Summary
Biallelic, truncating mutations of the hSNF5/INI1 gene have recently been documented in malignant rhabdoid tumor (MRT), one of the most aggressive human cancers. This finding suggests that hSNF5/INI1 is a new tumor-suppressor gene for which germline mutations might predispose to cancer. We now report the presence of loss-of-function mutations of this gene in the constitutional DNA from affected members but not from healthy relatives in cancer-prone families. Furthermore, a constitutional mutation is documented in a patient with two successive primary cancers. In agreement with the two-hit model, the wild-type hSNF5/INI1 allele is deleted in the tumor DNA from mutation carriers. In all tested cases, DNA from parents demonstrated normal hSNF5/INI1 sequences, therefore indicating the de novo occurrence of the mutation, which was shown to involve the maternal allele in one case and the paternal allele in two other cases. These data indicate that constitutional mutation of the hSNF5/INI1 gene defines a new hereditary syndrome predisposing to renal or extrarenal MRT and to a variety of tumors of the CNS, including choroid plexus carcinoma, medulloblastoma, and central primitive neuroectodermal tumor. This condition, which we propose to term “rhabdoid predisposition syndrome,” may account for previous observations of familial and multifocal cases of the aforementioned tumor types. It could also provide the molecular basis for cases of Li-Fraumeni syndrome without p53 germline mutations.
Introduction
Malignant rhabdoid tumor (MRT) was initially described in the kidney, as an aggressive variant of Wilms tumor (Beckwith and Palmer 1978). Subsequently, rhabdoid phenotypes were observed in a variety of neoplasia localized in the brain, the abdomen, or the soft tissue (Parham et al. 1994; Wick et al. 1995; Rorke et al. 1996; Burger et al. 1998). The observation of recurrent alterations of chromosome 22 in both renal and extrarenal MRTs has provided the first indication that these tumors could share a common genetic basis and constitute a distinct tumoral entity (Douglass et al. 1990; Biegel et al. 1992, 1996; Schofield et al. 1996; Rosty et al. 1998; White et al. 1999). Using a positional cloning strategy aimed at the characterization of these chromosome 22 alterations, we recently identified hSNF5/INI1, a gene encoding a member of the SWI/SNF ATP-dependent chromatin-remodeling complex, as the target of recurrent loss-of-function alterations in renal and extrarenal MRT (Kalpana et al. 1994; Muchardt et al. 1995; Versteege et al. 1998; Wade and Wolffe 1999). The observation of biallelic, truncating mutations in tumors strongly supported the hypothesis that hSNF5/INI1 is a new tumor-suppressor gene consistently inactivated in MRT. In addition to somatically acquired alterations, constitutional mutations of this gene also have been observed very recently in patients with MRT (Biegel et al. 1999).
To further delineate the spectrum of tumors with hSNF5/INI1 inactivation and to gain insights into a possible predisposition syndrome associated with germline mutations, we have screened a variety of neoplastic lesions, by direct sequencing or denaturing high-performance liquid chromatography (dHPLC) analysis of tumor DNA (Underhill et al. 1997). Surprisingly, in addition to MRTs, hSNF5/INI1 mutations were detected in most choroid plexus carcinomas (CPCs), highly malignant tumors arising from the cerebral ventricles, and in a subset of medulloblastomas and central primitive neuroectodermal tumors (PNETs) (Sévenet et al., in press). Moreover, during the course of this study, truncating mutations were identified in the tumors of patients that had individual or familial histories of cancer. Together with previous reports of familial cases of MRT and of germline mutations of hSNF5/INI1, this observation strongly suggested that constitutional mutations of hSNF5/INI1 could predispose to cancer (Lynch et al. 1983; Biegel et al. 1999). We now report the analysis of hSNF5/INI1-gene mutation in those individuals or families with multiple cases of cancer.
Patients and Methods
Patients
Tumors were obtained either as frozen material stored at −80°C or as paraffin-embedded tissue. Blood samples from affected and unaffected family members were collected. Informed consent was obtained from all individuals or parents.
