To the Editor:
Microcephaly is a condition in which the head circumference is smaller than <3 SD below the mean for age. Syndromic microcephaly is found in a number of environmental, chromosomal, or single-gene disorders. Nonsyndromic, isolated microcephaly is also etiologically heterogeneous, and micrencephaly—that is, a generalized reduction of the brain mass causing a small skull without craniosynostosis—appears as a distinct subtype within this group. When micrencephaly is the only or the leading pathological alteration, it is referred to as “primary microcephaly,” or “microcephalia vera” (Ross and Frias 1977; Baraitser 1997). Mental retardation ranges from moderate to severe in primary microcephaly, although motor development may be normal during the first years of life. Linear and ponderal growth is often impaired, and, although the cause for this finding is not clear, a deficiency of growth hormone (GH) has been implicated in some cases (Dacou-Voutetakis et al. 1974). When familial, primary microcephaly often appears to be transmitted as an autosomal recessive disorder (MIM 251200) with a significant proportion of cases associated with parental consanguinity and with an incidence of 1/30,000–1/250,000 (Van den Bosch 1959, and references therein). Microcephaly may not be present until late in the third trimester of pregnancy, so prenatal diagnosis is problematic (Tolmie et al. 1987). Genetic heterogeneity has long been suspected, on the basis of subtle phenotypic differences among families (Cowie 1960). Recently, a locus for primary microcephaly, MCPH1, has been identified at 8p22-pter by homozygosity mapping (see below), and evidence for locus heterogeneity has been shown (Jackson et al. 1998). MCPH2 is ascribed to 19q13 in the Human Gene Nomenclature Database, in which the as-yet-unpublished locus MCPH3 has also been registered.
Homozygosity mapping in consanguineous families is based on the assumption that a rare mutation is inherited from a common ancestor via both parents, so that affected siblings are homozygous by descent, for polymorphic markers close to the disease locus. Comparison of genotypic data, both between and within subjects, makes it a powerful strategy that needs only a few affected individuals in order to map a recessive disorder (Lander and Botstein 1987). We now report homozygosity mapping of a new locus, MCPH4, to chromosome 15, in a newly ascertained family.
The propositus is a male 22 years of age who presented, at age 4 mo, with microcephaly and left cryptorchidism. He was a first child born after an uneventful pregnancy and delivery to healthy young Moroccan parents, who are first cousins once removed and who both have normal head circumferences and an otherwise unremarkable family history. No craniosynostosis was present in the patient, and the initial psychomotor development was normal. In early childhood, growth was at the 3d centile for height and weight, and microcephaly was <6 SD below the mean for age (41.5 cm at age 4.0 years). Mental retardation was noted. In late childhood, IQ measures were consistently ∼⩽50. A brain computed-tomography scan showed large cerebral ventricles and no cerebral malformation or neuronal ectopia. A partial deficiency of GH secretion was demonstrated by dynamic testing with insulin, glucagon, and GH-releasing factor. Therapeutic GH supplementation at age 11–13 years produced no change in the growth curves. The pubertal development was normal. The patient now has a kind, collaborative, cheerful personality. A sister and two younger brothers presented with an identical picture of microcephaly and with height and weight growth at approximately the 3d centile (table 1). Minor malformations were noted in the youngest boy (epicanthal folds, single palmar creases, a left preauricular tag, and myopia) but not in the sister and other brother. GH treatment in the youngest brother, at age 4–7 years, yielded no appreciable effect on growth. Results of karyotypes of blood lymphocytes were normal. Extensive metabolic workups gave normal results. The levels of maternal blood glucose and phenylalaninemia were strictly normal.
Table 1.
Phenotypic Data of the Microcephaly Kindred
| Characteristics | Proband | AffectedSister | AffectedBrother | AffectedBrother |
| Age (years) | 22 | 21 | 19 | 16 |
| Height (cm) | 157 | 152 | 156 | 157 |
| Weight (kg) | 35 | 44 | 39 | 36 |
| Head circumference (cm) | 44.5 | 47 | 48 | 45 |
| Head circumference relative to mean for age (SD) | <−6 | <−5 | <−5 | <−6 |
The parents and patients gave informed consent to the genomic study, and DNA was extracted from peripheral-blood leukocytes. In a first analysis, we studied markers from the 8p region, where MCPH1 maps (Jackson et al. 1998), and found no evidence for linkage. A genomewide screen was then launched by use of a set of microsatellite markers from the Cooperative Human Linkage Center human screening set, Weber version 9 (Research Genetics). From this set of 386 markers, 239 (mainly tetranucleotides) were selected to span the entire genome in intervals of ⩽30 cM, since this spacing is sufficient for detection of homozygosity by descent in a family with this coefficient of inbreeding (Terwilliger et al. 1997). A pooling approach was employed for the initial screen. The DNAs of the parents were pooled in one sample, and the DNAs of the affected children were pooled in another (Arbour et al. 1997). Subsequently, these pools were typed by PCR amplification using 6 ng of each individual's DNA in a 15-μl final volume, followed by PAGE and silver staining (Budowle et al. 1988). Seven loci in the affected siblings—on 2q, 8p, 12p, 12q, 13q, and 15q—were identified as homozygous by state. These loci were then analyzed in the individual subjects, with additional, closely spaced markers (<2 cM apart). Marker order was obtained from the Center for Medical Genetics map, the Cooperative Human Linkage Center map, GeneMap '98, and the Genetic Location Database. When minor discrepancies between the various maps were observed, radiation-hybrid mapping was performed to determine the most probable order, by use of the GeneBridge 4 panel (Research Genetics) and the RH mapping program of the Whitehead Institute for Biomedical Research/MIT Center for Genome Research.
