Summary
Cleidocranial dysplasia (CCD) is a dominantly inherited disorder characterized by patent fontanelles, wide cranial sutures, hypoplasia of clavicles, short stature, supernumerary teeth, and other skeletal anomalies. We recently demonstrated that mutations in the transcription factor CBFA1, on chromosome 6p21, are associated with CCD. We have now analyzed the CBFA1 gene in 42 unrelated patients with CCD. In 18 patients, mutations were detected in the coding region of the CBFA1 gene, including 8 frameshift, 2 nonsense, and 9 missense mutations, as well as 2 novel polymorphisms. A cluster of missense mutations at arginine 225 (R225) identifies this residue as crucial for CBFA1 function. In vitro green fluorescent protein fusion studies show that R225 mutations interfere with nuclear accumulation of CBFA1 protein. There is no phenotypic difference between patients with deletions or frameshifts and those with other intragenic mutations, suggesting that CCD is generally caused by haploinsufficiency. However, we were able to extend the CCD phenotypic spectrum. A missense mutation identified in one family with supernumerary teeth and a radiologically normal skeleton indicates that mutations in CBFA1 can be associated exclusively with a dental phenotype. In addition, one patient with severe CCD and a frameshift mutation in codon 402 had osteoporosis leading to recurrent bone fractures and scoliosis, providing first evidence that CBFA1 may help maintain adult bone, in addition to its function in bone development.
Introduction
Cleidocranial dysplasia (CCD; MIM 119600) is a skeletal dysplasia characterized by persistently open or delayed closure of sutures, Wormian bones, hypoplastic and/or aplastic clavicles, wide pubic symphysis, dental anomalies, and short stature. A number of other radiological and clinical signs are characteristic and should be sought before diagnosis. The face and skull have a characteristic appearance with a relatively large cranium, frontal and parietal bossing, and a frontal groove originating from ossification defects within the metopic suture. The nose is short and anteverted, and the maxilla is hypoplastic, giving the impression of midface hypoplasia and relative prognathism. Nails and terminal phalanges are frequently hypoplastic, and the second metacarpal consistently has two ossification centers. Other radiological changes include a narrow bell-shaped thorax, cervical or missing ribs, characteristic changes in the pelvis, and large femoral epiphysis (for review, see Mundlos 1999). The condition is inherited as an autosomal dominant trait and was mapped to chromosome 6p21 (Mundlos et al. 1995). Recently, the human CBFA1 gene was identified as the CCD gene (Lee et al. 1997; Mundlos et al. 1997).
The CBFA1 gene encodes a transcription factor that is a member of the runt family of proteins (Levanon et al. 1994). These proteins contain a DNA-binding domain highly homologous to the Drosophila pair-rule gene runt (Nüsslein-Volhard and Wieschaus 1980). CBFA1 contains a region of glutamine and alanine repeats in the N-terminal region. Toward the C terminus is a region rich in proline-serine-threonine (PST), which is necessary for transcriptional activation of target genes. The CBFA1 gene consists of eight coding exons and spans a genomic region of 130 kb (Mundlos et al. 1997; Geoffroy et al. 1998). Several splice variants have been described recently (Mundlos et al. 1997; Geoffroy et al. 1998; Xiao et al. 1998).
Whereas the other two members of the mammalian runt family—Cbfa2 and Cbfa3—are expressed in cells of hematopoietic origin, the expression of Cbfa1 is restricted to bone and cartilage (Ducy et al. 1997; Otto et al. 1997; Kim et al. 1999). Cbfa2 is essential for the initiation of mature-type hematopoiesis (Okuda et al. 1996; Wang et al. 1996). In contrast, Cbfa1 is essential for the development of osteoblasts from their mesenchymal precursors (Ducy et al. 1997). Inactivation of Cbfa1 in mice by gene targeting leads to a complete absence of bone. Although the cartilage anlagen develop normally, there is no differentiation of mesenchymal stem cells into osteoblasts, no ossification, and no vascular invasion of cartilage (Komori et al. 1997; Otto et al. 1997). Such mice die shortly after birth because of impaired respiratory function. Heterozygous Cbfa1 mutant mice display all the hallmarks of CCD, including open fontanelles and hypoplastic clavicles, but not the dental anomalies (Huang et al. 1997; Otto et al. 1997). A radiation-induced mouse mutant exhibiting a CCD phenotype was shown to harbor a deletion on mouse chromosome 17, in a region exhibiting conserved synteny to human 6p21, containing the Cbfa1 gene (Otto et al. 1997).
We have analyzed the coding sequence of the CBFA1 gene in 42 individuals with the clinical diagnosis of CCD. The purpose of this study was to identify the spectrum of mutations in CBFA1 in this population and to determine whether any correlations between phenotype and genotype could be established.
