symmetry and lattice interactions of bovine gamma IVa-crystallin. J Mol Biol 207:217–235 [DOI] [PubMed]
- Wood S, Wetzel R, Martin J, Hurle M (1995) Prolines and amyloidogenicity in fragments of Alzeimer's peptide β/A4. Biochemistry 34:724–730 [DOI] [PubMed]