Mutation Analysis
DNA from blood and frozen tumor samples was extracted, by means of standard procedures, with phenol/chloroform extraction and ammonium acetate/ethanol precipitation. Paraffin-embedded fragments were deparaffinized by xylene and were rinsed in ethanol prior to DNA extraction by the QiaAmp Tissue Kit (Qiagen). RNA was isolated from frozen samples by means of Trizol reagent (GibcoBRL). For mutation detection, dHPLC (denaturing high-performance liquid chromatography) analysis using the Wave technology (Transgenomic) and direct sequencing were used.
In brief, for each tumor DNA, the nine hSNF5/INI1 exons were PCR amplified by means of primers localized in flanking intronic sequences. In addition, the complete hSNF5/INI1 transcript was reverse transcribed, and five overlapping PCR products were analyzed. For dHPLC analysis, PCR products were denatured for 10 min at 98°C, were submitted to a gradual reannealing from 98°C to 25°C over a period of 40 min, were loaded onto a DNAsep column, and were eluted, by means of an acetonitrile gradient, at a defined melting temperature. Wavemaker 1.2.2 software was used to predict both the mean melting temperature of each PCR fragment and the appropriate linear acetonitrile gradient necessary to differentiate hetero- and homoduplexes. These conditions were also evaluated experimentally by analysis of the elution time at different temperatures (melting curves) and by testing of the elution time of PCR products from known mutants (Versteege et al. 1998). The start and end points of the gradient were adjusted according to the size and the percentage of GC nucleotides of each PCR fragment. Sequencing was performed on purified PCR products by means of the BigDye Terminator method (PE Biosystems) and 373A or 377 automatic sequencers. The sequences of the intron-exon boundaries can be retrieved at the EMBL database, and the sequences of the primer pairs used for PCR, together with the dHPLC conditions, can be found at our web site at the Institut Curie.
Microsatellite Analysis
The loci used for haplotype determination have been described elsewhere (Versteege et al. 1998). Analysis at microsatellite loci was performed according to standard procedure. In brief, 40 ng of DNA were amplified by means of 6 pmol of each primer, by the GenAmp PCR core kit (PE Biosystems). After an initial denaturation at 96°C for 2 min, 35 cycles of PCR were performed. Each cycle consisted of denaturation at 96°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30s. Terminal extension was performed at 72°C for 5 min. PCR products were denatured, migrated on a 6% denaturing polyacrylamide gel, and then were transferred to a nylon N+ membrane (Amersham International) and hybridized with a 3′ end–labeled (CA)12 probe. The membranes were then washed three times with 2×SSC at room temperature and were subjected to autoradiography.
Results
Analysis of hSNF5/INI1 Sequences in Cancer-Prone Families
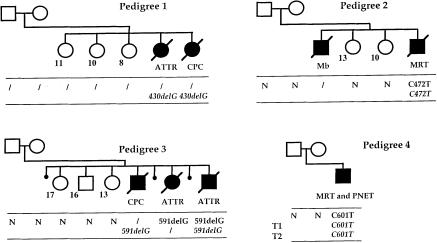
Three pedigrees with multiple cases of cancer demonstrating the presence of a hSNF5/INI1 point mutation in the tumor DNA and/or constitutional DNA of at least one member of the family were further analyzed. Pedigree 1 was characterized by two cases of CNS tumors: an atypical teratoid and rhabdoid tumor (ATTR) and a CPC, diagnosed at age 34 mo and 16 mo, respectively. Analysis of the sequence of the hSNF5/INI1 gene of the DNA of these two tumors demonstrated a deletion of one G at codon 144 (bp 430), resulting in a frameshift (fig. 1). The observation of the same mutation in these two tumors strongly suggested that it was constitutionally inherited. Constitutional DNA from members of this family were not available to confirm this hypothesis.
Figure 1.