Subjects and Methods
Patients
Forty-two unrelated families of different ethnic backgrounds and with the clinical diagnosis of CCD (for review of diagnostic criteria, see Mundlos 1999) were included in this study. Informed consent was obtained from all individuals. DNA was prepared from blood leukocytes by means of standard procedures.
Mutation Detection
Exons 0–7 of the CBFA1 gene were amplified by PCR with the primers given in table 1. PCR reactions were elicited in a total volume of 100 μl, each containing 200 ng DNA, 2 μl dNTP mix at 10 mM each, 0.2 μM each primer, 10 μl 10 × buffer, and 2.5 U Taq polymerase (TAQ PCR core kit, Qiagen). For exon 1, DMSO was added at a final concentration of 10%. Amplification was done in a thermal cycler (PE 2400, PE Biosystems). After denaturing at 94°C for 4 min, 32 cycles of 94°C for 20 s, 60°C for 30 s, and 72°C for 30 s were performed. The amplification products were checked by agarose gel electrophoresis and purified with the PCR purification kit (Qiagen) according to manufacturer's instructions. Sequencing reactions were performed with 5 μl purified PCR product, 0.2 μM primer, and 4 μl Big Dye Terminator Cycle sequencing mix with AmpliTaq DNA polymerase FS (PE Biosystems). Automated sequencing was performed on an ABI 377 sequencer. Sequences were analyzed by means of ABI View version 1.2.1 software. Every mutation was confirmed by sequencing the product of an independent PCR. Restriction digests were performed in those cases where the mutation introduced a new restriction enzyme site or abolished a site present in the wild-type DNA. To resolve ambiguities in the primary sequence, products of an independent PCR reaction were subcloned by use of the Original TA Cloning Kit (Invitrogen). On average, a total of 10 individual clones were sequenced. Base-pair numbers used refer to GenBank entries AF001443–AF001450.
Table 1.
Primers Used for Polymerase Chain and Sequencing Reactions
| Exon | Sense | Sequence | Antisense | Sequence |
| 0 | AML3E0F | TACCCAGCCACCGAGACCAACAGAG | AML3E0R | GTTTTGCTGACATGGTGTCAC |
| 1 | CBFA12 | ACTTGTGGCTGTTGTGATGC | CBFA17 | GCCGCCAAGGCAGGA |
| 2 | AML3I1F2 | CAGATGCTTGATTCCTGTCGG | AML3I2R1 | GTGCTGATTTGTATACAGACTAG |
| 3 | AML3I2F2 | TCATTGCCTCCTTAGAGATGC | AML3I3R2 | GGACATGAAAGTGACACTAAC |
| 4 | AML3I3F1 | TATAAAGCAATTTGAAATGCAAGG | AML3I4R1 | GTTTTGAAGTGAACACATCTCC |
| 5 | AML3I4F1 | TAAGGCTGCAATGGTTGCTAT | AML3I5R1 | GTCACTGTGAGCATGGATGAG |
| 6 | AML3I5F1 | TAGAACATTAGAGCTGGAAGG | AML3I6R1 | CGGACAGTAACAACCAGACAG |
| 7 | AML37F | TGTGGCTTGCTGTTCTTTATG | AML37R | GATACCACTGGGCCACTGCT |
Subcellular Localization of Mutant CBFA1 Protein
To transiently express green fluorescent protein fusions, full-length Cbfa1 cDNA (nt 1–1566) and the runt domain, including the putative nuclear localization signal (NLS) (nt 304–708), were subcloned into pEGFP-C1 (Clontech). The mutations R225Q and R225W were introduced into the resulting plasmids by site-directed mutagenesis (Sculptor in vitro mutagenesis system, Amersham Buchler). All plasmids were fully sequenced to exclude any additional mutations. National Institutes of Health (NIH) 3T3, C3H 10T1/2, and U2OS cells were transiently transfected by calcium phosphate coprecipitation by means of 30 μg of plasmid DNA per transfection. Fluorescence microscopy was performed on an Axioplan microscope (Zeiss). Laser-scan micrographs were taken with a confocal laser scan microscope (410; Zeiss).
Results
Of the 42 unrelated families investigated for alterations in the coding region of the CBFA1 gene, we were able to identify a total of 19 mutations in 18 patients and an additional 2 polymorphisms. Only one of the mutations has been described before. With the exception of mutations affecting codon 225, the mutations were evenly spread over the entire coding region. Mutations were detected in all exons except exons 0 and 6 (table 2 and fig. 1).
Table 2.
