Summary
We report the identification of a new locus for generalized epilepsy with febrile seizures plus (GEFS+). Six family members manifested isolated typical febrile seizures (FS), and five had typical FS associated with generalized epilepsy (FS+, generalized tonic/clonic seizures). Afebrile seizures occurred from childhood until the teenage years. The maximum two-point LOD score was 3.99 for markers D2S294 and D2S2314. Flanking markers place the GEFS+ locus between D2S141 and D2S116, with multipoint analysis favoring the 13-cM interval spanned by D2S294 and D2S364. This locus is the second GEFS+ locus to be reported, which suggests that this syndrome is genetically heterogeneous.
Introduction
Febrile seizures (FS [MIM 601087]) are common, affecting 2%–5% of children between 6 mo and 6 years of age. Seizures occur in normally healthy children, in cases of fever with body temperature >38°C. Seizures are not associated with developmental delay, intracranial infection, or any disease known to cause epilepsy, such as tumor, cranial traumatism, etc. (Lennox 1949). Most of the children present with a single episode. However, a small percentage of children experience recurrent FS (Berg 1997), and some of these go on to develop epilepsy, especially idiopathic generalized epilepsy (IGE [MIM 600669]) (Doose and Maurer 1997) and temporal lobe epilepsy (Maher and McLachlan 1995).
The precise cause of FS is unknown. However, the existence of familial cases supports the idea that FS have a genetic factor (Frantzen et al. 1970). The main genetic study was performed by Rich et al. (1987), who concluded that FS were transmitted as a dominant autosomal trait in 30% of cases and as a polygenic trait in other cases.
Some families appear to be clinically heterogeneous, with cases of isolated FS, isolated epilepsy, or FS associated with epilepsy, especially IGE. Genetic factors are also implicated in the etiology of IGE. Indeed, IGE is up to 95% concordant in monozygotic twins (Berkovic et al. 1998), and close relatives of IGE probands frequently present with IGE, with seizures similar or different from those of the probands (Tsuboi and Endo 1977). Genetic studies of IGE have shown that the risk to first-degree relatives of IGE probands is ∼5%–10%, versus <3% to controls (Beck-Mannagetta and Janz 1991). A family with several cases of generalized epilepsy with febrile seizures plus (GEFS+) has been extensively described in a recent article (Scheffer and Berkovic 1997), and this description was later completed by studies of nine further pedigrees (Singh et al. 1999).
One of these GEFS+ pedigrees has recently been linked to a missense mutation (C387G transversion in exon 3) in the sodium-channel β1-subunit gene (SCN1B [MIM 600235]), which maps to chromosome 19q (Wallace et al. 1998). This mutation results in the replacement of a highly conserved cysteine with a tryptophan (C121W).
Five putative loci for phenotypes, in which FS occur, have been published as being involved in six pedigrees: FEB1 on chromosome 8q13-q21 (Wallace et al. 1996), chromosome 2 (Lopes-Cendes et al. 1996), chromosome 11 (Anderson et al. 1995), FEB2 on chromosome 19p (Johnson et al. 1998; Kugler et al. 1998), and chromosome 19q (Wallace et al. 1998).
We performed a genomewide search for a novel FS locus in a French family with GEFS+ and observed linkage of the disorder to a group of microsatellite markers located on chromosome 2q24-q33.
Patients, Material, and Methods
Family Ascertainment and Diagnostic Classification
Each member was directly interviewed (by D.C.) to determine the occurrence and the type of seizure (table 1). Individuals were considered affected when either clinical records or at least one close relative (generally, the mother) confirmed that they presented between the ages of 6 mo and 6 years with clonic, tonic/clonic, or atonic seizures associated with fever. The diagnosis of other seizure types was made according to the classification of epileptic seizures (Commission on Classification and Terminology of the International League against Epilepsy 1981). Family members who had presented with signs or symptoms that were suggestive of epileptic seizures but were not confirmed by at least one relative or by clinical records from previous hospitalizations were considered to be of unknown disease status. Informed consent was obtained from all study participants. This study has been approved by the ethics committee of the Pediatric Department of the Geneva University Hospital.
Table 1.
Clinical Data[Note]
| Patient | Age at First FS (mo) | FS after 6 Years | Afebrile Seizures |
| I 01 | … | No | Unknowna |
| I 02 | Unknownb | No | No |
| II 01 | 12 | No | No |
| II 03 | 1 | Yes | GTCS, A |
| II 12 | 12 | No | No |
| II 13 | 36 | Yes | No |
| III 01 | 36 | Yes | No |
| III 03 | 7 | Yes | HCS |
| III 04 | 12 | Yes | No |
| III 05 | 7 | Yes | A, GTCS, AtS |
| III 09 | 7 | No | GTCS |
| III 10 | 7 | Yes | A, GTCS, AtS |
| III 11 | 36 | ? | No |
Note.—A = absence seizures, AtS = atonic seizures, and HCS = hemicorporeal clonic seizures.