Schematic representation of four pedigrees with mutations of hSNF5/INI1. The results of the molecular analysis are shown below each family member. Normal and italic characters indicate the results of the analysis of constitutional DNA and tumor DNA, respectively. N = normal sequence; a virgule (/) indicates that DNA was not available for analysis. Mb = medulloblastoma; other abbreviations are as defined in the text. The age (in years) of the healthy sibs is indicated. In pedigree 4, the results of the DNA analysis of the two tumors (T1 and T2) are given.
In pedigree 2, two of the four children died of aggressive cancers at a very young age. In the first child, a medulloblastoma with frontal metastasis was diagnosed at age 3 mo. The fourth child developed a congenital MRT of the soft tissues of the neck. DNA analysis of the latter tumor demonstrated a C→T transition resulting in a nonsense mutation at codon 158 (bp 472). The constitutional DNA from this patient displayed heterozygosity of the same mutant allele. Conversely, this mutation was not detected in the constitutional DNA of the healthy parents and unaffected sisters (fig. 1). The presence of the mutation at the constitutional and/or tumor level could not be documented for child II.1, because of the absence of stored biological material. However, the association, in this family, of a germline mutation in one sib (II.4) and of a rare CNS childhood tumor in another (II.1) , is certainly not coincidental, especially in light of our recent results showing that somatic mutations of hSNF5/INI1 can occur in medulloblastoma (Sévenet et al., in press).
Pedigree 3 was characterized by three cases of cancer, including one CPC at age 4 mo and two ATTR, one at age 2 mo and one at age 12 mo. Molecular analysis revealed a deletion of one G at codon 197 (bp 591), leading to a frameshift of the hSNF5/INI1 coding sequence, in all tested constitutional DNA and tumor DNA from affected members. In contrast, DNA from the healthy parents and from the three unaffected sibs demonstrated wild-type hSNF5/INI1 sequences (fig. 1).
Analysis of hSNF5/INI1 in One Patient with Bifocal Tumors
Constitutional DNA and tumor DNA from one patient with two successive tumors could be studied (fig. 1, pedigree 4); this patient was cured of a renal MRT resected at age 5 mo, then developed a central PNET 10 years later. Given the distinct phenotypes of these cancers and the long interval between their occurrences, this latter neoplasm was most probably a second tumor, rather than a late recurrence of the former. DNA from the two tumors demonstrated the presence of the same C→T transition, resulting in a nonsense mutation at codon 201 (bp 601). Constitutional DNA isolated from normal renal tissue and from blood exhibited heterozygosity for this mutation. Similar to what was observed in pedigrees 2 and 3, the DNA from the unaffected parents displayed wild-type sequences, indicating the de novo occurrence of this mutation.
Loss of the Wild-Type Allele in Tumors
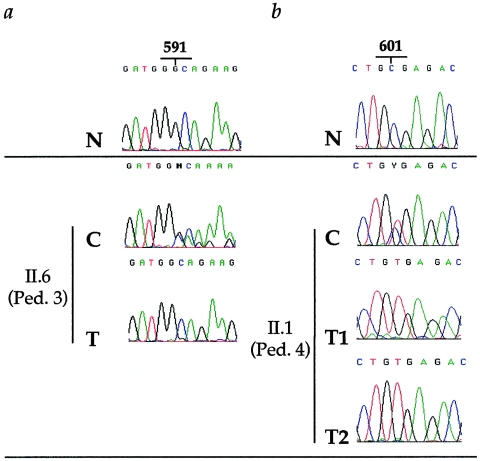
Three tumors displayed a high tumor-cell content, compatible with loss-of-heterozygosity (LOH) analysis at the hSNF5/INI1 locus. Indeed, comparison of the sequence profiles from constitutional DNA and tumor DNA from these cases clearly documented the complete loss of the wild-type hSNF5/INI1 allele in the tumor DNA (fig. 2). These data therefore provided evidence for a complete loss-of-function of hSNF5/INI1 in the tumors arising in the context of a constitutional mutation.
Figure 2.