CBFA1 Mutations Detected in CCD Patients
| Patient | Codon | Nucleotide | No. ofPatientsa | Mutation Type | Length ofPredictedFrameshift(aa) | Referenceb |
| 1 | 62 | 186ins(16) | 2 | Insertion/frameshift | 103 | This study, 1 |
| 2 | 69 | 206A→G | 1 | Polymorphism | This study | |
| 3 | 74 | 222ins(30) | 1 | Insertion | 1 | |
| 4 | L113R | 338T→G | 1* | Missense | This study | |
| 5 | S118R | 354C→G | 1 | Missense | This study | |
| 6 | F121C | 362T→G | 1+ | Missense | This study | |
| 7 | C123R | 366–367GT→TC | 1* | Missense | This study | |
| 8 | D161X | 481–482delGA | 1 | Deletion/nonsense | 0 | This study |
| 9 | M175R | 524T→G | 1 | Missense | 2 | |
| 10 | 180 | 540–549del(10) | 1 | Deletion/frameshift | 1 | 1 |
| 11 | S191N | 572G→A | 1 | Missense | 2 | |
| 12 | R193X | 577C→T | 1 | Nonsense | This study | |
| 13 | T205G | 614C→G | 1 | Missense | This study | |
| 14 | R225W | 673C→T | 1 | Missense | This study | |
| 15 | R225Q | 674G→A | 3 | Missense | This study | |
| 16 | 274 | 824delG | 1 | Deletion/frameshift | 32 | This study |
| 17 | 295 | 887delC | 1 | Deletion/frameshift | 11 | This study |
| 18 | W297X | 891G→A | 1 | Nonsense | 1 | |
| 19 | 305 | 915delC | 1 | Deletion/frameshift | 1 | This study |
| 20 | 376 | 1127–1128insT | 1 | Insertion/frameshift | 6 | This study |
| 21 | 385 | 1157delG | 1 | Deletion/frameshift | 97 | This study |
| 22 | 402 | 1205–1206insC | 1 | Insertion/frameshift | 87 | This study |
| 23 | 495 | 1379–1380insC | 1 | Insertion/frameshift | 26 | This study |
| 24 | G511S | 1531G→A | 1+ | Missense/polymorphism | This study |
Figure 1.
Summary of mutations identified in CCD patients. The bar depicts the CBFA1 protein. Glutamine-alanine repeat (Q/A), DNA binding runt homology (RUNT), putative NLS, and PST domains are indicated. The TLE corepressor protein interaction motif VWRPY is shown at the carboxy terminus. AD1–3 and RD denote transcriptional activation and repression domains as described by Thirunavukkarasu et al. (1998). Mutations are indicated as follows: circle, missense; octogon, nonsense; triangle, insertion; inverted triangle, deletion; and square, polymorphism.
Missense Mutations
Missense mutations were detected in eight patients. In four of these, arginine 225 (R225) was mutated; it is located at the C-terminal end of the runt domain. The exchange of glutamine for arginine (674G→A [R225Q]) occurred in three unrelated patients. A replacement by tryptophan (673C→T [R225W]) was identified in one patient. Both amino acid exchanges abolish the positive charge of the residue at this position. The high frequency of mutations affecting R225 identifies this codon as being either prone to mutagenic events—for example, by deamination of methyl-C within a CG dinucleotide in this codon or of important relevance for the normal function of CBFA1. One patient had two missense mutations within the same exon, separated by 27 bp of unaltered DNA (see table 2). These mutations convert both leucine 113 and cystine 123 to arginine residues (338T→G [L113R] and 366-367GT→TC[C123R]). Both mutations were passed on from the affected mother to the daughter and thus are located on the same allele. All missense mutations identified were located in the runt domain of CBFA1.
Nonsense Mutations
In addition to the nonsense mutation (891G→A [W297X]) described in a previous report (Mundlos et al. 1997), two other nonsense mutations were detected. A 2-bp deletion resulting in a stop codon (481–482delGA [D161X]) was identified in a family with three affected generations. The other mutation is predicted to result in a truncated protein, including the amino-terminal part of the runt domain. The 577C→T (R193X) mutation was passed on to the patient from his father, whose parents were unaffected.
Deletions and Insertions
Deletions were identified in five patients. Single base-pair deletions with subsequent frameshifts occurred in four patients. Whereas one patient (1157delG) had healthy parents, the other mutations were passed on for several generations. Radiographs of patient F (887delC) are shown in figure 2. The predicted proteins contain the entire runt domain.