May have presented with GTCS in childhood. Nobody was able to confirm these facts, and no clinical records were available.
Treated in childhood, with phenobarbital, for “nervous breakdown.” Nobody was able to confirm these facts, and no clinical records were available.
Genotyping and Linkage Analysis
DNA was extracted from peripheral blood leukocytes by use of the Nucleon BACC 2 kit (Amersham). Microsatellite DNA polymorphisms were amplified by PCR with the following conditions in a final volume of 25 ml: 100 ng DNA, 20 pmol each primer, 200 μM each dNTP, 1.5 mM MgCl2, and 1.5 U Taq polymerase with 2.5 ml of the correspondent buffer (Eurobio). Both the primers used and their annealing conditions were those reported by Dib et al. (1996). Samples were amplified in a thermocycler (Biometra), with the following conditions: 95°C for 3 min; then 30 cycles of 30 s at 92°C, 30 s of annealing, and 30 s at 72°C. PCR products were loaded with a 10-pb ladder, on a 10% polyacrylamide gel. The observed alleles were numbered according to Généthon.
Two-point linkage analyses were performed by the MLINK subroutine of the LINKAGE program, and multipoint linkage analyses were performed by the LINKMAP subroutine of the LINKAGE program (Lathrop et al. 1985), with the model used by Wallace et al. (1998): autosomal dominant model of disease inheritance (because of a male-to-male transmission), with a disease-allele frequency of .01, a phenocopy rate of .03, and a genetic penetrance of .90 for LOD-score calculations. Marker-allele frequencies were those estimated in the CEPH reference families.
Results
Clinical Analysis
Seizures were diagnosed in 12 living subjects, including 11 relatives and a spouse. Febrile seizures often lasted for >20 min, without focalization or motor deficiency (except in three relatives), and therefore did not meet complex FS criteria. The fever was often mild (38°C, 38.5°C). The mean number of attacks was ∼5 (range 2–20).
Five family members presented with afebrile seizures associated with FS until their teenage years (mean 11 years). These seizures were generalized or hemicorporeal, tonic/clonic or atonic absence attacks. Neurological examination showed normal findings. Interrictal electroencephalograms were normal except in three patients, in whom generalized spike-and-wave discharges arose a few years after the onset of FS.
The first seizure occurred sooner in patients with both FS and other types of seizures (mean 5.8 mo, range 1–7 mo) than in those with FS only (mean 24 mo, range 12–36 mo).
The paternal grandfather (I 01) may have had childhood generalized tonic/clonic seizures (GTCS), and the paternal grandmother (I 02) was treated with phenobarbital for “nervous breakdown” for a few months in childhood. However, nobody was able to confirm these facts, and no clinical records were available. Thus, these subjects were considered to be of unknown disease status.
Exclusion Analysis
We tested linkage to microsatellite polymorphisms located at known idiopathic epilepsy or FS loci (juvenile myoclonic epilepsy on chromosomes 6p (Greenberg et al. 1988) and 15q (Elmslie et al. 1997), benign familial neonatal convulsions on chromosomes 20q (Leppert et al. 1989) and 8q (Lewis et al. 1993), benign familial infantile convulsions on chromosome 19q (Guipponi et al. 1997), FS on chromosomes 8q, 11q, and 19) (Anderson et al. 1995; Wallace et al. 1996, 1998; Johnson et al. 1998; Steinlein 1998). We observed negative LOD scores for markers flanking and within these loci (data not shown). These data indicated that the disorder in this kindred was not linked to these known epilepsy loci.
Genomewide Search for a Novel GEFS+ Locus
We then undertook genomewide genetic-linkage analysis to identify the novel GEFS+ locus in this family. We analyzed 257 microsatellite polymorphisms from the Généthon linkage map (Dib et al. 1996), spaced 5–20 cM throughout the autosomal chromosomes, for linkage with the disorder in this GEFS+ kindred. We observed positive LOD scores for individual markers located on chromosomes 2 and 17: for D2S382, 1.41 (recombination fraction [θ] 0), and for D17S1824, 1.48 (θ=.05). Then, nine additional markers on chromosome 2 and four on chromosome 17 were studied. We observed significantly negative LOD scores for markers as close as 1–2 cM from the chromosome 17 locus (data not shown). These data indicated that the positive LOD score for D17S1824 was simply random association observed in a relatively small kindred.