Loss of the wild-type hSNF5/INI1 allele in tumors. The results for the male child in pedigree 3 who had ATTR (II.6) and for the patient in pedigree 4 who had bifocal tumors (II.1) are depicted. The normal sequences (N) are shown at the top; the constitutional sequences (C) that demonstrate heterozygosity between wild-type and mutated alleles are shown below them. a, Case II.6 from pedigree 3. The sequence of the ATTR DNA (T) shows the mutated 591delG allele with complete disappearance of the wild-type allele. b, Case II.1 from pedigree 4. DNA from the two successive tumors (T1 [renal MRT] and T2 [central PNET]) demonstrates the presence of the 601T mutated allele and the loss of the wild-type 601C allele. Therefore, in these three tumors, no functional allele of hSNF5/INI1 was retained.
Parental Origin of the Mutated Allele
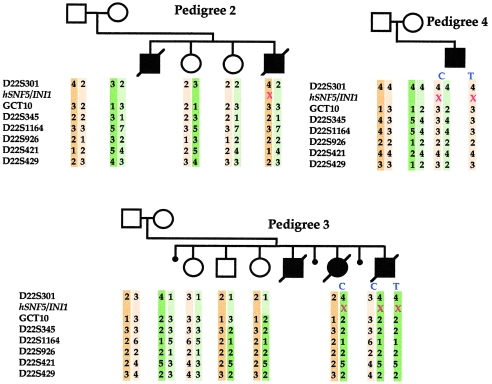
Microsatellite analysis at seven chromosome 22q polymorphic loci (D22S301, GCT10, D22S345, D22S1164, D22S926, D22S421, and D22S429) flanking the hSNF5/INI1 locus was performed on three pedigrees. Segregation and LOH analyses enabled us to determine the parental origin of lost and retained alleles in the tumor DNA and to reconstitute mutated and wild-type haplotypes. In pedigree 3, the mutation in cases II.5 and II.6 is inherited from the mother, which indicates the presence of a maternal mosaicism. Unaffected daughter II.1 has inherited the other maternal allele, and unaffected sibs II.2 and II.3 display crossing-over between maternal chromosomes excluding the hSNF5/INI1 mutation (fig. 3). The normal sequence of hSNF5/INI1 in both fibroblast DNA and blood DNA from the mother suggests that the mosaicism is restricted to her germinal lineage. A similar approach was performed for pedigrees 2 and 4 and showed that the mutation involved the paternal allele (fig. 3). However, for pedigree 4, characterized by a unique affected child, it could not be concluded whether the mutation was inherited from the father or occurred on the paternal chromosome at a postzygotic stage in the patient.
Figure 3.
Parental origin of the mutated allele. Haplotypes around the hSNF5/INI1 gene on chromosome 22 are displayed for pedigrees 2–4. Paternal and maternal haplotypes are indicated on the left and right, respectively. The seven microsatellite markers shown on the left are included within a 3-cM region (Dib et al. 1996). “C” and “T” denote constitutional DNA and tumor DNA, respectively. The point mutations of hSNF5/INI1 are depicted by red crosses. In pedigree 3, the mutation is carried by the maternal allele, whereas in pedigrees 2 and 4 it involves the paternal allele.
Discussion
The present report further strengthens the hypothesis that hSNF5/INI1 is a tumor-suppressor gene, since it fulfills the genetic features of this class of genes: biallelic, somatic loss-of-function mutations in sporadic tumor cases, and constitutional alterations causing a dominantly inherited cancer-predisposition syndrome associated with somatic loss of the wild-type allele in tumors (Knudson 1996). We propose to term this hereditary condition, caused by constitutional hSNF5/INI1 mutations, “rhabdoid predisposition syndrome” (RPS). At present, the spectrum of tumors observed in RPS includes renal and extrarenal MRT, CPC, central PNET, and medulloblastoma and fully overlaps with the spectrum of cancers that demonstrate acquired, biallelic mutations of this gene (Sévenet et al., in press). The observation of loss-of-function mutations of hSNF5/INI1 in these different neoplasia strongly suggests that they share common pathways of oncogenesis and that they might constitute a single family of tumors.