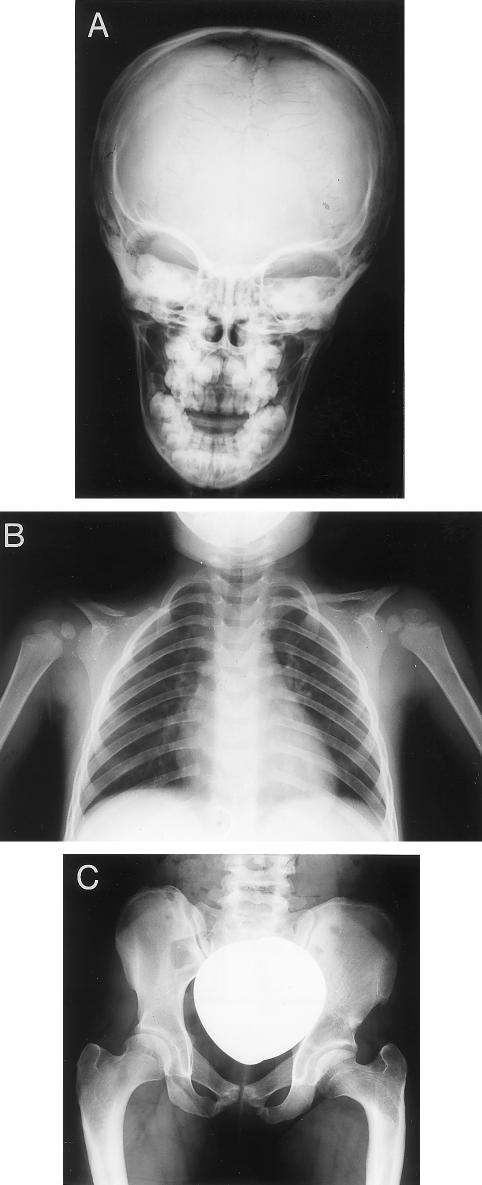
Figure 2.
Radiological findings in patient F (887delC). A, Anteroposterior radiograph of the skull showing multiple Wormian bones in lambdoid and sagittal sutures. B, Chest radiograph demonstrating cone-shaped thorax, right clavicular hypoplasia, and abnormally shaped left clavicle with lateral gap. C, Abnormal pelvis with hypoplastic iliac wings, reduced pelvic diameter necessitating cesarian section, and coxa vara.
Single base-pair insertions were found in three patients. In one patient, part of the CAG repeat within exon 1 was extended by 16 nucleotides. This mutation is identical to the one characterized in our previous report (Mundlos et al. 1997), describing an unrelated patient. A 2-bp deletion (481–482delGA [D161X]) is detailed in the Nonsense Mutations subsection, above.
Polymorphisms
Two novel polymorphisms were identified. An A→G transition (206G→A, silent) within the Q/A domain was found on one allele in patient A, whose healthy mother carried the transition on both alleles.
Patient B had two mutations within the coding region of the CBFA1 gene. The missense mutation 362T→G (F121C) was inherited from the mother. The healthy father of a patient carried a G→S change at position 511 (1531G→A [G511S]) that was passed on to patient B. Of interest, the mother and grandfather of the patient had very mild phenotypes, with supernumerary teeth but no clavicular involvement. Chest, skull, and pelvic radiographs showed none of the changes classically observed in CCD. In contrast, patient B had a one-sided pseudarthrosis of the clavicle and typical facial appearance. Thus, one might assume that the simultaneous occurrence of two mutations led to the typical CCD phenotype, although either one by itself produced a phenotype that was weak (in the case of the mother) or not discernible (in the case of the father). However, intrafamilial variation in expressivity, which is a common feature in CCD, may serve as an alternative explanation for the more-severe phenotype of patient B, with G551S being a polymorphism.
Genotype-Phenotype Correlation
To evaluate the functional significance of the mutations identified, we undertook a careful evaluation of phenotypes and compared them with the different types of mutations observed. Patients were evaluated for clavicular involvement, open fontanelles, presence of supernumerary teeth (if applicable), and height. Although penetrance was 100% in our cases, intrafamilial variation in expressivity was common. With the exception of the family discussed above, clavicular involvement was invariably present but was frequently confined to the most distal part of the clavicle, giving an almost normal impression upon inspection. The number and presence of supernumerary teeth was highly variable and did not correlate with the rest of the phenotype. In summary, we were not able to identify any significant genotype-phenotype correlation. Also, there was no difference in severity of phenotypes among patients with identified mutations and those in whom no mutations in CBFA1 had been found. However, the identification of mutations in patients with less-typical phenotypes allowed us to redefine the phenotypic spectrum of CCD.
Nonsense or frameshift mutations within or 5′ of the runt domain are expected to result in a truncated protein without DNA binding domain and thus without function. In contrast, premature stop codons 3′ of the runt domain coding region could have antagonistic effects, leading to a truncated protein that binds DNA but has no activation potential. Depending on the stability of mRNA and protein, such mutations could have a dominant negative effect. Therefore, we were particularly interested in mutations affecting the last exon and thus having a greater probability of giving rise to a stable mRNA product. We identified a total of four mutations in this part of the gene. The patients with the mutation 1205–1206insC and 1379–1380insC are remarkable in that they represent the extremely severe and the mild ends of the phenotypic spectrum, respectively.