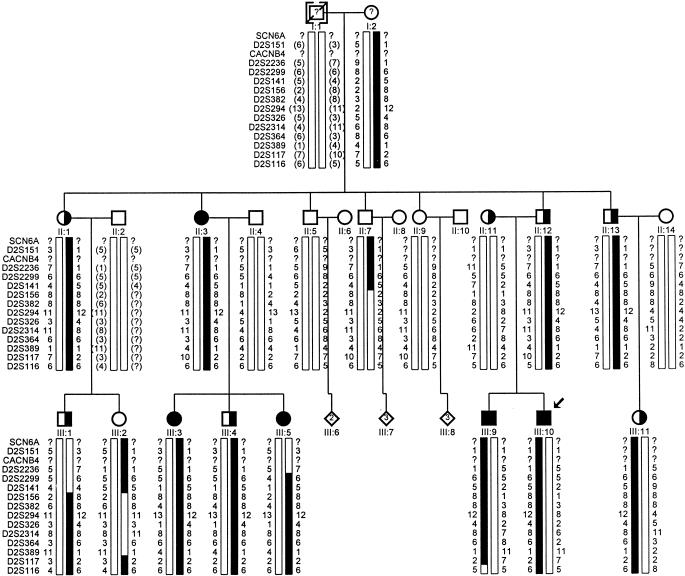
Four of the nine additional markers on chromosome 2—namely, D2S294, D2S326, D2S2314, and D2S364—yielded two-point LOD scores >3.0 (table 2). The genotypes for all markers with no recombination and for the two adjacent flanking markers showing obligate recombinants at D2S141 and D2S117 are shown with the pedigree in figure 1.
Table 2.
Chromosome 2 Linkage Analysis Results
|
LOD Score at θ = |
|||||||||
| Marker | 0 | .01 | .05 | .1 | .2 | .3 | .4 | Maximum LOD Score | Maximum θ |
| D2S151 | −1.49 | −1.43 | −.88 | −.38 | .10 | .19 | .11 | .19 | .3 |
| D2S2236 | −1.00 | −.94 | −.38 | .10 | .48 | .44 | .23 | .46 | .2 |
| D2S2299 | .62 | .63 | .66 | .64 | .52 | .33 | .11 | .66 | .05 |
| D2S141 | −.85 | −.09 | .59 | .88 | .92 | .71 | .35 | .92 | .2 |
| D2S156 | 1.16 | 1.14 | 1.05 | .93 | .69 | .41 | .14 | 1.16 | 0 |
| D2S382 | 1.94 | 1.90 | 1.76 | 1.58 | 1.19 | .76 | .29 | 1.94 | 0 |
| D2S294 | 3.99 | 3.93 | 3.67 | 3.33 | 2.58 | 1.75 | .82 | 3.99 | 0 |
| D2S326 | 3.63 | 3.57 | 3.33 | 3.01 | 2.33 | 1.57 | .72 | 3.63 | 0 |
| D2S2314 | 3.99 | 3.93 | 3.67 | 3.33 | 2.58 | 1.75 | .82 | 3.99 | 0 |
| D2S364 | 3.13 | 3.08 | 2.87 | 2.60 | 2.00 | 1.33 | .60 | 3.13 | 0 |
| D2S389 | 2.31 | 2.27 | 2.07 | 1.82 | 1.28 | .70 | .19 | 2.31 | 0 |
| D2S117 | 2.99 | 2.97 | 2.85 | 2.65 | 2.12 | 1.45 | .67 | 2.99 | 0 |
| D2S116 | −2.09 | −.14 | .43 | .58 | .55 | .37 | .15 | .58 | .1 |
Figure 1.
Pedigree of French GEFS+ family showing haplotype analysis for 13 markers on chromosome 2q. Markers are listed top to bottom from centromere to telomere. For each marker, the alleles are numbered on the basis of sizes reported on CEPH families (Généthon). Novel alleles were assigned numbers after the last reported in the Généthon database. Circles denote females, squares denote males, a slashed symbol denotes a deceased subject, blackened symbols denote GEFS+ patients, half-blackened symbols denote isolated FS patients, a question mark (?) denotes possible seizures reported by relatives (no medical records), a bracketed subject denotes an adopted child, and bracketed allele numbers denote deduced alleles. The boldfaced bars represent the GEFS+ haplotype. An arrow indicates the proband.