The present study suggests that the penetrance of this syndrome is high at a very young age, since all first neoplasia occurred at age <3 years in mutation carriers and since no hSNF5/INI1 mutation was detected in the DNA of the 11 healthy sibs or parents who were analyzed. This high penetrance, together with the frequently fatal outcome in affected members, is expected to account for the rarity of large pedigrees with transmission of the mutation across multiple generations.
In all instances in which DNA from healthy parents could be studied, it displayed a normal hSNF5/INI1 sequence, therefore demonstrating the de novo origin of the mutation. Such new mutations can occur either prezygotically, during oogenesis or spermatogenesis, or postzygotically, during early steps of embryogenesis. Our study documents that, in pedigree 3, the mutation was inherited from the mother and probably occurred during oogenesis, since both maternal-fibroblast DNA and maternal-blood DNA displayed normal hSNF5/INI1 sequences. In contrast, the mutation in pedigree 2 was inherited from the father and thus probably arose during spermatogenesis. In pedigree 4, the mutation also involved the paternal allele; however, at present, the pre- or postzygotic step of its occurrence cannot be documented in this pedigree with a single child. The analysis of sperm from the father might enable us to document the presence of the mutation at the germinal level. Although definitive conclusions cannot be drawn from these cases, our study suggests that RPS de novo mutations, like those of von Hippel-Lindau syndrome, do not demonstrate the preferential or exclusive involvement of the paternal allele, which is observed in other cancer-predisposition syndromes, such as MEN2A, MEN2B, neurofibromatosis type 1, and retinoblastoma (Dryja et al. 1989; Zhu et al. 1989; Jadayel et al. 1990; Carlson et al. 1994; Richards et al. 1995; Schuffenecker et al. 1997).
The present study demonstrates that a predisposition to a variety of cancer is linked to hSNF5/INI1 mutation. This alteration probably provides the molecular basis for familial and multifocal cases of the various tumors that are part of the RPS spectrum that has been reported elsewhere (Bonnin et al. 1984; Tijssen 1991; Fort et al. 1994; Matsumura et al. 1997; Chidambaram et al. 1998; Moschovi et al. 1998; Parellada et al. 1998; Raila et al. 1998). In particular, since the spectrum of tumors observed in RPS, especially brain tumors, largely overlaps with that of Li-Fraumeni syndrome (LFS), a cancer-predisposition syndrome frequently associated with p53 germline mutations, the possibility that a number of p53 gene mutation–negative LFS cases harbor hSNF5/INI1 mutations can now be tested (Kleihues et al. 1997; Varley et al. 1997; Hisada et al. 1998).
Acknowledgments
This work was supported by grants from the Association pour la Recherche contre le Cancer, the Institut Curie, the Institut National de la Santé et de la Recherche Médicale, and the Programme Hospitalier de Recherche Clinique. N.S. is the recipient of a fellowship from the Ministère de l'Education Nationale, de la Recherche et de la Technologie. We thank André Nicolas for technical help, and we thank the following clinicians for providing samples used in this study: A. Bornemann, V. Costes, T. Klingebiel, A. Lacquerrière, G. Marguerite, R. Meyermann, P. Ruck, N. Saran, and J.-P. Vannier.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- EMBL database, http://www.ebi.ac.uk/cgi-bin/emblfetch (for sequences of the intron-exon boundaries [Y17118-Y17126])