Because of his unusual clinical features, patient C (with the mutation 1205–1206insC) is of particular interest. Patient C is the offspring of healthy parents. On a radiograph taken in utero, a transverse fracture of the left femur was noted. X-rays taken 2 days after birth demonstrated greenstick fractures of the tibia and fibula bilaterally. At birth, the skull was almost unossified, both clavicles were absent, and distal hypoplasia of phalanges with partial absence of the nails was noted. On the basis of these observations, the preliminary diagnosis of CCD was made. During childhood and adolescence, a total of nine fractures of long bones occurred after minor trauma. The patient developed a pseudarthrosis after a fracture of the femur with lack of callus formation, necessitating fixation with an intramedullary nail. The patient suffers from partial deafness due to conduction hearing impairment. A number of supernumerary teeth were extracted. Progressive severe scoliosis and lordosis of the spine developed, necessitating the implantation of a Harrington rod for spinal support (fig. 3). At the age of 23 years, patient C had a body height of 129 cm. The long bones of both legs display increasing bending toward the midline. X-rays taken during childhood and adolescence demonstrated severe osteoporosis as the cause of the recurrent fractures and the spinal deformity. DEXA scans performed at the age of 24 years revealed an extreme form of osteoporosis, with −3.8 for the lumbar spine (L2/L3), −6.09 for the hip, and −6.34 for the left forearm. (Values are SDs, with 0 representing the mean for age and sex. Values <−1.5 represent osteopenia.)
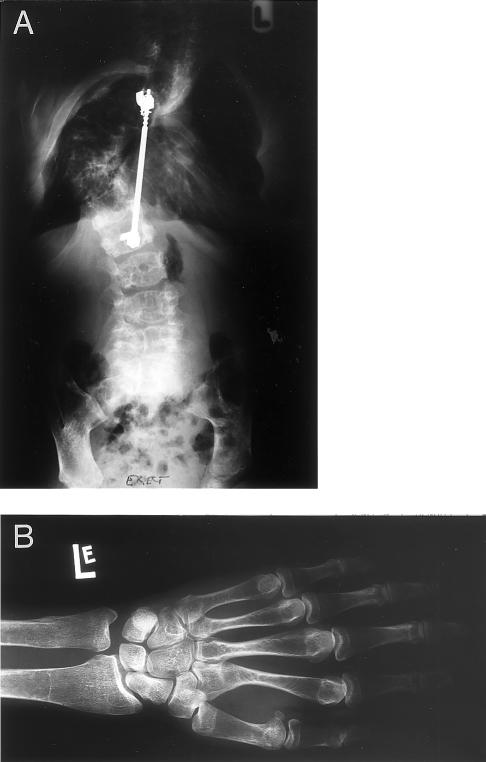
Figure 3.
Radiological findings in patient C. A, Anteroposterior radiograph of patient C, carrying the mutation 1205insC. The chest is severely deformed because of compression fractures of the thoracic vertebrae and the subsequent development of a kyphoscoliosis. A Harrington rod was implanted for stabilization. Both clavicles are absent. The lumbar vertebrae have an irregular structure, are severely osteopenic, and are partially collapsed. The entire skeleton is extremely osteopenic, giving an almost radiotranslucent appearance especially of the ribs and the thoracic spine. The iliac wings are hypoplastic. B, Radiograph of the left hand and the distal part of the radius and the ulna of patient C. Note severe osteoporosis and greatly reduced trabecular and cortical bone. There is also hypoplasia of the terminal phalanges.
In contrast, patient D exhibited a very mild phenotype. He was brought to medical attention only because of supernumerary teeth. His mutation, 1379–1380insC, causes a frameshift at the very 3′ end of the coding region, leaving the runt domain, the NLS, and putative activation domains intact, but inactivating the valine-tryptophan-arginine-proline-tyrosine (VWRPY) f, a region suspected to interact with the transducin-like enhancer of split (TLE) family of transcriptional corepressors. Patient D is of normal height and excellent physical health. The clinical signs of CCD are restricted to a gap in the most lateral part of the clavicle bilaterally and supernumerary teeth.
Subcellular Localization of NLS Mutant CBFA1 Protein
The amino acid R225, which was the residue most frequently affected by mutations in our series, resides within a stretch of basic amino acids at the carboxy terminus of the runt domain. Since NLSs necessary for the function of transcription factors usually contain a number of basic residues, we suspected that R225 mutations might affect the subcellular distribution of CBFA1. To test this hypothesis, we constructed fusion proteins between green fluorescent protein and CBFA1. The constructs were transiently transfected into human osteosarcoma U2OS, mouse fibroblast NIH 3T3, and mesenchymal stem cell C3H 10T1/2 lines. Similar results were obtained with all three cell lines. In contrast to a GFP control, which was evenly distributed throughout cytoplasm and nucleus, CBFA1-GFP fusion protein was detected exclusively in the nucleus (fig. 4). Similar results were obtained when the runt domain of CBFA1, including the putative NLS, was expressed as GFP fusion (fig. 5). Thus, amino acids 102–236 were sufficient to control nuclear accumulation of CBFA1.