Multipoint linkage analysis was performed for the disease locus and two adjacent markers at a time, sequentially, for all markers from D2S382 to D2S117. Published distances between these markers were obtained from Généthon (Dib et al. 1996). Multipoint linkage analysis produced a maximum LOD score of 4.16 at D2S294 and favored the 13-cM interval spanned by D2S294 and D2S364.
Discussion
We identified a new locus for GEFS+ on chromosome 2q within a French family. Previous physical mapping of the microsatellite markers allows us to assign this locus to chromosome 2q24-q33. At present, our data localize this region to a 30-cM interval between D2S141 and D2S117. This interval seems to overlap with that of the chromosome 2–linked GEFS+ Australian pedigree (Lopes-Cendes et al. 1996; G. Rouleau, personal communication).
With identification of FS loci on chromosome 8q (Wallace et al. 1996), 19p (Johnson et al. 1998; Kugler et al. 1998), and 19q (Wallace et al. 1998), it is possible to compare phenotypes in families in which the disorder is linked to each of these loci. In our pedigree, almost half the affected relatives (5/11) presented with afebrile seizures (atonic seizures, hemicorporeal seizures, absences, and GTCS) with FS, whereas, in 19p-linked families, FS were isolated. In the 8q-linked pedigree, very few family members (3/19) have afebrile seizures (including one case of temporal lobe epilepsy with hippocampal sclerosis). The penetrance of the trait seems to be higher (∼90%) in 2q-linked GEFS+ pedigrees (Lopes-Cendes et al. 1996; the present one) than in other families affected by GEFS+ (Scheffer and Berkovic 1997; Wallace et al. 1998; Singh et al. 1999) or by FS (Johnson et al. 1998; Kugler et al. 1998).
On the contrary, the frequency of isolated FS (11% in the Australian family vs. 50% in the French family) and the severity of the epileptic syndromes seems to be different between 2q-linked GEFS+ families. This must be confirmed by analyzing new 2q-linked GEFS+ pedigrees.
Within the candidate region there is a three-gene cluster of great interest. The three genes of this cluster encode sodium-channel α-subunits (SCN1A, 2q24; SCN3A, 2q24.3; and SCN2A1, 2q31). Two other potential candidate channel subunit genes (sodium-channel α6-subunit gene SCN6A, 2q21-q23 and calcium-channel β4-subunit gene CACNB4, 2q22-q23) were excluded by centromeric recombinants. One sodium channel subunit is already known to be involved in GEFS+ (SCN1B, Wallace et al. 1998). Thus, the analysis of this three-gene cluster is under way in the French family with FC+.
In conclusion, we identified, in a French pedigree, a new locus for GEFS+. It maps to chromosome 2q24-q33 and spans a 30-cM candidate region bounded by markers D2S141 and D2S116. In the near future, this candidate region will be tested in other families with FS and GEFS+, and the sodium-channel gene cluster mapping to 2q24-q33 will be screened for mutations in those chromosome 2q–linked pedigrees.
Acknowledgments
We would like to thank the family studied and all subjects who agreed to consult D.C. and then consented to the genetic study. We also thank Dr. G. Rouleau for providing unpublished information about the second chromosome 2–linked pedigree and Dr. B. Echenne for providing data for some relatives. The study was supported by PNR38 grant 4038-044050 from the Swiss FNRS (National Research Science Foundation). M.G. was supported by the Groupement de Recherche et d'Etude des Génome (GREG).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Généthon, http://www.genethon.fr
- Genome Database, http://gdbwww.gdb.org
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for FS [MIM 601087], IGE [MIM 600669], and sodium-channel β1-subunit gene [MIM 600235])
References
- Anderson VE, Rich SS, Wilcox KJ, Ahrens MJ, Weber JL, Dubowsky J (1995) Gene mapping studies in febrile convulsions. Epilepsia 36:S215 [Google Scholar]
- Beck-Mannagetta G, Janz D (1991) Syndrome-related genetics in generalized epilepsy. Epilepsy Res Suppl 4:105–111 [PubMed]
- Berg AT, Shinnar S, Darefsky AF, Holford TR, Shapiro ED, Salomon ME, Crain EF, et al (1997) Predictors of recurrent febrile seizures: a prospective cohort study. Arch Pediatr Adolesc Med 151: 371–378 [DOI] [PubMed]