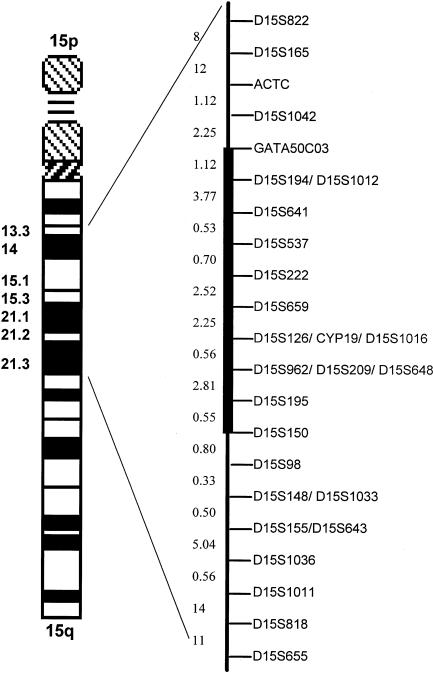
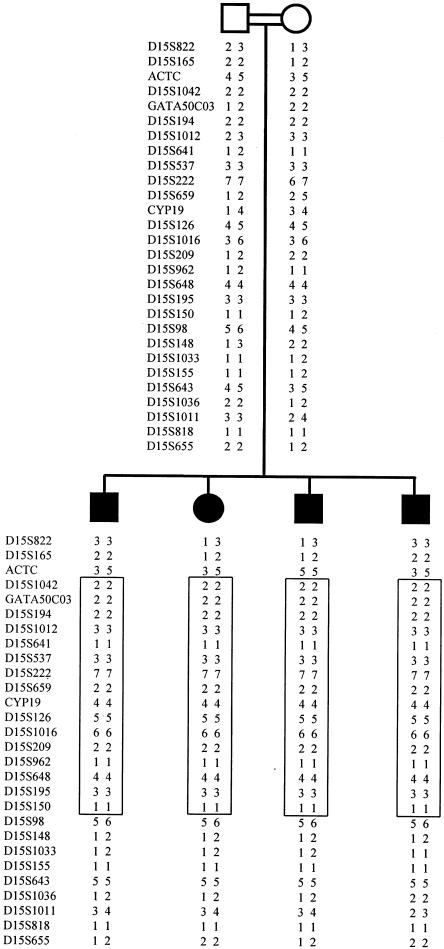
Six of the seven initial regions (listed above) were not consistently homozygous at each polymorphic locus, indicating, for the initial marker locus, identity by state rather than identity by descent. Conversely, one of the seven markers initially identified on chromosome 15q was found to be part of a genomic segment (fig. 1) where all informative markers were homozygous in the affected siblings but were heterozygous in the parents. This was consistent with identity by descent and homozygosity for a disease haplotype (fig. 2).
Figure 1.
Genetic map of chromosome 15 markers. Distances are shown in centimorgans. The distance between markers ACTC and D15S98 is 19 cM. The blackened bar indicates the MCPH4 candidate region.
Figure 2.
Haplotypes in the microcephaly kindred. Blackened symbols represent affected individuals. The region of homozygosity is boxed.
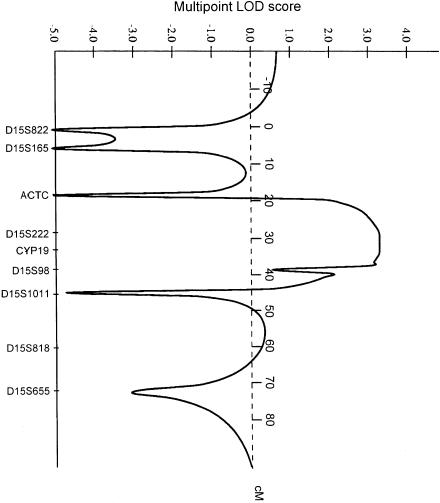
Multipoint linkage analysis was performed by use of the MAPMAKER/HOMOZ algorithm software (Kruglyak et al. 1995), under the assumption of a fully penetrant disease with an allele frequency of .002. Allele frequencies for each polymorphic marker of the candidate region were evaluated by genotyping 30 unrelated individuals from the same ethnic population. This analysis provided a maximum multipoint LOD score of 3.29 (fig. 3). Heterozygosity was found in one of the affected siblings for marker ACTC and in all four affected siblings for marker D15S98 (fig. 2), indicating that recombination events had occurred at both loci. Thus, a minimal critical region—that is, the smallest region found to be identical by descent, in all affected siblings—of 19 cM was observed between markers ACTC and D15S98. Because of uninformativeness of parental markers for its boundaries, however, the critical region might be as small as 5.3 cM, encompassing D15S222 and D15S962.