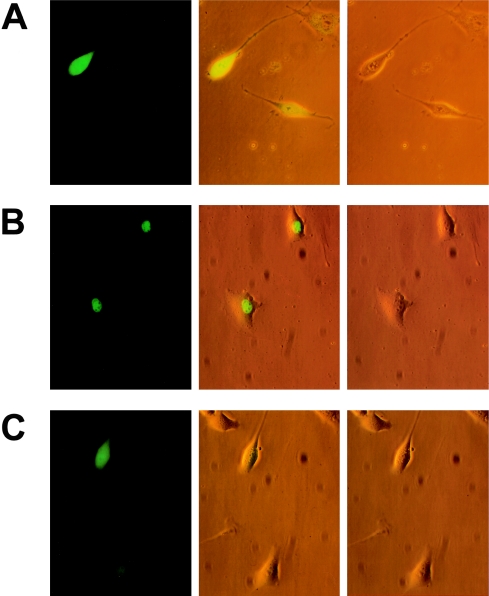
Figure 4.
Subcellular localization of mutant CBFA1. Fluorescence (left), visible-light (right), and double-projection (middle) micrographs of CBFA1-GFP fusion proteins expressed in NIH 3T3 cells. Note the different subcellular localization of control GFP (A), wild-type CBFA1 (B), and mutant R225Q CBFA1 (C).
Figure 5.
Subcellular localization of runt domain-GFP fusion protein in NIH 3T3 cells. Laser-scan micrographs show the intracellular distribution of control GFP (A), wild-type CBFA1-runt (B), R225Q (C), and R225W (D) mutant CBFA1-runt proteins.
When glutamine (R225Q) or tryptophan (R225W) was substituted for R225, CBFA1 was unable to quantitatively accumulate in the nucleus. The intracellular distribution pattern of mutated CBFA1, as well as isolated runt/NLS, was indistinguishable from GFP control (fig. 4 and 5). Thus, mutations in R225 completely abolish the function of the NLS in accumulating CBFA1 in the nucleus.
Discussion
Mutations in CBFA1, a transcription factor of the runt family, have been identified as a cause of the autosomal dominant skeletal disorder CCD (Lee et al. 1997; Mundlos et al. 1997). In the present study, we have identified new mutations in 18 unrelated families of different ethnic backgrounds. All of the identified mutations were unique, except for those in three patients who had the same 674G→A (R225Q) mutation and in patient E, who had a 16-bp insertion at bp 186 of exon 1. The latter mutation is identical to a mutation in an unrelated patient described in our earlier report (Mundlos et al. 1997). An imperfect 69-bp GAG repeat exists at this site in exon 1, with both patients having insertions of small parts of the repeat sequence. The existence of these repetitive sequences may predispose this site to errors during recombination or DNA replication and may account for the recurrent insertion seen in these two unrelated patients.
The mutations identified in this study were evenly distributed along the coding region of CBFA1. No mutations were found in exons 0 and 6, perhaps because of their small size. However, an alternative splice product containing all exons but exon 6 has been identified (Mundlos et al. 1997). Thus, exon 6 might be dispensable for CBFA1 function, and mutations involving exon 6 might not produce a phenotype. Likewise, the functional significance of exon 0, although expressed in osteoblasts, remains unknown, since alternative start sites are likely to exist (Mundlos et al. 1997). Genetic heterogeneity is a possible explanation for the relatively low frequency of mutations detected in this study. However, all CCD families tested so far map to the 6p21 locus. Other explanations are mutations within yet unknown regulatory sequences or smaller deletions involving single exons. Deletion testing with YACs from the critical region in 10 patients without identifiable mutations revealed no deletion (C. Mundlos, unpublished data), indicating that large genomic deletions, as described by Mundlos et al. (1997), are not a common feature of CCD.
For CBFA1, as well as for the closely related transcription factor CBFA2, a number of functional domains have been defined (Kanno et al. 1998; Thirunavukkarasu et al. 1998). CBFA1 contains an amino-terminal stretch of polyglutamine and polyalanine repeats (Q/A domain), followed by the DNA-binding runt homology domain and the carboxy-terminal PST-rich domain. The PST domain consists of two subdomains, the amino terminal having transactivating capabilities and the carboxy terminal being able to repress transactivation. A short stretch of amino acids between runt homology and PST domains is responsible for nuclear localization of the CBFA1 protein. The carboxy terminus of CBFA1 consists of the VWRPY motif that is conserved in all runt homology proteins in every species analyzed so far. Its function might be to bind the corepressor proteins of the TLE/Grg family homologous to Drosophila groucho (Stifani et al. 1992).
The identification of a number of nonsense and frameshift mutations in this study poses the question of whether the predicted proteins derived from the mutant alleles have a function of their own. With the exception of one patient, however, the phenotypes of all patients analyzed were within the range of variation known to occur in CCD, and no systematic effect relating the length of the hypothetical truncated protein to the severity of the clinical features was seen. Thus it seems reasonable to conclude that these mutations cause functional haploinsufficiency. Truncated proteins may be unstable in the cellular environment and can be degraded rapidly (Akeson et al. 1989; Hashimoto et al. 1996). Moreover, there are examples of instability and altered nuclear export of mRNA derived from mutant alleles (Redford-Badwal et al. 1996). Thus it may be speculated that the predicted protein products are not present in adequate levels to produce distinguishable effects that exceed the effect caused by the mere absence of one copy of the CBFA1 gene.