- Institut Curie, http://www.curie.fr/sr/unites/u509 (for sequences of the primer pairs used for PCR together with the dHPLC conditions)
References
- Beckwith JB, Palmer NF (1978) Histopathology and prognosis of Wilms tumor. Cancer 41:1937–1948 [DOI] [PubMed]
- Biegel JA, Allen CS, Kawasaki K, Shimizu N, Budarf ML, Bell CJ (1996) Narrowing the critical region for a rhabdoid tumor locus in 22q11. Genes Chromosomes Cancer 16:94–105 [DOI] [PubMed]
- Biegel JA, Burk CD, Parmiter AH, Emanuel BS (1992) Molecular analysis of a partial deletion of 22q in a central nervous system rhabdoid tumor. Genes Chromosomes Cancer 5:104–108 [DOI] [PubMed]
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B (1999) Germ-line and acquired mutations of INI1 in atypical teratoid rhabdoid tumors. Cancer Res 59:74–79 [PubMed]
- Bonnin JM, Rubinstein LJ, Palmer NF, Beckwith JB (1984) The association of embryonal tumors originating in the kidney and in the brain: a report of seven cases. Cancer 54:2137–2146 [DOI] [PubMed]
- Burger PC, Yu IT, Tihan T, Friedman HS, Strother DR, Kepner JL, Duffner PK, et al (1998) Atypical teratoid/rhabdoid tumor of the central nervous system: a highly malignant tumor of infancy and childhood frequently mistaken for medulloblastoma, a pediatric oncology group study. Am J Surg Pathol 22:1083–1092 [DOI] [PubMed]
- Carlson KM, Bracamontes J, Jackson CE, Clark R, Lacroix A, Wells SA Jr, Goodfellow PJ (1994) Parent-of-origin effects in multiple endocrine neoplasia type 2B. Am J Hum Genet 55:1076–1082 [PMC free article] [PubMed]
- Chidambaram B, Santhosh V, Shankar SK (1998) Identical twins with medulloblastoma occurring in infancy. Childs Nerv Syst 14:421–425 [DOI] [PubMed]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Douglass EC, Valentine M, Rowe ST, Parham DM, Wilimas JA, Sanders JM, Houghton PJ (1990) Malignant rhabdoid tumor: a highly malignant childhood tumor with minimal karyotypic changes. Genes Chromosomes Cancer 2:210–216 [DOI] [PubMed]
- Dryja TP, Mukai S, Petersen R, Rapaport JM, Walton D, Yandell DW (1989) Parental origin of mutations of the retinoblastoma gene. Nature 339:556–558 [DOI] [PubMed]
- Fort DW, Tonk VS, Tomlinson GE, Timmons CF, Schneider NR (1994) Rhabdoid tumor of the kidney with primitive neuroectodermal tumor of the central nervous system: associated tumors with different histologic, cytogenetic and molecular findings. Genes Chromosomes Cancer 11:146–152 [DOI] [PubMed]
- Hisada M, Garber JE, Fung CY, Fraumeni JF, Li FP (1998) Multiple primary cancers in families with Li-Fraumeni syndrome. J Natl Cancer Inst 90:606–611 [DOI] [PubMed]
- Jadayel D, Fain P, Upadhyaya M, Ponder MA, Huson SM, Carey J, Fryer A, et al (1990) Paternal origin of new mutations in von Recklinghausen neurofibromatosis. Nature 343:558–559 [DOI] [PubMed]
- Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP (1994) Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002–2006 [DOI] [PubMed]
- Kleihues P, Schäuble B, Hausen AZ, Esteve J, Ohgaki H (1997) Tumours associated with p53 germline mutations, a synopsis of 91 families. Am J Pathol 150:1–13 [PMC free article] [PubMed]
- Knudson AG (1996) Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol 122:135–140 [DOI] [PubMed]
- Lynch HT, Shurin SB, Dahms BD, Izant RJ, Lynch J, Danes BS (1983) Paravertebral malignant rhabdoid tumor in infancy, in vitro studies of a familial tumor. Cancer 52:290–296 [DOI] [PubMed]
- Matsumura H, Yoshimine T, Yamamoto S, Maruno M, Hayakawa T, Ono Y, Kondoh N, et al (1997) Single solitary metastasis of the slowly progressive type of renal cell carcinoma to the choroid plexus—case report. Neurol Med Chir (Tokyo) 37:916–919 [DOI] [PubMed]