- Berkovic SF, Howell RA, Hay DA, Hopper JL (1998) Epilepsies in twins: genetics of the major epilepsy syndromes. Ann Neurol 43:435–445 [DOI] [PubMed]
- Commission on Classification and Terminology of the International League Against Epilepsy (1981) Proposal for a revised clinical and electroencephalographic classification of epileptic seizures. Epilepsia 22:489–501 [DOI] [PubMed]
- Dib C, Fauré C, Fizames D, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Doose H, Maurer A (1997) Seizure risk in offspring of individuals with a history of febrile convulsions. Eur J Pediatr 156:476–481 [DOI] [PubMed]
- Elmslie FV, Rees M, Williamson MP, Kern M, Kjeldsen MJ, Pang KA, Sundqvist A, et al (1997) Genetic mapping of a major susceptibility locus for juvenile myoclonic epilepsy on chromosome 15q. Hum Mol Genet 6:1329–1334 [DOI] [PubMed]
- Frantzen E, Lennox-Buchtal M, Nygaard A, Stene J (1970) A genetic study of febrile convulsions. Neurology 20:909–917 [DOI] [PubMed]
- Greenberg DA, Delgado-Escueta AV, Widelitz H, Sparkes RS, Treiman L, Maldonado HM, Park MS, et al (1988) Juvenile myoclonic epilepsy (JME) may be linked to the BF and HLA loci on human chromosome 6. Am J Med Genet 31:185–192 [DOI] [PubMed]
- Guipponi M, Rivier F, Vigevano F, Beck C, Crespel A, Echenne B, Lucchini P, et al (1997) Linkage mapping of benign familial infantile convulsions (BFIC) to chromosome 19q. Hum Mol Genet 6:473–477 [DOI] [PubMed]
- Johnson EW, Dubovsky J, O'Donovan CA, Orr HT, Anderson VE, Gil-Nagel A, Ahmann P, et al (1998) Evidence for a novel gene for familial febrile convulsions, FEB2, linked to chromosome 19p in an extended family from the Midwest. Hum Mol Genet 7:63–67 [DOI] [PubMed]
- Kugler SL, Stenroos ES, Mandelbaum DE, Lehner T, McKoy VV, Prossick T, Sasvari J, et al (1998) Hereditary febrile convulsions: phenotype and evidence for a chromosome 19p locus. Am J Med Genet 79:354–361 [DOI] [PubMed]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multipoint linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed]
- Lennox MA (1949) Febrile convulsions in childhood: a clinical and electroencephalographic study. Am J Dis Child 78:868–882 [DOI] [PubMed] [Google Scholar]
- Leppert M, Anderson VE, Quattlebaum T, Stauffer D, O'Connell P, Nakamura Y, Lalouel JM, et al (1989) Benign familial neonatal convulsions linked to markers on chromosome 20. Nature 337:647–648 [DOI] [PubMed]
- Lewis TB, Leach RJ, Ward K, O'Connell P, Ryan SJ (1993) Genetic heterogeneity in benign familial neonatal convulsions: identification of a new locus on chromosome 8q. Am J Hum Genet 53:670–675 [PMC free article] [PubMed]
- Lopes-Cendes I, Scheffer IE, Berkovic SF, Rousseau M, Andermann E, Rouleau GA (1996) Mapping a locus for idiopathic generalised epilepsy in a large multiplex family. Epilepsia 37:127 [Google Scholar]
- Maher J, McLachlan RS (1995) Febrile convulsions: is seizure duration the most important predictor of temporal lobe epilepsy? Brain 118:1521–1528 [DOI] [PubMed]
- Rich SS, Annegers JF, Hauser WA, Anderson VE (1987) Complex segregation analysis of febrile convulsions. Am J Hum Genet 41:249–257 [PMC free article] [PubMed]
- Scheffer IE, Berkovic SF (1997) Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain 120:479–490 [DOI] [PubMed]
- Singh R, Scheffer IE, Crossland K, Berkovic SF (1999) Generalized epilepsy with febrile seizures plus: a common childhood-onset genetic epilepsy syndrome. Ann Neurol 45:75–81 [DOI] [PubMed]
- Steinlein OK (1998) New insights into the molecular and genetic mechanisms underlying idiopathic epilepsies. Clin Genet 54:169–175 [DOI] [PubMed]
- Tsuboi T, Endo S (1977) Incidence of seizures and EEG abnormalities among offspring of epileptic patients. Hum Genet 36:173–189 [DOI] [PubMed]
- Wallace RH, Berkovic SF, Howell RA, Sutherland GR, Mulley JC (1996) Suggestion for a major gene for familial febrile convulsions mapping to 8q13q21. J Med Genet 33:308–312 [DOI] [PMC free article] [PubMed]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George AL Jr, Phillips HA, Saar K, et al (1998) Febrile seizures and generalized epilepsy associated with a mutation in the Na+ - channel beta 1 subunit gene SCN1B. Nat Genet 19:366–370 [DOI] [PubMed]