One possible exception to this is the frameshift mutation in codon 402 (patient C). The protein product of this allele is predicted to contain the first 401 amino acids of CBFA1, followed by 87 additional novel amino acids. The patient carrying this mutation exhibits features beyond the spectrum typically seen in CCD. In addition to the classical features of CCD, which are remarkably severe, bone fragility and severe osteoporosis are present—findings not generally considered part of the CCD spectrum. The presence of intrauterine bone fractures indicates a congenital defect in bone stability. X-rays and DEXA scans of the spine and radius revealed an extreme form of bone loss. Cortical bone was very thin and undermineralized, whereas trabecular bone was dramatically reduced and almost absent in some parts. This form of bone loss is indistinguishable from other forms of osteoporosis and explains the recurrent fractures and the spinal deformity observed in this patient.
Mice carrying one mutated Cbfa1 allele show all the hallmarks of CCD. When both copies of the Cbfa1 gene are inactivated, bone development cannot proceed, because of a block in differentiation of mesenchymal precursor cells toward osteoblasts (Komori et al. 1997; Otto et al. 1997). The phenotype exhibited by patient C could be an intermediate form between these two extremes. Retarded osteoblast differentiation and/or the production of a lower number of osteoblasts from precursor cells leading to an overall reduced bone mass could explain this exceptional phenotype. The mutated protein containing the DNA binding domain, but not the transactivation domains, might act as a dominant negative form of CBFA1 by binding to the consensus binding site on target gene promoters without being able to transactivate or repress. An alternative explanation is the possibility that the 87 novel amino acids appended to the carboxy terminus provide novel repressive properties to the CBFA1 protein. In either case, the wild-type protein transcribed from the second allele will not be able to access these promoters at the same rate as in patients in whom one allele is effectively inactivated. If this conclusion is correct, one would expect CBFA1 to play a role not only during embryonic development, but also during bone remodeling in adulthood. This is likely, since bone is continuously renewed by differentiation of marrow precursor cells into osteoblasts and the subsequent production of new bone matrix. As in early development, CBFA1 is likely to be the rate-controlling factor in this differentiation process. This conclusion is supported by the observation that transgenic mice overexpressing a dominant negative form of Cbfa1 develop an osteopenic phenotype (Ducy et al. 1999). Haploinsufficiency, however, seems not to be sufficient to reduce bone mass, as first preliminary DEXA scan experiments in a small number of CCD patients have shown (S. Mundlos, unpublished data). Since patient C has unaffected parents and no children, a modification of the CCD phenotype by an additional mutation in another bone-related gene cannot be excluded. In either case, the data identify CBFA1 as a gene involved not only in skeletal development and growth but also in the regulation of bone mass.
The missense mutations identified in this study showed an obvious clustering at R225. R225 resides within an area of the CBFA1 protein thought to be responsible for nuclear localization. Whereas Thirunavukkarasu et al. (1998) describe a myc-related NLS in CBFA1 starting three amino acids carboxy-terminal to R225 and not including this residue, Kanno et al. (1998) identify an NLS in a region of CBFA2 with sequence identity to CBFA1, which contains R225 as one of six basic amino acids crucial for nuclear import. To test whether R225 is indeed a functional part of an NLS, we analyzed the subcellular distribution of CBFA1-GFP fusion proteins after transient transfection into different cell lines. There was no apparent difference in intracellular distribution between CBFA1 and the isolated runt domain. Thus, the nuclear matrix–targeting signal located in the carboxy-terminal region of CBFA1 does not seem to influence nuclear accumulation (Lindenmuth et al. 1997; Thirunavukkarasu et al. 1998). Both mutations—R225Q and R225W—completely abolished the function of the NLS, rendering the protein unable to quantitatively accumulate in the nucleus. Although our in vitro data may only qualitatively reflect the in vivo situation in the respective CCD patients because of unphysiologically high intracellular protein levels in transient transfection assays, these results suggest that a lack of nuclear CBFA1 accumulation is the cause of haploinsufficiency in these cases. This finding represents a novel mechanism by which mutations can lead to a disease phenotype.
All missense mutations identified reside within either the NLS or the DNA-binding runt homology domain. It is possible that the latter abolish DNA-binding ability of the transcription factor CBFA1. On the other hand, the runt homology domain is responsible for heterodimerization with Cbfβ, a cofactor that enhances the affinity of the α subunit to DNA (Kagoshima et al. 1996). Although Thirunavukkarasu et al. (1998) show in vitro evidence that the QA domain prevents binding of CBFβ to CBFA1, this might not necessarily be true for the in vivo situation. There is a high degree of sequence homology between the DNA binding domains of the mammalian members of the runt homology family of proteins and the Drosophila transcription factors runt and lozenge. The evolutionary conservation in this protein domain indicates that only a small degree of sequence variation is allowed when function is to be retained. Thus, a high percentage of missense mutations involving the runt domain is likely to affect protein function and lead to haploinsufficiency. Further collection of mutation data of CBFA1, their analysis with the use of in vitro studies, and the determination of the three-dimensional structure of CBFA1 will contribute to a better understanding of the function of this protein.