- Moschovi M, Sotiris Y, Prodromou N, Tsangaris GT, Constantinidou Van-Vliet C, Kalpini-Mavrou A, Tzortzatou-Stathopoulou F (1998) Familial medulloblastoma. Pediatr Hematol Oncol 15:421–424 [DOI] [PubMed]
- Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M (1995) A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res 23:1127–1132 [DOI] [PMC free article] [PubMed]
- Parellada JA, Gupta P, Rose W (1998) Rhabdoid tumor of the kidney with primitive neuroectodermal tumor of the central nervous system. Eur J Radiol 28:219–221 [DOI] [PubMed]
- Parham DM, Weeks DA, Beckwith JB (1994) The clinicopathologic spectrum of putative extrarenal rhabdoid tumors. Am J Surg Pathol 18:1010–1029 [DOI] [PubMed]
- Raila FA, Bottoms WT, Fratkin JD (1998) Solitary choroid plexus metastasis from a renal cell carcinoma. South Med J 91:1159–1162 [DOI] [PubMed]
- Richards FM, Payne SJ, Zbar B, Affara NA, Ferguson-Smith MA, Maher ER (1995) Molecular analysis of de novo germline mutations in the von Hippel-Lindau disease gene. Hum Mol Genet 4:2139–2143 [DOI] [PubMed]
- Rorke LB, Packer RJ, Biegel JA (1996) Central nervous system atypical teratoid/rhabdoid tumours of infancy and childhood: definition of an entity. J Neurosurg 85:56–65 [DOI] [PubMed]
- Rosty C, Peter M, Zucman J, Validire P, Delattre O, Aurias A (1998) Cytogenetic and molecular analysis of a t(1;22)(p36;q11.2) in a rhabdoid tumor with a putative homozygous deletion of chromosome 22. Genes Chromosomes Cancer 21:82–89 [PubMed]
- Schofield DE, Beckwith JB, Sklar J (1996) Loss of heterozygosity at chromosome regions 22q11-12 and 11p15.5 in renal rhabdoid tumors. Genes Chromosomes Cancer 15:10–17 [DOI] [PubMed]
- Schuffenecker I, Ginet N, Goldgar D, Eng C, Chambe B, Boneu A, Houdent C, et al (1997) Prevalence and parental origin of de novo RET mutations in multiple endocrine neoplasia type 2A and familial medullary thyroid carcinoma. Am J Hum Genet 60:233–237 [PMC free article] [PubMed]
- Sévenet N, Lellouch-Tubiana A, Schofield D, Hoang-Xuan K, Gessler M, Birnbaum D, Jeanpierre C, Jouvet A, Delattre O. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet (in press) [DOI] [PubMed] [Google Scholar]
- Tijssen CC (1991) Familial medulloblastoma in siblings: report in one family and review of the literature. Surg Neurol 36:234 [DOI] [PubMed]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, et al (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed]
- Varley JM, Evans DGR, Birch JM (1997) Li-Fraumeni syndrome—a molecular and clinical review. Br J Cancer 76:1–14 [DOI] [PMC free article] [PubMed]
- Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, et al (1998) Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203–206 [DOI] [PubMed]
- Wade PA, Wolffe AP (1999) Transcriptional regulation: SWItching circuitry. Curr Biol 25:R221–R224 [DOI] [PubMed]
- White FV, Dehner LP, Belchis DA, Conard K, Davis MM, Stocker JT, Zuppan CW, et al (1999) Congenital disseminated malignant rhabdoid tumor: a distinct clinicopathologic entity demonstrating abnormalities of chromosome 22q11. Am J Surg Pathol 23:249–256 [DOI] [PubMed]
- Wick MR, Ritter JH, Dehner LP (1995) Malignant rhabdoid tumors: a clinicopathologic review and conceptual discussion. Sem Diagn Pathol 12:233–248 [PubMed]
- Zhu XP, Dunn JM, Phillips RA, Goddard AD, Paton KE, Becker A, Gallie BL (1989) Preferential germline mutation of the paternal allele in retinoblastoma. Nature 340:312–313 [DOI] [PubMed]