Figure 3.
Results of MAPMAKER/HOMOZ multipoint linkage analysis. A maximum multipoint LOD score of 3.29 is observed at CYP19.
Although linkage to this candidate region should be confirmed in additional families, our results present strong evidence for the presence of a new gene, MCPH4, at15q15-q21, a mutation of which presumably affects an aspect of neuronal proliferation. Although, during the past few years, knowledge of neuronal migration and brain-patterning defects such as holoprosencephaly or schizencephaly has increased, the molecular defects of neuronal proliferation are still poorly known. Considering the complexity of this process, locus heterogeneity is not unexpected (Walsh 1999). Identifying the genes implicated in primary microcephaly may prove particularly useful, since proper animal models for the developmental defects affecting the growth of the hemispheres are lacking, in part because of its human-specific nature.
Acknowledgments
This work was supported by a grant from the Fonds A & J Forton, Belgium. Cedric Govaerts is a research fellow of the Fonds pour la Recherche dans l'Industrie et l'Agriculture. We thank G. Vassart for constant advice, M. Georges for support, P. Vanderhaeghen for helpul discussions, and J. Richelle for informatics.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield, Medical Research Foundation, http://www.marshmed.org/genetics (for order and distances of markers on chromosome 15)
- Cooperative Human Linkage Center, http://lpg.nci.nih.gov/CHLC (for microsatellite markers used for the genome screen)
- GeneMap'98, http://www.ncbi.nlm.nih.gov/genemap98/ (for order and distances of markers on chromosome 15)
- Genetic Location Database, http://cedar.genetics.soton.ac.uk/ (for order and distances of markers on chromosome 15)
- Human Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for microcephaly [MIM 251200])
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu/ (for RH mapping of markers on chromosome 15)
References
- Arbour NC, Zlotogora J, Knowlton RG, Merin S, Rosenmann A, Kanis AB, Rokhlina T, et al (1997) Homozygosity mapping of achromatopsia to chromosome 2 using DNA pooling. Hum Mol Genet 6:689–694 [DOI] [PubMed]
- Baraitser M (1997) Microcephaly. In: Motulsky AG, Bobrow M, Harper PS, Scriver C (eds) The genetics of neurological disorders. Oxford University Press, Oxford, pp 17–18 [Google Scholar]
- Budowle B, Chakraborty R, Giusti AW, Eisenberg AJ, Allen RC (1991) Analysis of the VNTR locus D1S80 by PCR followed by high-resolution PAGE. Am J Hum Genet 48:137–144 [PMC free article] [PubMed]
- Cowie V (1960) The genetics and sub-classification of microcephaly. J Ment Defic Res 4:42–47 [DOI] [PubMed] [Google Scholar]
- Dacou-Voutetakis C, Carpathios T, Logothetis N, Constantinidis M, Matsaniotis N (1974) Defective growth hormone secretion in primary microcephaly. J Pediatr 85:498–502 [DOI] [PubMed]
- Jackson AP, McHale DP, Campbell DA, Jafri H, Rashid Y, Mannan J, Karbani G, et al (1998) Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet 63:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Lander ES (1995) Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am J Hum Genet 56:519–527 [PMC free article] [PubMed]
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed]
- Ross JJ, Frias JL (1977) Microcephaly. In: Vinken PJ, Bruyn GW (eds) Congenital malformations of the brain and skull. Vol. 30: Handbook of clinical neurology. Elsevier Holland Biomedical, Amsterdam, pp 507–524 [Google Scholar]
- Terwilliger JD, Shannon WD, Lathrop GM, Nolan JP, Goldin LR, Chase GA, Weeks DE (1997) True and false positive peaks in genomewide scans: applications of length-biased sampling to linkage mapping. Am J Hum Genet 61:430–438 [DOI] [PMC free article] [PubMed]
- Tolmie JL, McNay M, Stephenson JB, Doyle D, Connor JM (1987) Microcephaly: genetic counselling and antenatal diagnosis after the birth of an affected child. Am J Med Genet 27:583–594 [DOI] [PubMed]
- Van den Bosche J (1959) Microcephaly in the Netherlands: a clinical and genetical study. Ann Hum Genet 23:91–116 [DOI] [PubMed] [Google Scholar]
- Walsh CA (1999) Genetic malformations of the human cerebral cortex. Neuron 23:19–29 [DOI] [PubMed]