Acknowledgments
The project was supported by Deutsche Forschungsgemeinschaft grants Ot 134/2-1 (to F.O.) and Mu 880/3-1 (to S.M.). We thank M. Follo and K. Geiger for their help with automated sequencing and laser scan microscopy; O. Alkan, D. Moradpur, and G. Gross for cell lines; and C. Mundlos for sharing unpublished data. D. Levanon is acknowledged for her helpful comments. We are grateful to all patients and family members who participated in the study.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Entrez/index.html (for sequences analyzed in this report [accession numbers AF001443–AF001450])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for CCD [MIM 119600]) [PubMed]
References
- Akeson AL, Wiginton DA, Hutton JJ (1989) Normal and mutant adenosine deaminase genes. J Cell Biochem 39:217–228 [DOI] [PubMed]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, et al (1999) A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev 13:1025–1036 [DOI] [PMC free article] [PubMed]
- Ducy P, Zhang R, Geoffroy V, Ridell AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754 [DOI] [PubMed]
- Geoffroy V, Corral DA, Zhou L, Lee B, Karsenty G (1998) Genomic organization, expression of the human CBFA1 gene, and evidence for an alternative splicing event affecting protein function. Mamm Genome 9:54–57 [DOI] [PubMed]
- Hashimoto S, Tsukada S, Matsushita M, Miyawaki T, Niida Y, Yachie A, Kobayashi S, et al (1996) Identification of Bruton's tyrosine kinase (Btk) gene mutations and characterization of the derived proteins in 35 X-linked agammaglobulinemia families: a nationwide study of Btk deficiency in Japan. Blood 88:561–573 [PubMed]
- Huang LF, Fukai N, Selby PB, Olsen BR, Mundlos S (1997) Mouse clavicular development: analysis of wild-type and cleidocranial dysplasia mutant mice. Dev Dyn 210:33–40 [DOI] [PubMed]
- Kagoshima H, Akamatsu Y, Ito Y, Shigesada K (1996) Functional dissection of the a and b subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J Biol Chem 271:33074–33082 [DOI] [PubMed]
- Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y (1998) Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor a subunit revealed in the presence of the β subunit. Mol Cell Biol 18:2444–2454 [DOI] [PMC free article] [PubMed]
- Kim IS, Otto F, Zabel B, Mundlos S (1999) Regulation of chondrocyte differentiation by Cbfa1. Mech Dev 80:159–170 [DOI] [PubMed]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, et al (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764 [DOI] [PubMed]
- Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, et al (1997) Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet 16:307–310 [DOI] [PubMed]
- Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y (1994) AML1, AML2 and AML3 the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 23:425–432 [DOI] [PubMed]
- Lindenmuth DM, van Wijnen AJ, Hiebert S, Stein JL, Lian JB, Stein GS (1997) Subcellular partitioning of transcription factors during osteoblast differentiation: developmental association of the AML/CBFa/PEBP2a-related transcription factor-NMP-2 with the nuclear matrix. J Cell Biochem 66:123–132 [PubMed]
- Mundlos S (1999) Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet 36:177–182 [PMC free article] [PubMed]
- Mundlos S, Mulliken JB, Abramsom DL, Warman ML, Knoll JH, Olsen BR (1995) Genetic mapping of cleidocranial dysplasia and evidence of a microdeletion in one family. Hum Mol Genet 4:71–75 [DOI] [PubMed]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, et al (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773–779 [DOI] [PubMed]
- Nüsslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801 [DOI] [PubMed]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321–330 [DOI] [PubMed]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, et al (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771 [DOI] [PubMed]
- Redford-Badwal DA, Stover ML, Valli M, McKinstry MB, Rowe DW (1996) Nuclear retention of COL1A1 messenger RNA identifies null alelles causing mild osteogenesis imperfecta. J Clin Invest 97:1035–1040 [DOI] [PMC free article] [PubMed]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S (1992) Human homologs of a Drosophila enhancer of split gene product define a novel family of nuclear proteins. Nat Genet 2:119–127 [DOI] [PubMed]
- Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G (1998) Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfβ. Mol Cell Biol 18:4197–4208 [DOI] [PMC free article] [PubMed]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA 93:3444–3449 [DOI] [PMC free article] [PubMed]
- Xiao ZS, Thomas R, Hinson TK, Quarles LD (1998) Genomic structure and isoform expression of the mouse, rat and human Cbfa1/Osf2 transcription factor. Gene 214:187–197 [DOI] [PubMed]